Abstract
Stat proteins are latent cytoplasmic transcription factors that are crucial in many aspects of mammalian development. In the immune system, Stat3 has distinct roles in T-cell, neutrophil, and macrophage function, but a role for Stat3 in B-cell development, particularly in the terminal differentiation of B cells into antibody-secreting plasma cells, has never been directly tested. In this study, we used the Cre/lox system to generate a mouse strain in which Stat3 was conditionally deleted in the B-cell lineage (Stat3fl/flCD19Cre/+). B-cell development, establishment of the peripheral B-cell compartment, and baseline serum antibody levels were unperturbed in Stat3fl/flCD19Cre/+ mice. Strikingly, Stat3fl/flCD19Cre/+ mice displayed profound defects in T-dependent (TD) IgG responses, but normal TD IgM, IgE, and IgA responses and T-independent (TI) IgM and IgG3 responses. In addition, germinal center (GC) formation, isotype switching, and generation of memory B cells, including IgG+ memory cells, were all intact in Stat3fl/flCD19Cre/+ mice, indicating that the requirement for Stat3 was limited to plasma cell differentiation. These results demonstrate a profound yet highly selective role for Stat3 in TD IgG plasma cell differentiation, and therefore represent a unique example of a transcription factor regulating isotype-specific terminal B-cell differentiation.
Introduction
Members of the signal transducer and activator of transcription (Stat) family are pivotal players in multiple developmental processes. Stat proteins are latent cytoplasmic transcription factors that are activated by numerous cytokines and growth factors. Upon activation, tyrosine-phosphorylated Stats dimerize and translocate to the nucleus where they accumulate and activate transcription of specific target genes.1 In the murine system, targeted gene deletion has been used to understand roles for each of the 7 Stat proteins (Stat1, Stat2, Stat3, Stat4, Stat5a, Stat5b, and Stat6). Of these, only Stat3 deletion was shown to be embryonically lethal.2
Since nullizygosity of Stat3 leads to early embryonic lethality,2 diverse roles of Stat3 in different tissues have been studied by conditional deletion of Stat3 in the cell type of interest using Cre/lox technology. In the skin, Stat3 is necessary for keratinocyte migration, wound repair, and the second hair cycle.3 Deletion of Stat3 in mammary epithelial cells leads to a delay in mammary gland evolution in part due to a decrease in mammary epithelial cell apoptosis.4 Mice in which Stat3 has been deleted in the liver have defective acute-phase responses.5 Loss of Stat3 in motor neurons leads to a decrease in a survival of these cells upon facial injury.6 Mice lacking Stat3 in cardiomyocytes have an increase in myocyte apoptosis in response to treatment with LPS, likely due to an increase in TNFα secretion upon exposure to LPS.7 As these mice age, they experience an increase in cardiac fibrosis.7 Hypothalamic deletion of Stat3 resulted in an increase in body weight and body fat percentage.8
In the immune system, selective deletion of Stat3 in cells of different hematopoietic lineages demonstrated a diverse role for Stat3 in different cell types. Mice that lack Stat3 in T cells have a decrease in IL-6-induced proliferation due to an impairment in IL-6-mediated survival.9 These mice also have a decrease in IL-2-mediated proliferation, though not as severe as that of IL-6-induced proliferation, which is caused by a defect in IL-2-induced IL-2Rα expression.9 Disruption of Stat3 in neutrophils and macrophages leads to an increased susceptibility to LPS-induced endotoxic shock concomitant with an increase in several proinflammatory cytokines including IL-6, TNFα, and IFN-γ.10 Additionally, as these mice aged, they developed chronic enterocolitis that may have been the result of skewed Th1 response.10 Inducible deletion of Stat3 in hematopoietic progenitor cells resulted in neutrophilia, due to misregulation of SOCS3.11 Deletion of Stat3 in hematopoietic precursors results in Crohn disease-like pathogenesis, a skewing of cells toward the myeloid lineage, and an increase in inflammatory responses.12 These mice also demonstrate a decrease in total dendritic cells (DCs).13 Treatment of these mice with Flt3L resulted in an increased frequency of BM-derived common myeloid progenitor (CMP)/common lymphoid progenitor (CLP) cells and impaired formation of BM-derived CD11c+CD11b- DCs.13
While the role of Stat3 has been studied in multiple immune cells, its role in B cells has not been directly examined. Stat3 is activated by numerous cytokines and growth factors, including IL-6, IL-10, leukemia inhibitory factor (LIF), Oncostatin M (OSM), ciliary neurotrophic factor (CNTF), and cardiotrophin-1, whose heterodimeric receptors include the gp130 chain.14,15 Stat3 is also activated by IL-4, IL-13, IL-2, and IL-21, which signal through the common γ chain.16-18 Many of these Stat3-activating cytokines, such as IL-2, IL-10, IL-6, and IL-21, have been implicated in the terminal differentiation of B cells into antibody-secreting plasma cells.18-21 We recently showed by transcriptional profiling that, compared with B cells, plasma cells selectively retain Stat3 and both chains of the IL-6 receptor, IL-6Rα, and gp130, whereas inhibitors of Stat3 signaling, including PIAS3 and SOCS3, are absent or down-regulated in plasma cells, as were Stat4, Stat5a/b, and Stat6.22 These results strongly implied an important role for Stat3 in plasma cell formation. In this report, we used the Cre/lox system to create a mouse strain in which Stat3 is specifically deleted in the B-cell lineage. We demonstrate that Stat3 plays a highly selective role in TD IgG immune responses, which appears to be limited to terminal differentiation of already class-switched IgG B cells to plasma cells. These results define a unique role for Stat3 in the terminal differentiation of a specific B-cell subset.
Materials and methods
Mice
Mice heterozygous for Cre recombinase inserted into the CD19 locus (Stat3+/+CD19Cre/+)23 were intercrossed to mice in which the DNA-binding domain of Stat3 was flanked by lox P sites (Stat3fl/flCD19+/+)5 through 3 generations to yield litters in which mice were Stat3fl/flCD19Cre/+ (deleted) or Stat3fl/flCD19+/+ (littermate controls). Mice used in this study were 2 to 6 months of age and were maintained at the Feinberg School of Medicine at Northwestern University according to university guidelines.
Cell isolation and B-cell activation
B cells, T cells, and myeloid cells were isolated from spleens (B and T cells) and bone marrow (myeloid cells) of Stat3fl/flCD19Cre/+ and Stat3fl/flCD19+/+ mice using anti-CD19 microbeads, anti-CD4 + anti-CD8 microbeads, or anti-CD11b microbeads (Miltenyi Biotech, Auburn, CA), respectively, followed by positive selection on LS+ columns (Miltenyi Biotech) according to the manufacturer's protocol. B cells were cultured in flat-bottom wells with 10 μg/mL LPS, either alone or with 10 ng/mL IL-4 or IFNγ (Biosource, Camarillo, CA), for 4 days, and total isotype-specific enzyme-linked immunospot (ELISPOT) assays were performed as described in “ELISPOT analysis.”
Genomic DNA isolation and PCR analysis
Genomic DNA was isolated from Stat3fl/flCD19Cre/+ and Stat3fl/flCD19+/+ B, T, and myeloid cells using Cell Lysis Solution and Protein Precipitation Solution from Gentra Systems (Minneapolis, MN) according to the manufacturer's protocol. For polymerase chain reaction (PCR), the following primers (provided by V.P.) were used: sense, 5′CACCAACACATGCTATTTGTAGG-3′; antisense, 5′GCAGCAGAATACTCTACAGCTC-3′. The floxed allele yields a 371-bp band and the deleted allele yields a 310-bp band.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis
Cell lysates were made in high-salt RIPA buffer (50 mM Tris [pH 8], 150 mM NaCl, Triton X-100, 1% DOC, 0.1% SDS, 1 mM EDTA) with protease inhibitors (Sigma, St Louis, MO) and clarified by centrifugation at 15 000g. Samples were separated on 12% polyacrylamide gels. After transferring to nitrocellulose, the membrane was blocked in Tris-buffered saline containing 0.05% Tween 20 (TBST) + 5% dry milk overnight, probed with rabbit anti-mouse Stat3 (K-15) (Santa Cruz Biotechnology, Santa Cruz, CA) followed by horseradish-peroxidase-conjugated anti-rabbit Ig second step (Biosource), and developed using ECL reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
Immunizations
For TD immunizations, mice were immunized intraperitoneally with 5 μg TNP12-CGG (Biosearch Technologies, Novato, CA) in CFA (Difco, San Jose, CA). Mice were immunized intraperitoneally with 5 μg TNP12-CGG in Imject alum (Pierce, Rockford, IL) for TD IgE immunizations or 5 μg TNP77-AECM-Ficoll (Biosearch Technologies) in PBS for TI immunizations. Immune sera were obtained by retro-orbital bleed on the days indicated. To elicit GC formation, mice were immunized intraperitoneally with 200 μL human red blood cells (RBCs), and spleens were isolated 10 to 12 days later for immunohistochemical staining. For memory B-cell analysis, mice were immunized intraperitoneally with 100 μg R-PE (Cyanotech, Kailua-Kona, HI) in CFA, and spleens were isolated at least 3 to 4 weeks later for fluorescence-activated cell sorter (FACS) analysis.24
FACS and immunohistochemistry
FACS analysis was performed as previously described.25 FITC-conjugated anti-IgD, APC-conjugated anti-B220, FITC-conjugated anti-GL7, PE-conjugated anti-CD23, biotinylated anti-CD5, biotinylated anti-IgG1, and PerCP-conjugated streptavidin were from Pharmingen (San Jose, CA). Biotinylated and FITC-conjugated anti-IgM (b76) were produced by T.J.W. PE-conjugated streptavidin was from Southern Biotechnology (Birmingham, AL). All data were collected using either a FACSort or FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA). To detect GC formation, paraffin-embedded spleens were stained with peanut agglutinin (PNA) by standard methods.
ELISA analysis
MaxiSorp plates (Nalge Nunc, Naperville, IL) were incubated overnight at 4°C with 5 μg/mL TNP18-BSA (Biosearch Technologies), washed 3 times in PBS/Tween + Kathon CG/KP (Supelco, Bellefonte, PA) (PBST), and blocked for more than 1 hour with PBS + 2% BSA at 37°C. Serial dilutions of serum samples were analyzed in triplicate. TNP-specific IgM, IgG1, and IgA standards (Pharmingen) were included in all enzyme-linked immunosorbent assays (ELISAs). For TNP-specific IgG2a and IgG2b ELISAs, concentrations were calculated using TNP-specific standards (Pharmingen and provided by T.J.W., respectively). Plates were incubated at 37°C for 1.5 to 2 hours; washed 3 × with PBST; incubated with biotinylated anti-IgM, anti-IgG1, IgG2a, IgG2b, or anti-IgA (Pharmingen) at 37°C for 1.5 to 2 hours; washed 3 × with PBST; incubated with streptavidin-conjugated alkaline phosphatase (Southern Biotechnology) for 30 minutes at 37°C; washed 6 × with PBST; and developed with PNPP (Sigma). Plates were read at A405 using Softmax Pro software (Sunnyvale, CA). IgE anti-TNP ELISAs were performed as previously described26 with EM95 (rat anti-mouse IgE mAb) used as capture Ab and A3B1 (mouse IgE anti-TNP mAb) used as reference Ab to generate standard curves. ELISA analysis for total baseline serum antibody was carried out in the same manner as TNP-specific ELISA analysis except that plates were coated with anti-mouse IgG (Jackson Immunoresearch Laboratories, West Grove, PA), anti-mouse IgM (Pharmingen), or anti-mouse IgA (Pharmingen) at 2 μg/mL in coating buffer. Purified mouse isotype standards were from Pharmingen.
ELISPOT analysis
Unifilter plates (Whatman, Clifton, NJ) were incubated overnight with 10 μg/mL TNP18-BSA in PBS in a humidified chamber at 4°C, washed 3 times with PBS, and blocked with RPMI + 1% BSA at room temperature. Triplicate serial dilutions of unfractionated spleen cells from immunized mice were made in complete RPMI. Plates were incubated at 37°C for 3 to 4 hours; washed 4 × with PBST; incubated with biotinylated anti-IgM, anti-IgG1, IgG2a, or IgG2b (Pharmingen) overnight at 4°C in a humidified chamber; washed 4 × with PBST; incubated with anti-biotin alkaline phosphatase (Vector, Burlingame, CA) for 2 hours at room temperature; and developed with the Alkaline Phosphatase Conjugate substrate kit from Bio-Rad (Hercules, CA).
Statistical analysis
All statistical analyses were performed using GraphPad InStat Version 3.0 software (GraphPad Software, San Diego, CA), using the Mann-Whitney nonparametric test, with P values less than .05 considered significant.
Results
Deletion of Stat3 in the B-cell lineage and analysis of B-cell development in Stat3fl/flCD19Cre/+ mice
The Stat3 floxed allele generates a null mutation in the presence of Cre recombinase.5 We therefore crossed Stat3fl/flCD19+/+5 mice with Stat3+/+CD19Cre/+ mice23 to generate mice that were Stat3fl/flCD19Cre/+. As expected, Stat3 was efficiently deleted in the B-cell lineage of Stat3fl/flCD19Cre/+ (KO) but not in other lineages tested or in Stat3fl/flCD19+/+ (Wt) controls (Figure 1A). Total cell numbers (data not shown) and B-cell numbers in the bone marrow, spleen, and lymph nodes (Figure 1B) were equivalent in Stat3fl/flCD19Cre/+ (KO) mice and Stat3fl/flCD19+/+ (Wt) controls. Bone marrow cells from Stat3fl/flCD19Cre/+ (KO) mice were able to form similar numbers of pre-B-cell colony-forming units in culture as controls (data not shown).
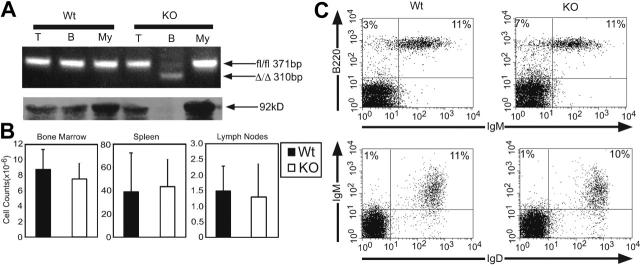
Figure 1.
Deletion of Stat3 in the B-cell lineage does not disrupt B-cell development. (A) Top row: Genomic DNA from T cells, B cells, and myeloid cells from Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) mice was subjected to PCR analysis using primers that yield bands corresponding to both the Stat3fl/fl and Stat3Δ/Δ alleles. Bottom row: Whole-cell lysates were prepared from the same cell types as in panel A and were subjected to Western blot analysis for total Stat3. (B) B-cell numbers in the bone marrow, spleen, and lymph nodes of Wt (▪) and KO (□) mice were calculated based on percentages of total cells that stained as B220+ upon flow cytometric analysis. Error bars represent the SD of at least 3 mice. (C) Flow cytometric analysis was performed on lymph nodes from Wt and KO mice for canonical B-cell markers. Flow cytometric results are representative of at least 3 experiments.
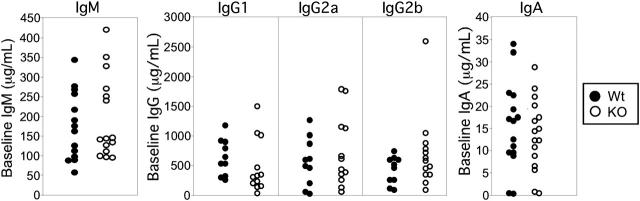
FACS analysis was performed on whole bone marrow, spleen, and lymph nodes for canonical B-cell markers. The percentages of B220+IgM- pre-B and B220+IgM+ immature B cells in the bone marrow were indistinguishable between Stat3fl/flCD19Cre/+ (KO) mice and littermate controls (data not shown). B cells from Stat3fl/flCD19Cre/+ (KO) mice exhibited normal phenotypes in the spleen (data not shown) and fully mature B cells (IgM-lo IgD-hi) were present in the lymph nodes in normal frequencies (Figure 1C). Baseline serum antibody levels for all isotypes tested were equivalent between Stat3fl/flCD19Cre/+ (KO) and Stat3fl/flCD19+/+ (Wt) controls (Figure 2). These results demonstrate that deletion of Stat3 by Cre recombinase driven by the CD19 locus did not affect B-cell development.
Figure 2.
Baseline serum antibody levels are unperturbed in Stat3fl/flCD19Cre/+ mice. Preimmune serum was taken from Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) mice and ELISAs were performed for baseline IgM, IgG1, IgG2a, IgG2b, and IgA secretion. • indicates Wt mice; ○, KO mice. Each circle represents one mouse.
Stat3 is selectively essential for TD IgG formation
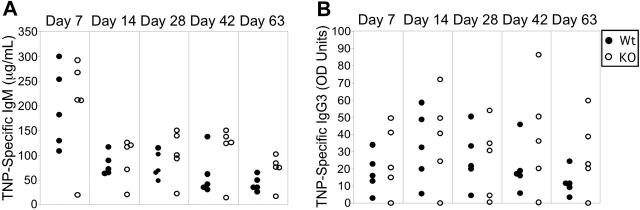
Although baseline serum antibody levels were equivalent between Stat3fl/flCD19Cre/+ (KO) and Stat3fl/flCD19+/+ (Wt) controls (Figure 2), these results do not exclude a role for Stat3 in the development of discrete plasma cell subsets. Since Stat3 is activated in response to a number of cytokines implicated in plasma cell development, we next determined if Stat3 is necessary for the generation of specific antibody. B-cell responses to different types of antigen can be divided into those requiring T-cell help (T-dependent, TD) and those that function independently of T cells (TI).27 To identify a role for Stat3 in TD antibody formation, Stat3fl/flCD19+/+ and Stat3fl/flCD19Cre/+ mice were immunized with the TD antigen TNP-CGG, and sera were collected over time. Stat3fl/flCD19Cre/+ (KO) mice exhibited no defects in TD IgM responses (Figure 3A, left panel). In sharp contrast, they exhibited a severe decrease in TD TNP-specific IgG1, IgG2a, and IgG2b levels (Figure 3B-D, left panels). In agreement with the ELISA results, Stat3fl/flCD19Cre/+ (KO) mice exhibited a profound decrease in TD IgG1, IgG2a, and IgG2b antibody-secreting cell (ASC) formation (Figure 3B-D, right panels), but no differences in TD IgM ASC formation (Figure 3A, right panel). In contrast, anti-TNP responses of Stat3fl/flCD19Cre/+ (KO) mice to TNP-Ficoll, a TI2 antigen, were not different for either IgM (Figure 4A) or IgG3 (Figure 4B). Additionally, purified B cells from Stat3fl/flCD19+/+ and Stat3fl/flCD19Cre/+ mice yielded equivalent numbers of IgG1 ASC when cultured with LPS + IL-4 (Wt 95.6 ± 21; KO, 70 ± 19; NS), and IgG2a and IgG2b ASC when cultured with LPS + IFNγ (IgG2a: Wt, 66.3 ± 6; KO, 67.3 ± 6; NS; and IgG2b Wt, 31.7 ± 1.1; KO 43 ± 7; NS). Together, these data suggest a highly selective role for Stat3 in TD IgG plasma cell development and antibody formation.
Figure 3.
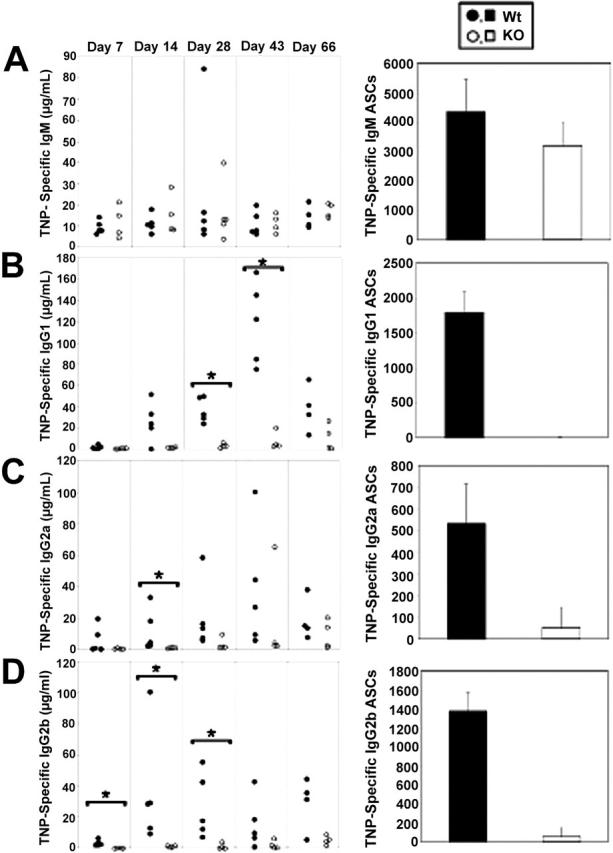
Selective impairment of TD IgG responses in Stat3fl/flCD19Cre/+ mice. TD antibody responses (left panels) and ASC formation (right panels) were assessed in Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) mice immunized with TNP-CGG in CFA by TNP-specific ELISAs or ELISPOTs of splenocytes, respectively. (A) TD IgM antibody responses and ASC formation were equivalent between Wt and KO mice. TD IgG1 (B), IgG2a (C), and IgG2b (D) antibody responses and ASC formation were sharply diminished in KO mice. Filled symbols represent Wt mice; open symbols, KO mice. For TD antibody response data, each circle represents one mouse. TD ELISPOT data are representative of multiple experiments. Error bars represent SD. *P = .016 based on Mann-Whitney analysis.
Figure 4.
TI responses are unperturbed in Stat3fl/fl CD19Cre/+ mice. Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) mice were immunized with TNP-Ficoll in PBS, and TNP-specific antibody responses were measured on the days indicated. TI TNP-specific IgM (A) or IgG3 (B) levels were equivalent between Wt (•) and KO (○) mice.
GC formation is unperturbed in Stat3fl/flCD19Cre/+ mice
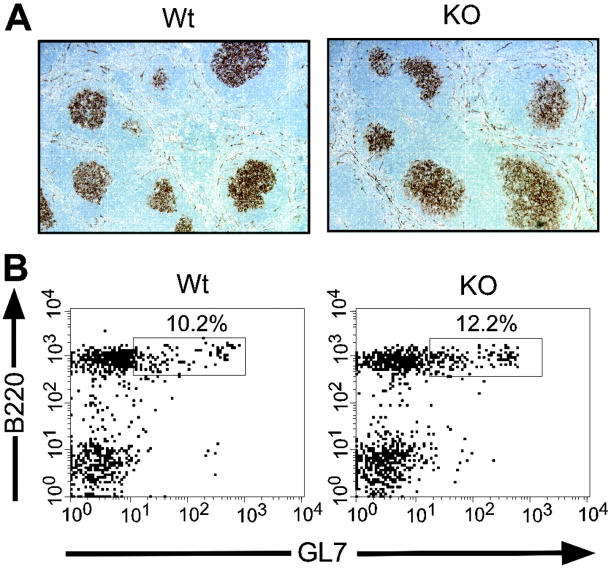
TD immune responses rely on effective germinal center (GC) reactions, whereas TI responses do not.27 The TD IgG defects that we observed in Stat3fl/flCD19Cre/+ mice could be secondary to a defect in GC formation. Therefore, to determine if the defects in TD IgG responses in Stat3fl/flCD19Cre/+ mice were due to impaired GC formation, mice were immunized with hRBCs, and spleens were isolated 10 to 12 days after immunization and stained with peanut agglutinin (PNA) to visualize GCs. Stat3fl/flCD19Cre/+ (KO) mice and littermate controls formed GCs that were similar in number, size, and morphology in response to immunization with hRBCs (Figure 5A).Additionally, expression of GL7, a GC marker, was equivalent between Stat3fl/flCD19Cre/+ (KO) mice and littermate controls (Figure 5B). These data indicate that Stat3fl/flCD19Cre/+ mice are able to form GCs and that the observed defects in TD IgG responses are not due to a grossly impaired GC reaction.
Figure 5.
GC formation is intact in Stat3fl/flCD19Cre/+ mice. (A) PNA staining of spleens from Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) hRBC-immunized mice shows equivalent GC formation. (B) Spleen cells from hRBC-immunized Wt and KO mice were analyzed by flow cytometry for GL7 expression to confirm equivalent GC formation. After mounting in Permamount solution (Sigma, St. Louis, MO), images were visualized using a Leica DMR microscope (Leica, Heidelberg, Germany) equipped with a 10×/0.3 objective lens and a 10× eyepiece, for a total original magnification of ×100. A Diagnostic CE230 SPOT camera (Diagnostic Instruments, Sterling Heights, MI) was used to capture images, and SPOT acquisition software (Diagnostic Instruments) was used to process and export them in Tagged Image File format (TIFF) to the canvas program (ACD Systems, Miami, FL) for further processing.
Isotype switching is unimpaired in Stat3fl/flCD19Cre/+ mice
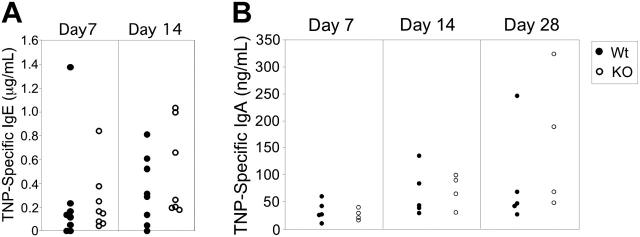
Although GC formation was not impaired in Stat3fl/flCD19Cre/+ mice, and baseline serum antibody data indicated that Stat3fl/flCD19Cre/+ (KO) mice are capable of producing equivalent amounts of all antibody isotypes tested, including class switched isotypes (IgG1, IgG2a, IgG2b, and IgA), this does not exclude the possibility that a defect in isotype switching is responsible for the severe TD IgG defects in these mice. To ascertain if Stat3fl/flCD19Cre/+ mice displayed more subtle defects in isotype switching, we assessed IgE and IgA formation in response to a TD antigen. Mice were immunized with TNP-CGG in alum in order to measure TD IgE responses.26 We also measured TD IgA responses in mice that had been immunized with TNP-CGG in CFA. Stat3fl/flCD19Cre/+ (KO) mice produced equivalent TNP-specific IgE (Figure 6A) and IgA responses (Figure 6B) compared with Stat3fl/flCD19+/+ (Wt) mice. Therefore, no generalized defects in TD isotype switching are present in Stat3fl/flCD19Cre/+ mice.
Figure 6.
Isotype switching is not disrupted in Stat3fl/flCD19Cre/+ mice. Stat3fl/flCD19Cre/+ (KO) mice (○) were able to elicit equivalent TD IgE responses (A) and TD IgA responses (B) compared with Stat3fl/flCD19+/+ (Wt) mice (•) in response to TD Ag. Mice were immunized with TNP-CGG as before, except that for induction of IgE, alum was used instead of CFA.
IgG memory B cells are formed in Stat3fl/flCD19Cre/+ mice
The data in Figure 6 do not exclude a defect in isotype switching specifically to IgG subclasses. In order to assess isotype switching to IgG in a way that does not depend on antibody formation, we measured the generation of IgG memory B cells, which by definition have undergone successful isotype switching to IgG. Mice were immunized with phycoerythrin (PE), which induces the development of PE-binding, antigen-specific B cells, some of which have undergone isotype switching.24 Both Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) mice formed B220-hi PE+ B cells upon immunization with PE (Figure 7A, lower panels), while few B220-hi PE+ B cells were present in naive controls (Figure 7A, upper panels). While Stat3fl/flCD19Cre/+ (KO) mice had a decreased percentage of B220-hi PE+ cells compared with Stat3fl/flCD19+/+ (Wt) mice in the experiment shown (Figure 7A, lower panels), this decrease was not reproducible throughout all experiments (Figure 7B-C). We therefore determined the percentage of PE+ cells among B220+IgM+, B220+IgG1+, B220+IgG2a+, or B220+IgG2b+ cells, and the percentage of cells expressing each isotype among B220+PE+ cells upon immunization of Stat3fl/flCD19Cre/+ mice and littermate controls. Measurement of PE+ cells within the various B220+Ig+ subsets revealed that there were equivalent percentages of PE+ cells for all isotypes tested in Stat3fl/flCD19Cre/+ (KO) mice compared with littermate controls (Figure 7B). In agreement with these data, there were similar percentages of IgM+, IgG1+, IgG2a+, and IgG2b+ cells among B220+PE+ cells in both Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) mice (Figure 7C). Therefore, TD isotype switching to IgG and formation of IgG memory B cells is intact in Stat3fl/flCD19Cre/+ mice. These findings cumulatively exclude defects in isotype switching as the basis for defects in TD IgG formation in Stat3fl/flCD19Cre/+ mice.
Figure 7.
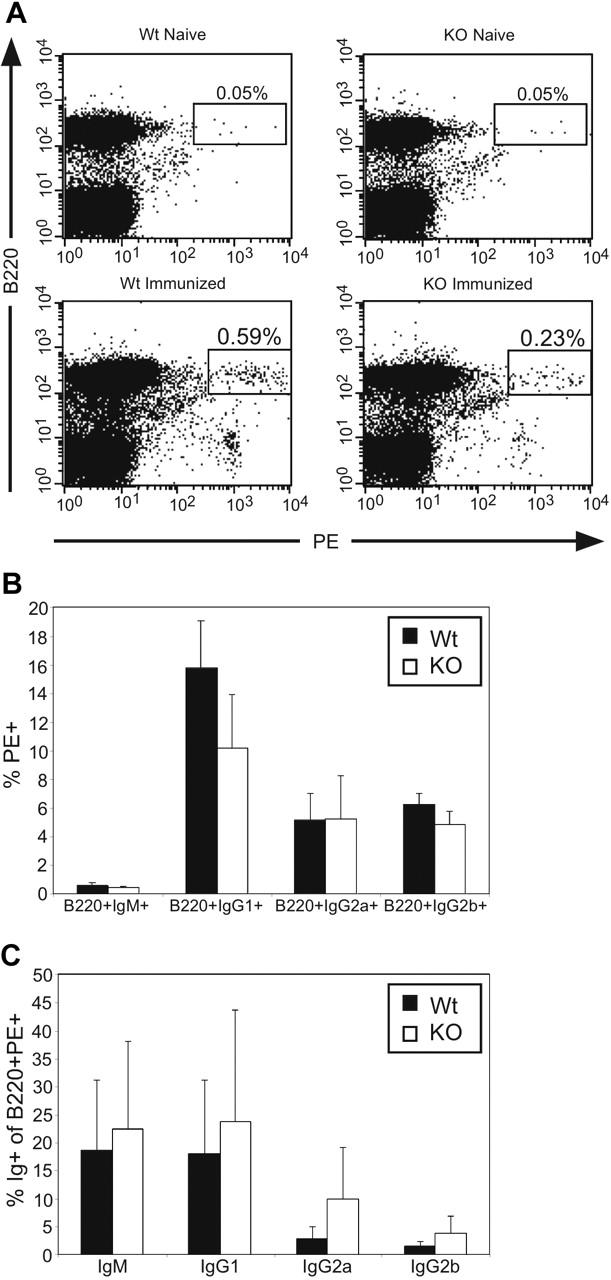
Stat3fl/flCD19Cre/+ mice are able to form IgG+ memory B cells. (A) Splenocytes from Stat3fl/flCD19+/+ (Wt) and Stat3fl/flCD19Cre/+ (KO) mice immunized with PE in CFA or naive controls were analyzed at least 2 months after immunization by flow cytometry for B220+PE+ cells. (B) Percentage of PE+ cells in the various B220+Ig+ subsets was calculated based on flow cytometric results for both PE-immunized Wt (▪) and KO (□) mice. (C) The percentage of IgM+, IgG1+, IgG2a+, or IgG2b+ cells in the B220+PE+ subset was calculated for both Wt and KO mice. Error bars represent the SEM of at least 2 experiments.
In summary, the lack of any detectable effect of Stat3 deletion in B cells on any aspect of B-cell development, germinal center formation, isotype switching, or generation of memory B cells, in combination with severe defects in production of IgG1, IgG2a, and IgG2b, and plasma cells secreting these isotypes, cumulatively demonstrates that B-cell Stat3 is selectively required for TD IgG plasma cell formation.
Discussion
Stat3 is a multifunctional transcription factor that plays a role in multiple biologic systems, including the immune system. A role for Stat3 in multiple immune cell types, including T cells, neutrophils, and macrophages, has been identified, but its role in B cells has never been directly studied. Numerous studies have indicated that Stat3 might play a crucial role in B-cell terminal differentiation. Most of these studies have focused on IL-6 or the signaling component of its receptor, gp130. For instance, mice lacking IL-6 have a decrease in neutralizing IgG upon infection with vesicular stomatitis virus, but also have other defects, extrinsic to B cells, that could underlie defects in antibody formation.28 Mice reconstituted with fetal liver cells from mice expressing a mutant form of gp130 incapable of activating Stat3 demonstrate a decrease in basal and TD IgG2a and IgG2b levels.29 Similarly, mice expressing a dominant-negative form of gp130 display a decrease in TD IgG responses.30 Here, we demonstrate, by selective deletion of Stat3 in the murine B-cell lineage, that Stat3 is necessary for TD IgG plasma cell differentiation. To our knowledge, this represents the first example of a transcription factor selectively required for the differentiation of a specific plasma cell subset.
B-cell development and maturation are intact in Stat3fl/flCD19Cre/+ mice (Figure 1 and data not shown), demonstrating that the profound defects in TD IgG antibody production (Figure 3) are not due to a defect at an earlier developmental stage. The defect in TD IgG antibody responses is also not secondary to a defect in GC formation. Stat3fl/flCD19Cre/+ mice produced GCs equivalent in size and morphology to that of littermate controls (Figure 5A-B). Several lines of evidence show that the defects in TD IgG are also not explained by a defect in isotype switching, a key function of the GC. First, Stat3fl/flCD19Cre/+ mice display similar amounts of baseline class-switched antibody levels compared with littermate controls (Figure 2). Second, Stat3fl/flCD19Cre/+ mice were capable of producing equivalent levels of TNP-specific IgE (Figure 6A) and IgA (Figure 6B) as littermate controls in response to immunization with a TD antigen. Third, Stat3fl/flCD19Cre/+ mice produced TNP-specific IgG3 responses at similar levels to that of littermate controls (Figure 4A) in response to immunization with a TI2 antigen. Similarly, induction of IgG1, IgG2a, and IgG2b ASC by LPS plus cytokines was not different between Stat3fl/flCD19Cr+/+ and Stat3fl/flCD19Cre/+ mice. Finally, class-switched IgG memory B cells of all isotypes tested (IgG1, IgG2a, and IgG2b) were induced at equivalent levels in Stat3fl/flCD19Cr+/+ and Stat3fl/flCD19Cre/+ mice (Figure 7B-C). Taken together, these data demonstrate that there is no general defect in class-switch recombination in Stat3fl/flCD19Cre/+ mice, nor is there a specific defect in TD IgG class-switch recombination. These findings directly exclude defective isotype switching as a potential explanation for the TD IgG defect observed in these mice. Combined with the sharply diminished levels of IgG plasma cells following immunization, these observations demonstrate that the TD IgG defect seen in these mice is due to a defect in the formation of IgG plasma cells from already class-switched IgG B cells (Figure 8).
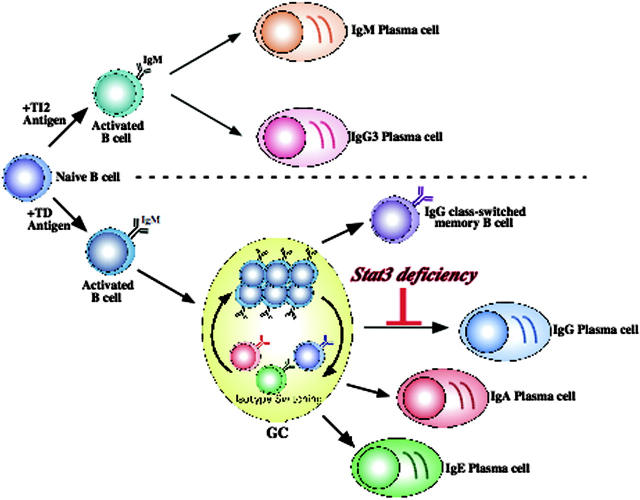
Figure 8.
Stat3 is necessary for TD IgG plasma cell formation. Stat3fl/flCD19Cre/+ mice are capable of forming IgM plasma cells in response to either a TD or TI antigen. In addition, germinal center formation proceeds normally, leading to successful isotype switching to all 3 classes (IgG, IgA, IgE). It is likely that this occurs in distinct anatomic locations for each isotype (not depicted), with IgG predominating in the spleen and lymph nodes; IgA, in the gut and related mucosal tissues such as appendix, Peyer patch, and tonsils; and IgE, in still-unknown sites. These class-switched B cells (of any isotype) can subsequently be selected into the memory pool or terminally differentiate into plasma cells. However, in the absence of B-cell Stat3, IgG B cells are unable to respond to T-cell-derived cytokines that drive terminal differentiation. Thus, Stat3 is uniquely required only for terminal differentiation and only for IgG plasma cells.
Stat3 is well known to function as a survival factor for multiple cell types, including plasma cells, suggesting that defects in survival could contribute to the deficiency in TD IgG in immunized Stat3fl/flCD19Cre/+ mice. Consistent with this, we found a 1.5- to 2-fold reduction in total IgG1-, IgG2a-, and IgG2b-ASC in bone marrow but not spleen of these mice (data not shown). However, this apparent impairment of survival is significantly smaller than the defects in TD IgG production that we found in our Stat3fl/flCD19Cre/+ mice, making it unlikely that impaired survival significantly accounts for our findings, particularly since no significant amounts of Ag-specific IgG were detected at any time point following immunization. Normal formation but impaired survival would be expected to show equivalent early levels of IgG with a more rapid decline, but this is not observed. Hence, these data are most consistent with a severe defect in the initial formation of plasma cells, with any possible survival defects being secondary.
It is interesting that our B-cell Stat3-deficient mice showed a profound defect in TD IgG responses despite normal total serum IgG levels. Which B cells and B-cell subsets contribute to the level of serum Ig remains unclear, but discrepancies between total serum Ig and Ab responses following immunization have been previously noted. For example, mice with engineered null mutations in CD19 show nearly normal levels of serum IgG of all subclasses but sharply impaired TD IgG responses to immunization.23 Thus, humoral responses of mutant mice to experimental immunization cannot be reliably predicted based on the total serum antibody profile.
A model that accounts for the defects in TD IgG plasma cell formation that we observe in Stat3fl/flCD19Cre/+ mice is that activated IgG B cells use Stat3 in a nonredundant fashion to respond to T-cell-derived cytokines that trigger the plasma cell genetic program and hence drive plasma cell formation. As mentioned in “Introduction,” a variety of cytokines that have unique signaling properties nonetheless share the property of activation of Stat3. We postulate that activation of Stat3 by one or more of these cytokines (IL-6, IL-2, IL-10, IL-21, and perhaps others) is an essential step in driving plasma cell formation, and that other signals cannot compensate for this activation step in IgG B cells. The lack of a compensating signal may be in part a function of distinct anatomic sites and/or microenvironments in which class-switched B cells of different isotypes differentiate, or may be an intrinsic property of IgG B cells.
Our data show that Stat3 is unique among transcription factors in its highly selective role in plasma cell development. Unlike transcription factors such as Blimp-131 and XBP-1,32 which are universally required for plasma cell development, Stat3 is the only transcription factor identified to date that has a selective role limited to TD terminal differentiation of a specific B-cell subset. Our data suggest that there may be distinct transcriptional control mechanisms governing different humoral immune responses originating with distinct B-cell subsets.
Prepublished online as Blood First Edition Paper, October 13, 2005; DOI 10.1182/blood-2005-07-2871.
Supported by a National Institutes of Health (NIH) Training Grant GM08061 appointment (J.L.F.), the Concern Foundation and a Research Scholar Award from the American Cancer Society (R.C.R.), the Italian Ministry of University and Research (MIUR) PRIN (V.P.), and the NIH AA014400 (T.J.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Levy DE, Darnell JE Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3: 651-662. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Noguchi K, Shi W, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94: 3801-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sano S, Itami T, Takeda K, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling but does not affect skin morphogenesis. EMBO J. 1999;18: 4657-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman RS, Lourenco PC, Tonner E, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13: 2604-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonzi T, Maritano D, Gorgoni B, Rizzuto G, Libert C, Poli V. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol Cell Biol. 2001;21: 1621-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweizer U, Gunnersen J, Karch C, et al. Conditional gene ablation of Stat3 reveals differential signaling requirements for survival of motoneurons during development and after nerve injury in the adult. J Cell Biol. 2002;156: 287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby JJ, Kalinowski A, Liu MG, et al. Cardiomyocyte-restricted knockout of STAT3 results in higher sensitivity to inflammation, cardiac fibrosis, and heart failure with advanced age. Proc Natl Acad Sci U S A. 2003;100: 12929-12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui Y, Huang L, Elefteriou F, et al. Essential role of STAT3 in body weight and glucose homeostasis. Mol Cell Biol. 2004;24: 258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161: 4652-4660. [PubMed] [Google Scholar]
- 10.Takeda K, Clausen BE, Kaisho T, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10: 39-49. [DOI] [PubMed] [Google Scholar]
- 11.Lee CK, Raz R, Gimeno R, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17: 63-72. [DOI] [PubMed] [Google Scholar]
- 12.Welte T, Zhang SS, Wang T, et al. STAT3 deletion during hematopoiesis causes Crohn's disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci U S A. 2003;100: 1879-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19: 903-912. [DOI] [PubMed] [Google Scholar]
- 14.Zhong Z, Wen Z, Darnell JE Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264: 95-98. [DOI] [PubMed] [Google Scholar]
- 15.Boulton TG, Zhong Z, Wen Z, Darnell JE Jr, Stahl N, Yancopoulos GD. STAT3 activation by cytokines utilizing gp130 and related transducers involves a secondary modification requiring an H7-sensitive kinase. Proc Natl Acad Sci U S A. 1995;92: 6915-6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolling C, Treton D, Pellegrini S, Galanaud P, Richard Y. IL4 and IL13 receptors share the gamma c chain and activate STAT6, STAT3 and STAT5 proteins in normal human B cells. FEBS Lett. 1996;393: 53-56. [DOI] [PubMed] [Google Scholar]
- 17.Johnston JA, Bacon CM, Finbloom DS, et al. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15 Proc Natl Acad Sci U S A. 1995;92: 8705-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asao H, Okuyama C, Kumaki S, et al. Cutting edge: the common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J Immunol. 2001;167: 1-5. [DOI] [PubMed] [Google Scholar]
- 19.Arpin C, Dechanet J, Van Kooten C, et al. Generation of memory B cells and plasma cells in vitro. Science. 1995;268: 720-722. [DOI] [PubMed] [Google Scholar]
- 20.Okada M, Sakaguchi N, Yoshimura N, et al. B cell growth factors and B cell differentiation factor from human T hybridomas: two distinct kinds of B cell growth factor and their synergism in B cell proliferation. J Exp Med. 1983;157: 583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano T, Yasukawa K, Harada H, et al. Complementary DNA for a novel human interleukin (BSF-2) that induces B lymphocytes to produce immunoglobulin. Nature. 1986;324: 73-76. [DOI] [PubMed] [Google Scholar]
- 22.Underhill GH, George D, Bremer EG, Kansas GS. Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood. 2003;101: 4013-4021. [DOI] [PubMed] [Google Scholar]
- 23.Rickert RC, Rajewsky K, Roes J. Impairment of T cell dependent B cell responses and B-1 development in CD19-deficient mice. Nature. 1995;376: 352-355. [DOI] [PubMed] [Google Scholar]
- 24.Hayakawa K, Ishii R, Yamasaki K, Kishimoto T, Hardy RR. Isolation of high-affinity memory B cells: phycoerythrin as a probe for antigen-binding cells. Proc Natl Acad Sci U S A. 1987;84: 1379-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagers AJ, Stoolman LM, Kannagi R, Craig R, Kansas GS. Expression of leukocyte fucosyl-transferases regulates binding to E-selectin: relationship to previously implicated carbohydrate epitopes. J Immunol. 1997;159: 1917-1929. [PubMed] [Google Scholar]
- 26.Waldschmidt TJ, Panoskaltsis-Mortari A, McEl-murry RT, Tygrett LT, Taylor PA, Blazar BR. Abnormal T cell-dependent B-cell responses in SCID mice receiving allogeneic bone marrow in utero: severe combined immune deficiency. Blood. 2002;100: 4557-4564. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5: 230-242. [DOI] [PubMed] [Google Scholar]
- 28.Kopf M, Baumann H, Freer G, et al. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368: 339-342. [DOI] [PubMed] [Google Scholar]
- 29.Ohtani T, Ishihara K, Atsumi T, et al. Dissection of signaling cascades through gp130 in vivo: reciprocal roles for STAT3- and SHP2-mediated signals in immune responses. Immunity. 2000;12: 95-105. [DOI] [PubMed] [Google Scholar]
- 30.Kumanogoh A, Marukawa S, Kumanogoh T, et al. Impairment of antigen-specific antibody production in transgenic mice expressing a dominant-negative form of gp130. Proc Natl Acad Sci U S A. 1997;94: 2478-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelin-Duclos C, Cattoretti G, Lin KL, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with blimp-1 expression in vivo. J Immunol. 2000;165: 5462-5471. [DOI] [PubMed] [Google Scholar]
- 32.Reimold AM, Iwakoshi NN, Manis J, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412: 300-307. [DOI] [PubMed] [Google Scholar]