Abstract
The tremendous dynamics of HIV infection finds expression in the tempo of sequence diversification. Genetic diversity calculations require the clearance of a majority of infected cells, the obvious predator being anti-HIV immune responses. Indeed, infiltration of germinal centers (GCs) by HIV-specific CD8+ cytotoxic T lymphocytes has been described. A corollary to this description would be limited diffusion of virus within lymphoid structures. HIV efficiently infects and replicates mainly in activated CD4+ T lymphoblasts. These cells are found within GCs after their activation in the adjacent periarteriolar lymphoid sheath (PALS). Here GCs and PALS have been dissected from consecutive 10-μm sections through splenic tissue from three HIV-1-infected patients. Nested PCR amplification of the two first hypervariable regions of the env gene indicated that 38–78% of sections contained HIV-infected cells. Since there are several hundred CD4+ T cells per GC section, approximately 0.09–0.64% harbor proviral DNA. Such a low frequency not only suggests that virions on the follicular dendritic cell surfaces do not readily infect adjacent T cells but also indicates highly restricted spread of HIV within GCs and the PALS. Sections were heavily infiltrated by CD8+ cells, which, together with a large body of extant data, suggests that the majority of infected cells are destroyed by HIV-specific cytotoxic T lymphocytes before becoming productively infected. Finally, sequence analysis revealed that those HIV-positive cells were multiply infected, which helps explain widespread recombination despite a low overall frequency of infected cells.
Keywords: spleen, pathogenesis, viral dynamics, multiple infection, recombination
Productive HIV replication requires activation of resting T cells. As antigenic stimulation is the driving force, secondary lymphoid organs are the main sites for HIV replication in vivo (1, 2). CD4 T lymphocytes are the principal targets for HIV, although macrophages and dendritic cells are infected at lower frequencies (3). Splenic white pulps are sites of intense immunological activity. HIV and T cell founder effects within white pulps suggested the local recruitment of antigen-specific T cells paralleled by de novo replication of HIV (1).
White pulps consist of T cell-rich areas, the periarteriolar lymphoid sheaths (PALS), and the B cell-rich follicles, including the germinal centers (GCs). T cells are stimulated in the PALS upon activation by their specific antigen presented by interdigitating dendritic cells. Together with activated B cells, they induce the formation of antigen-specific follicles. GCs are secondary follicles responsible for B cell memory formation and plasmocyte maturation. By immunohistological staining GCs are easily recognized by their follicular dendritic cell (FDC) networks (4). Under physiological conditions, most T cells within a GC are antigen specific and express CD4 and specific markers for activated and memory cells (5, 6). Within GCs, HIV proviruses are mainly present within CD4 T lymphocytes, whereas the FDC surfaces are abundantly coated with virions in the form of immune complexes (7–12).
Anti-HIV CD8+ cytotoxic T lymphocytes (CTLs) are found in the peripheral blood and lymphoid organs of patients until the latest stages of the disease. Such T cells have been shown to infiltrate white pulps and GCs in HIV-infected lymphoid organs (13). They are thought to be HIV specific, drawn in by local proliferation of HIV, and contribute to the destruction of these immunological structures.
Intrapatient HIV genetic variation is both dynamic in time (14) and variable in space (13, 15–17). Viral mutation and fixation rates have been exploited to draw conclusions about viral dissemination, notably the important role of antigenic activation of latently infected T cells (1, 2). To better define the dynamics of HIV replication in the spleen, GC and PALS regions were dissected out from consecutive cryostat sections, and the presence of HIV provirus was assessed by PCR.
Methods
Immunohistochemistry.
A brief clinical profile of patients A, C, and R is given in Table 1A. Small blocks of spleen were frozen in liquid nitrogen immediately after splenectomy and kept at −80°C until use. The top and bottom of the block were discarded to avoid edge effects. Consecutive 10-μm cryostat sections were dried overnight and fixed in acetone at −20°C for 20 min. The sections were stained with the FDC-specific DRC-1 mAb (Dako) followed by alkaline phosphatase-conjugated AffiniPure goat anti-mouse IgM (Jackson ImmunoResearch Laboratories), FDC-specific mAb KiM-4, CD3-specific mAb Leu4 (Becton Dickinson) followed by a rabbit anti-mouse Ig (Dako) and APAAP (alkaline phosphatase/anti-alkaline phosphatase; Dako), CD8-specific mAb Leu2a (Becton Dickinson), or CD4-specific mAb Leu3a (Becton Dickinson) followed by rabbit anti-mouse Ig (Dako) and PAP (peroxidase/anti-peroxidase; Dako) according to the suppliers' instructions. Staining was revealed by using either an alkaline phosphatase substrate kit or a diaminobenzidine substrate kit (Vector) according to the supplier's instructions. Endogenous phosphatase and peroxidase activities were blocked by levamisole or peroxidase-blocking reagent (Dako), respectively. Sections were counterstained in hematoxylin and mounted in Kaiser's glycerol gelatin.
Table 1.
Clinical and GC data for three HIV-1-positive splenectomized patients
| Data | Patient
|
||
|---|---|---|---|
| A | C | R | |
| Clinical data | |||
| Clinical stage | IVA | C2 | C2 |
| Symptoms | Thrombocytopenia | Tuberculosis, chronic hepatitis B | Thrombocytopenia |
| Histopathology | Normal splenic architecture | Burkitt's lymphoma, slightly hyperplastic white pulp | Castelman's disease, hyperplastic white pulp |
| Blood CD4/μl | 129 | 250 | 317 |
| Blood CD8/μl | — | 1,010 | 780 |
| Plasma viremia/ml | 14,900 | ND | 126,000 |
| GC data | |||
| GC area, mm2 | 0.24 ± 0.07 | 0.34 ± 0.13 | 0.54 ± 0.16 |
| CD4+ cells/GC section | 484 ± 311 | 804 ± 31 | 291 ± 66 |
| CD8+ cells/GC section | 533 ± 208 | 911 ± 76 | 348 ± 79 |
| HIV+ sections (GC), % | 38 (n = 65) | 78 (n = 23) | 76 (n = 29) |
| HIV+ sections (PALS), % | 58 (n = 28) | ND | ND |
| HIV DNA/CD4+ cells, % | |||
| Assumption 1 | 0.08 | 0.09 | 0.26 |
| Assumption 2 | 0.09 | 0.37 | 0.64 |
Patients A, C, and R were splenectomized in August 1994, March 1995, and May 1996, respectively. Therapy was zidovudine (AZT) then didanosine (ddI) for patient A, AZT for patient C, and AZT plus ddI for patient R. For the bottom two lines of the table, an estimation of the fraction of HIV DNA-positive CD4 T cells was made in two different ways. The first assumed a single HIV-positive CD4 cell per section. The second assumed that a distinct sequence motif characterizes an infected CD4 T cell. Given the existence of recombinants, the real values lies somewhere between the two. ND, not done.
Dissection, PCR, and Sequencing.
Dissection of GCs and PALS was performed manually by using a 10× stereomicroscope. Instruments were rinsed regularly in 1 M HCl followed by distilled water to prevent DNA cross-contamination. The sections were covered by several microliters of 3% BSA and 1 unit/ml collagenase in PBS. Dissected tissue was picked up in 5–30 μl of PBS/BSA/collagenase by using a microcapillary with the aid of a peristaltic pump and stored in PCR tubes at −20°C. This procedure was made in a DNA-free room to avoid PCR contamination. Protocols for nested PCR of the HIV-1 env V1 and V2 regions, cloning, and sequencing have been described (15, 18). Between 15 and 25 clones per PCR product were sequenced. To ensure that there were no PCR inhibitors in tissue, GC and PALS from an HIV-1-negative spleen were dissected and added to a PCR tube containing a single acetone-fixed 8E5 cell (20-min fixation at −20°C followed by extensive washing in PBS/3% BSA). This cell line contains a single defective HIV-1 provirus and serves as a control for sensitivity (19). Nested PCR picked up 10/10 single-cell samples, 9/10 and 8/10 when a dissected GC or PALS was added to the tube. Hence PCR sensitivity is close to 1 DNA copy per sample. All sequences were screened against those already handled in the lab. No contamination was detected. Phylogenetic trees were made on aligned sequences with unweighted indel coding (unpublished work), using the SplitsTREE2 program (ref. 20 and http://www.mathematik.uni-bielefeld.de/∼huson/phylogenetics/splitstree.html or http://www.bibiserv.techfak.uni-bielefeld.de/splits). Indels were coded according to their position and length. All were given a weight of 1, hence “mutations” means a single event: substitution, insertion, or deletion. Only trees with a “fit” of 100% were used. To achieve this a few sequences were discarded. Hamming distance matrices were used. All sequences are available at ftp://www.pasteur.fr/pub/retromol/through anonymous user.
Results
Histology.
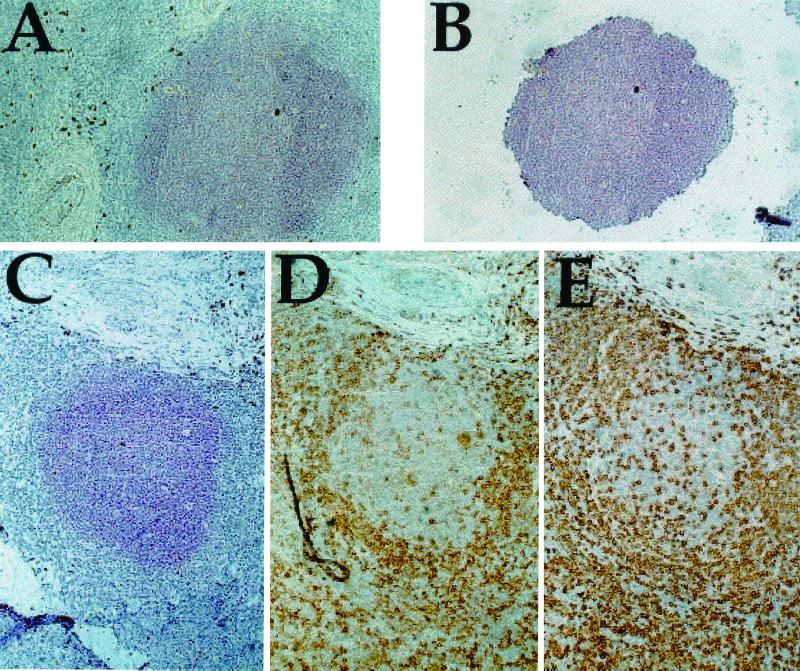
In HIV-1 infection, splenectomy is sometimes necessary for patients with untreatable immune thrombocytopenic purpura or as a diagnostic procedure. The present study was made on spleens from three HIV-1-positive individuals, pertinent clinical information being found in Table 1. Consecutive 10-μm cryostat sections were generated for each patient, spanning a total of 100–300 μm of tissue. This thickness of each section was comparable to the diameter of T lymphoblasts, typically 10–12 μm. Sections were stained with antibodies specific for either FDCs or T cells (CD3, CD4, and CD8) to define clearly the germinal centers and the PALS regions, respectively (Fig. 1). In total, 10 GCs and 3 PALS were analyzed. GC architecture for patient A was normal (microdissected sections ≈0.24 mm2), whereas patients C (≈0.34 mm2) and R (≈0.54 mm2) showed signs of hyperplasia, particularly the latter (Table 1). CD4+ cells, ranging from 300 to 800 per section, were present in every GC section (Table 1). As previously described in ref. 21, the GCs from all of the patients were extensively infiltrated by CD8+ T cells, which outnumbered the CD4+ cells (Fig. 1 and Table 1). Presumably some of them were HIV-specific CTLs (13, 21).
Figure 1.
Tissue sections were treated as described in the text. Here are shown sections from patient R. (A) DRC-1-stained tissue before dissection. (B) The same GC after dissection, showing the amount of tissue picked up for analysis. (C–E) DRC-1 (C), CD4 (D), and CD8 (E) stained GC. (×10.)
Scant and Inhomogeneous Distribution of HIV DNA.
For purposes of dissection, GCs were taken as the FDC-stained areas and the PALS were defined as the CD3+ areas surrounding arteriolae (Fig. 1). For patient A, tissue from two distinct parts of the spleen was analyzed, whereas data for patients C and R were derived from a single series of sections. Between 12 and 30 consecutive sections were studied per GC or PALS. Microdissection and PCR were performed in DNA-free rooms to avoid contamination. Transfer of dissected samples to PCR tubes was always verified visually, a necessary precaution as just occasionally samples can be lost, perhaps because of an interaction between high static on the sample and omnipresent plastic.
DNA corresponding to the first and second hypervariable regions (V1V2) of the gp120 envelope coding region was amplified by nested PCR (15) from each dissected section. These regions of the HIV genome are fixing substitutions at approximately 1–3% per site per year in a near-neutral manner, providing a sensitive marker of the tempo of viral replication (1, 14). As demonstrated by amplification of individual 8E5 cells (19), the nested PCR protocol was capable of detecting a single proviral copy in the presence of GC or PALS sections from an HIV-negative spleen (see Methods). The overall frequencies of HIV DNA-positive sections are given in Table 1. For patient A, 38% and 58% of GC and PALS sections, respectively, were positive. Only GCs were dissected from the spleens of patients C and R, the proportion of positive sections being 78% and 76%, respectively.
The distribution of HIV DNA-positive sections within both GCs and PALS was highly irregular, with adjacent sections being positive and negative (Figs. 2A and 3, vertical series of sections). This was true not only for sequential GC sections from the three spleens but also for PALS sections and laterally between the GCs and PALS from spleen A (Fig. 3, GC1/PALS1). That not all of the sections were positive for HIV DNA argues for a trivial number of HIV-infected cells per section—if there were just two infected cells per section, virtually all would have been positive, given that the nested pairs of primers can detect ≈1 genome. Clusters of infected cells are excluded by the fact that the 10-μm cuts effectively represent cell monolayers, there being no reason why infection should spread solely in the arbitrarily defined cutting plane.
Figure 2.
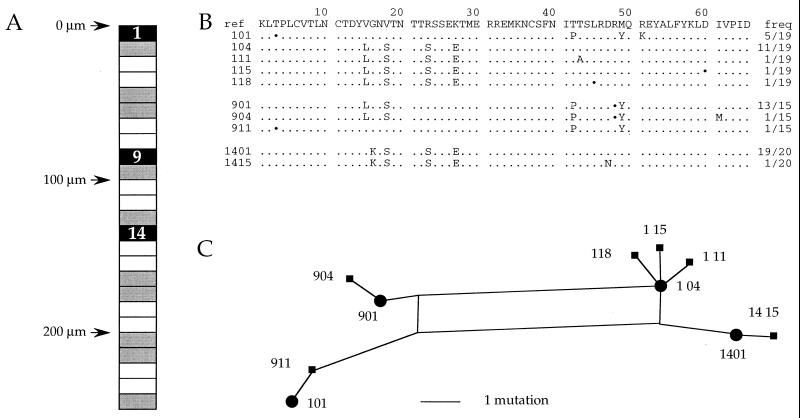
Spatial distribution of HIV DNA, sequences, and phylogenetic analysis from patient A-GC-6. (A) The GC is represented as a long rectangle, boxes representing the different sections. Black and gray indicate HIV-1 DNA-positive and -negative sections, respectively. The white sections in between were stained for CD4, CD3, or CD8. Sections are numbered from top to bottom. The vertical scale is given in micrometers. (B) Sequences present in the HIV-positive cuts. All sequences were aligned to that of an internal reference. Only sequence differences are shown; ⋅s represent synonymous substitutions; and frequencies (freq) of the sequences are given on the right. The sequence code is appended to the section number. For example, 901 refers to sequence 01 from section 9. (C) Phylogenic relationship of A-GC6 sequences determined by the SplitsTree2 program. The distances separating sequences in the trees are strictly proportional to the number of mutations: a single substitution, insertion, or deletion.
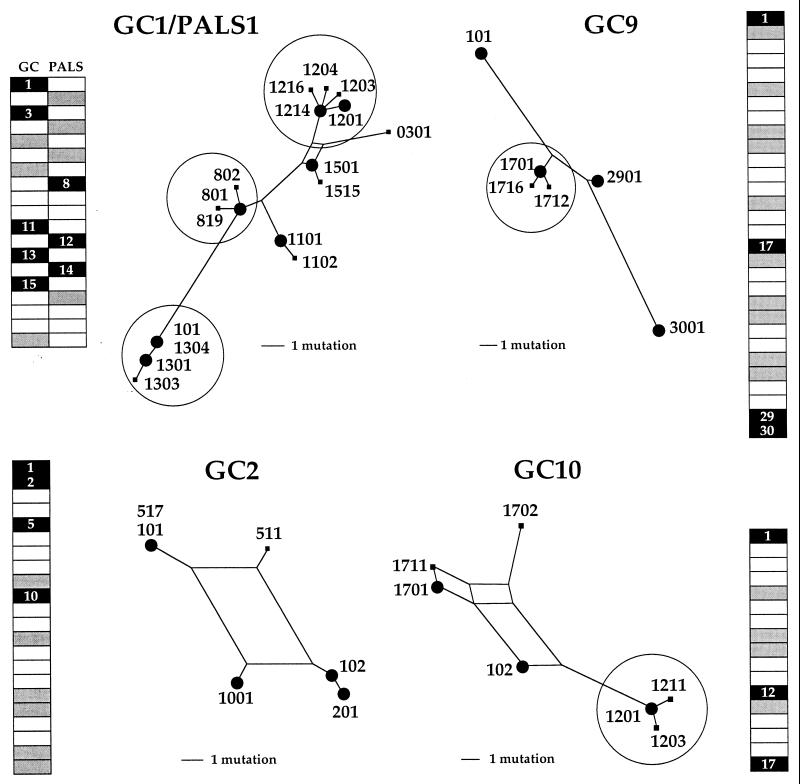
Figure 3.
Spatial distribution of HIV DNA and phylogenetic analysis of sequences from four GCs from patient A. Three GCS and a GC/PALS pair from patient A were analyzed. They are represented by long rectangles at the periphery. Black and gray indicate HIV DNA-positive and -negative sections, respectively. White sections in between were stained for CD4, CD3, or CD8. Phylogenetic analyses corresponding to HIV-positive sections are presented toward the center of the figure. Clusters of sequences differing by a single event are encircled. Large dots indicate a sequence found at a frequency of >20% of all those derived from the section. For the record, one sequence (GC9–3001) is a defective G → A hypermutant, demonstrating that it was amplified from a single cell, as there was no other sequence in the section that could play the role of helper virus. It also reiterates the sensitivity of env V1V2 nested PCR. A few identical sequences were found in different GCs, notably: GC2-101 and GC6-101; GC2-102 and GC6-104; GC2-511 and GC6-901; and GC3-602 and GC6-911.
It will therefore be assumed, for the present, that there is a single infected cell per PCR-positive section. Accordingly, the proportion of HIV DNA-positive CD4 cells would be ≈0.08%, 0.09%, and 0.26% for patients A, C, and R, respectively (Table 1, bottom two lines). These findings agree well with HIV-1 proviral loads in bulk splenic mononuclear cells (SMCs) (patient A and C, 0.03%; patient R, not done), or CD4 cells purified therefrom (patient A, 1%; patient C, 0.6%; patient R, not done) (3). Given that the CD4 T cell-rich regions surround GCs, it is logical that the proviral loads were lower in the GCs than in bulk SMCs. These numbers agree well with previous work on fractionated bulk SMCs from patient A, as well as from other HIV-infected patients—infection frequencies were rarely greater than a few percent. Furthermore, the vast majority of cells harboring HIV DNA were CD4+ T lymphocytes (3).
Clusters of Genomes Despite Restricted Spreading.
PCR products obtained from the different GCs and PALS sections were cloned in M13mp18 replicative form DNA, and approximately 20 clones per sample were sequenced. Each GC harbored a different set of sequences, a typical example being shown in Fig. 2. For sections 1, 9, and 14 there was a major sequence accompanied by a variant(s) differing by a single point mutation (Fig. 2B). A single section could harbor sequences bearing different motifs such that they were not related to one another by a trivial number of rounds of replication—e.g., sequences 101 and 104. The sequences could be more succinctly represented in the form of a phylogenetic tree compiled by using the SplitsTREE2 program (20), in which branch lengths are proportional to the number of mutations separating sequences. The major form and attendant variants were encircled. By contrast, sequences 101 and 911, differing by a single purine transition, were spatially distinct, suggesting that they represent unrelated foci of infection, at least in the short term.
Phylogenetic trees for three more microdissected GCs, as well as one GC and adjacent PALS, are shown in Fig. 3. Sometimes adjacent positive sections were observed, which might be taken as prima facie evidence of local spreading (i.e., GC2 sections 1 and 2 and GC9 sections 29 and 30, Fig. 3). If there was spread, then sequences in adjacent sections should be identical—one cycle of infection being insufficient to produce any noticeable substitutions among the 15–25 clones analyzed for each sample. As can be seen the sequences proved to be different, indicating ruling out the notion of infection spreading from an infected cell in an adjacent section. Equally, sequences derived from a PALS section were distinguishable from those in adjacent GC sections. Once again virus spread was strongly curtailed, despite the finding of clusters of genomes differing by a single point mutation.
Fully 32% of all patient A amplifications yielded distinct yet perfectly homogenous HIV-1 V1V2 sequences, indicating that the vast majority of the observed substitutions results from sequence diversity generated during reverse transcription and not through PCR. Such a low PCR error is typical of the Taq/Vent polymerase pair using XL amplification conditions (ref. 18 and data not shown). A number of sequence clusters were in evidence (encircled, Fig. 3). Each was characterized by a major form and two or three variants differing by a single nucleotide. Although one variant might occasionally be ascribed to PCR error, two or three variants per sample are too many.
Multiply Infected Cells and Recombination.
Three issues bear on the above observations. (i) Given the mutation rate of HIV (22), a variant bearing a single base substitution out of 20 V1V2 clones sampled would require ≈10 replication cycles, yet (ii) viral spread is highly curtailed. Finally, (iii) as mentioned above, they cannot represent tight clusters of infected cells because the thickness of the sections (10 μm) is comparable to the diameter of T lymphoblasts (10–12 μm), and there is no reason why a cluster would be confined to the section plane. A coherent explanation would be that an individual cell becomes infected by many virions, eminently possible if infection occurs by cell-to-cell spread. In a chain where cells are infected by many virions, the founder variant will pass, and it will slowly be accompanied by the accumulation of base substitutions.
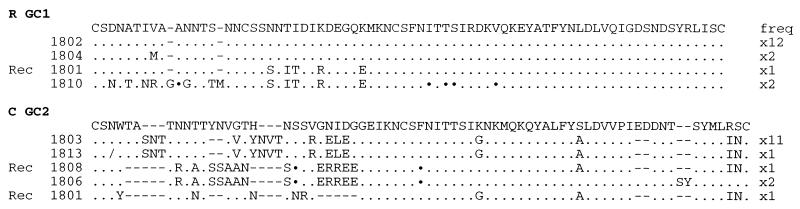
If cells were multiply infected by HIV, then recombinants would be anticipated. Some clear examples of recombinants—e.g., sequences R-GC1-1801, C-GC2-1808, and C-GC2-1801 (Fig. 4), were found in some dissected GC sections, supporting this hypothesis. The frequency of PCR-mediated recombination for the env V1V2 locus is <0.5% (23). As the proportions of HIV recombinants were 6% and 18% for these two GC sections, respectively, they cannot be attributed to PCR.
Figure 4.
Recombinants within GC sections. Recombinant genome (Rec 1801) within GC1 section 18 from patient R and a collection of recombinants (Recs 1808 and 1801) found within GC2 section 18 from patient C are shown. Sequences span the hypervariable V1V2 regions from gp120 and are given in the one-letter amino acid code. Only differences with respect to a reference sequence are shown. Synonymous substitutions are indicated with ⋅ and single base deletions with/; hyphens were introduced to maximize alignments. Sequence identifier codes are given on the left and their frequencies (freq) are given on the right.
Up to this point, the frequency of HIV DNA-positive CD4 T cells was calculated assuming one infected cell per PCR-positive section (assumption 1 in Table 1). However, the sequence data permit an alternative means of calculating the number of infected CD4 T cells. Given what is clearly a low level of infection, one may postulate that a particular sequence motif, including variants with one mutation, defines a single multiply infected cell; for example, for A-GC6 section 1 there were sequences with two distinct motifs, and one each for A-GC9 section 17 and A-GC10 section 12 (Figs. 3 and 4) which would translate into as many infected cells. Accordingly, the fraction of infected CD4 T cells increases a little to between 0.09% and 0.64% for the three spleens (Table 1, assumption 2). This means of calculating is somewhat liberal, as the existence of a few HIV recombinants indicates that a single cell can be infected by viruses bearing distinct sequence motifs (Fig. 4). It is to be expected that the real frequencies are somewhere between those calculated according to the two assumptions (Table 1). Either way, the infected cell frequency remains low, in agreement with other observations (24).
Discussion
The finding of HIV DNA-negative GC sections and the qualitatively and quantitatively similar distributions of HIV within dissected PALS would suggest that HIV, in the form of immune complexes on FDC surfaces (7–12), does not fuel infection of juxtaposed CD4 T lymphoblasts (25) or other antigen-presenting cells (26). The low frequency of HIV-infected CD4 T cells also indicates that virus spread is not limited by the supply of target CD4 T lymphoblasts. In turn it helps explain why GCs, albeit sometimes hyperplastic, remain functional despite HIV within.
The control of HIV spread in vivo is presumably due to a ferocious immune response. Indeed, the GCs were heavily infiltrated by CD8 cells (Table 1), so much so as to outnumber the CD4 cells, reiterating previous observations (12, 18). Because under normal circumstances CD8 cells rarely infiltrate functional GCs (27), it is considered that they are mainly HIV-specific CTLs (13, 18), recruited by HIV infection. Infiltration is substantial, allowing detection of HIV-specific CTL activity in microdissected splenic white pulps by means of a direct lysis assay (unpublished data). Immune restriction of HIV-infected cells must be very efficient, otherwise the virus would spread rapidly in a milieu rich in CD4 T lymphoblasts, the preferred target for the virus. Note that for infected cells to be restricted by HIV-specific CTLs, they must be producing HIV mRNA and antigens but not necessarily viral particles. It would seem, therefore, that equating viral mRNA production, as evidenced by in situ hybridization, with productively infected cells is not axiomatic.
The finding of multiply infected cells has at least two implications. The first is that even if a mutation in a CTL epitope were to occur, epitope-specific CTLs would still recognize the infected cell by means of the nonmutated epitope encoded by other proviruses. This conclusion is encouraging and is coherent with the highly restricted spread of HIV despite substantial genetic variation (Figs. 2 and 3). The second is less so, for multiinfection is a prerequisite for recombination. Recently the recombination rate of HIV-1 has been found to be of the order of three crossovers per genome per cycle (28). The importance of recombination in vivo has already been shown for the simian immunodeficiency virus (SIV) model, whereby coinfection of an animal by two recombinants each harboring a lesion in a different gene resulted in the emergence of wild-type virus by 15 days after infection (29). The present findings provide the background for understanding the emergence of multidrug resistant HIV (30).
The highly restricted spread of HIV highlighted above was predicted from the tempo of HIV/SIV sequence variation (14, 31). Much work has shown the control exerted by CTLs on HIV/SIV replication (32, 33). The ability of HIV to remain transcriptionally inactive in resting CD4 T cells (34) means that despite the wrath of the immune system the virus cannot be cleared. Activation of these latently infected specific T cells not only ensures viral replication but concentrates the virus within antigen-specific T cell subsets. Playing out this cat-and-mouse game will result in their depletion. However, this has to be a slow process, as a minority of antigen-specific T cells are infected at any moment.
Acknowledgments
We thank Dr. Thierry Debord at Hôpital Begin for clinical data pertaining to one patient and Dr. Andreas Meyerhans for critiquing the manuscript. S.G. was supported by bursaries from the Institut Pasteur, the Agence Nationale de Recherches sur le SIDA, and the National Health Research and Development Program (Canada), and M.-J.D. was supported by a bursary from the Agence Nationale de Recherches sur le SIDA. This work was supported by Grants from the Institut Pasteur, the Agence Nationale de Recherches sur le SIDA, and SIDACTION.
Abbreviations
- PALS
periarteriolar lymphoid sheath
- GC
germinal center
- FDC
follicular dendritic cell
- CTL
cytotoxic T lymphocyte
Footnotes
References
- 1.Cheynier R, Gratton S, Halloran M, Stahmer I, Letvin N L, Wain-Hobson S. Nat Med. 1998;4:421–427. doi: 10.1038/nm0498-421. [DOI] [PubMed] [Google Scholar]
- 2.Ostrowski M A, Krakauer D C, Li Y, Justement S J, Learn G, Ehler L A, Stanley S K, Nowak M, Fauci A S. J Virol. 1998;72:7772–7784. doi: 10.1128/jvi.72.10.7772-7784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIlroy D, Autran B, Cheynier R, Wain-Hobson S, Clauvel J P, Oksenhendler E, Debré P, Hosmalin A. J Virol. 1995;69:4737–4745. doi: 10.1128/jvi.69.8.4737-4745.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satoh T, Takeda R, Oikawa H, Satodate R. Anat Rec. 1997;249:486–494. doi: 10.1002/(SICI)1097-0185(199712)249:4<486::AID-AR8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 5.Bowen M B, Butch A W, Parvin C A, Levine A, Nahm M H. Hum Immunol. 1991;31:67–75. doi: 10.1016/0198-8859(91)90050-j. [DOI] [PubMed] [Google Scholar]
- 6.Zheng B, Han S, Kelsoe G. J Exp Med. 1996;184:1083–1091. doi: 10.1084/jem.184.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong J A, Horne R. Lancet. 1984;2:370–372. doi: 10.1016/s0140-6736(84)90540-3. [DOI] [PubMed] [Google Scholar]
- 8.Cavert W, Notermans D W, Staskus K, Wietgrefe S W, Zupancic M, Gebhard K, Henry K, Zhang Z Q, Mills R, McDade H, et al. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 9.Haase A T, Henry K, Zupancic M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z Q, et al. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 10.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. Nature (London) 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 11.Racz P, Tenner-Racz K, Kahl C, Feller A C, Kern P, Dietrich M. Prog Allergy. 1986;37:81–181. doi: 10.1159/000318442. [DOI] [PubMed] [Google Scholar]
- 12.Tenner-Racz K, Racz P, Dietrich M, Kern P. Lancet. 1985;i:105–106. doi: 10.1016/s0140-6736(85)91994-4. [DOI] [PubMed] [Google Scholar]
- 13.Cheynier R, Henrichwark S, Hadida F, Pelletier E, Oksenhendler E, Autran B, Wain-Hobson S. Cell. 1994;78:373–387. doi: 10.1016/0092-8674(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 14.Pelletier E, Saurin W, Cheynier R, Letvin N L, Wain-Hobson S. Virology. 1995;208:644–652. doi: 10.1006/viro.1995.1195. [DOI] [PubMed] [Google Scholar]
- 15.Delassus S, Cheynier R, Wain-Hobson S. J Virol. 1992;66:5642–5645. doi: 10.1128/jvi.66.9.5642-5645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epstein L G, Kuiken C, Blumberg B M, Hartman S, Sharer L R, Clement M, Goudsmit J. Virology. 1991;180:583–590. doi: 10.1016/0042-6822(91)90072-j. [DOI] [PubMed] [Google Scholar]
- 17.Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- 18.Cheng S, Fockler C, Barnes W M, Higuchi R. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gendelman H E, Theodore T S, Willey R, McCoy J, Adachi A, Mervis R J, Venkatesan S, Martin M A. Virology. 1987;160:323–329. doi: 10.1016/0042-6822(87)90002-x. [DOI] [PubMed] [Google Scholar]
- 20.Bandelt H J, Dress A W M. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 21.Tenner-Racz K, Racz P, Thomé C, Meyer C G, Anderson P J, Schlossman S F, Letvin N L. Am J Pathol. 1993;142:1750–1758. [PMC free article] [PubMed] [Google Scholar]
- 22.Mansky L M, Temin H M. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyerhans A, Vartanian J P, Wain-Hobson S. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, et al. Nature (London) 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 25.Heath S L, Tew J G, Tew J G, Szakal A K, Burton G F. Nature (London) 1995;377:740–744. doi: 10.1038/377740a0. [DOI] [PubMed] [Google Scholar]
- 26.Frank I, Kacani L, Stoiber H, Stossel H, Spruth M, Steindl F, Romani N, Dierich M P. J Virol. 1999;73:3449–3454. doi: 10.1128/jvi.73.4.3449-3454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willfuhr K U, Westermann J, Pabst R. Eur J Immunol. 1990;20:903–911. doi: 10.1002/eji.1830200428. [DOI] [PubMed] [Google Scholar]
- 28.Jetzt A E, Yu H, Klarmann G J, Ron Y, Preston B D, Dougherty J P. J Virol. 2000;74:1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wooley D P, Smith R A, Czajak S, Desrosiers R C. J Virol. 1997;71:9650–9653. doi: 10.1128/jvi.71.12.9650-9653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kellam P, Larder B A. J Virol. 1995;69:669–674. doi: 10.1128/jvi.69.2.669-674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wain-Hobson S. Nature (London) 1993;366:22. doi: 10.1038/366022b0. (lett.). [DOI] [PubMed] [Google Scholar]
- 32.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, et al. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 33.Jin X, Bauer D E, Tuttleton S E, Lewin S, Gettie A, Blanchard J, Irwin C E, Safrit J T, Mittler J, Weinberger L, et al. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]