Abstract
TEL2/ETV7 is highly homologous to the ETS transcription factor TEL/ETV6, a frequent target of chromosome translocation in human leukemia. Although both proteins are transcriptional inhibitors binding similar DNA recognition sequences, they have opposite biologic effects: TEL inhibits proliferation while TEL2 promotes it. In addition, forced expression of TEL2 but not TEL blocks vitamin D3–induced differentiation of U937 and HL60 myeloid cells. TEL2 is expressed in the hematopoietic system, and its expression is up-regulated in bone marrow samples of some patients with leukemia, suggesting a role in oncogenesis. Recently we also showed that TEL2 cooperates with Myc in B lymphomagenesis in mice. Here we show that forced expression of TEL2 alone in mouse bone marrow causes a myeloproliferative disease with a long latency period but with high penetrance. This suggested that secondary mutations are necessary for disease development. Treating mice receiving transplants with TEL2-expressing bone marrow with the chemical carcinogen N-ethyl-N-nitrosourea (ENU) resulted in significantly accelerated disease onset. Although the mice developed a GFP-positive myeloid disease with 30% of the mice showing elevated white blood counts, they all died of T-cell lymphoma, which was GFP negative. Together our data identify TEL2 as a bona fide oncogene, but leukemic transformation is dependent on secondary mutations.
Introduction
The ETS (E26 transformation specific) proteins belong to a large family of eukaryotic transcription factors (TFs) unique to the metazoan lineage and highly conserved throughout evolution.1,2 Some ETS proteins are expressed ubiquitously while others show a restricted tissue distribution.3,4 All ETS proteins possess a highly conserved 85–amino acid (aa) ETS domain that binds a purine-rich GGAA/T core motif present in promoters and enhancers of different target genes.4,5 A subgroup of ETS TFs, including the TEL proteins, also contains a conserved pointed (PNT) protein-protein interaction domain, which mediates the formation of homodimers/oligomers6,7 and heterodimers/oligomers.8,9 In TEL this domain is involved in transcriptional repression.10
Vertebrate ETS TFs are implicated in many aspects of normal development and differentiation, including that of the hematopoietic system.11 For example, Ets-1, Fli-1, and Erg are expressed early during mouse development in the blood islands of the yolk sac where hemangioblasts are present.12 A high level of Pu-1 expression in CD34+ hematopoietic progenitors directs their differentiation toward the macrophage lineage, while low Pu-1 expression induces B-cell differentiation.13 The expression of IL-7Rα, a receptor essential for pro-B-cell development, is directly regulated by Pu-1 in lymphoid progenitors.3,14 The ubiquitously expressed TEL/ETV6 is essential for normal embryonic yolk sac angiogenesis and is important for the maintenance of the adult bone marrow microenvironment.15
Several ETS genes, including ETS1, ETS2, PU1, FLI1, TEL/ETV6, and ERG, possess oncogenic properties.4 Human TEL/ETV6 is a frequent target of chromosome translocations16 both in hematopoietic malignancies as well as in some solid tumors.17 TEL/ETV6 translocations mostly encode oncogenic fusion proteins16,18-21 with some exceptions.22-24 In addition, TEL might also have tumor suppressor activity because the gene is often deleted during later stages of t(12;21) childhood pre–B-cell acute lymphoblastic leukemia (pre–B-ALL).25 This hypothesis is supported by the observation that TEL expression inhibits Ras-induced transformation of NIH3T3 fibroblasts.26,27
Recently, a novel ETS gene highly related to TEL1 was isolated and coined TEL2.9,19,28 TEL2 is expressed predominantly in human hematopoietic tissues,9 contains a PNT domain and an ETS DNA binding domain, and localizes to the nucleus. TEL2 self-associates via its PNT domain but can also form heterodimers/oligomers with TEL1, suggesting that these proteins might affect each other's function in vivo.9,29 Although both proteins function as transcriptional repressors in transient transfection assays,19,29 their biologic effects appear distinct. For example, TEL1 inhibits colony formation of Ras-transformed NIH3T3 fibroblasts, while TEL2 slightly stimulates colony formation.29 During vitamin D3–induced differentiation of the promonocytic cell line U937, TEL2 but not TEL1 expression is down-regulated, and forced expression of TEL2 blocks differentiation. In contrast, overexpression of TEL2 mutants containing an impaired ETS or PNT domain had no effect on vitamin D3–induced differentiation of U937 cells.29 Also TEL2, but not its PNT or ETS mutant versions, cooperates with Myc in B-lymphomagenesis in mice,30 and its expression is up-regulated in some adult human leukemia samples29 and in more than 30% of pediatric ALL patients.30 Moreover, TEL2 is expressed in many human tumor cell lines.31 Together these data suggest a role for TEL2 in hematopoietic malignancy. Herein we show that expression of TEL2 alone in the mouse hematopoietic system causes a nontransplantable myeloproliferative disease (MPD) after a long latency period. This suggests that secondary mutations are necessary for disease development. Indeed, treatment with the DNA-damaging agent N-ethyl-N-nitrosourea (ENU) in mice receiving transplants greatly accelerated tumorigenesis.
Materials and methods
Plasmid constructs, retroviral production, and viral titer determination
TEL2 cDNA was cloned upstream of the IRES element into the unique EcoRI site of the murine stem cell virus (MSCV) IRES-GFP vector.29,32-35 Retroviral constructs were used to generate replication-incompetent retroviral stocks by transient transfection of the Ecotropic Phoenix 293T packaging cells line36 using Fugene 6 (Roche, Indianapolis, IN). Viral titers were determined by infection of NIH3T3 cells in the presence of Polybrene (Sigma Chemical, St. Louis, MO) (8 μg/mL) and varied between 4 × 105 to 1 × 106 CFU/mL.
Bone marrow extraction, Lin- isolation, and retroviral transduction
Bone marrow (BM) cells were harvested by flushing the femurs and tibiae of FVB-J male donor mice 3 days after intraperitoneal injection of 150 μg/g body weight 5-fluorouracil (5FU) (Sigma Chemical). Lineage-negative (Lin-) cells were purified by immunodepletion of cells presenting myeloid, erythroid, and lymphoid differentiation markers using biotinylated mouse antibodies Ly-6G (Pharmingen, San Diego, CA), Cd11b (Pharmingen), Cd45R/B220 (Pharmingen), CD5 (Pharmingen) and TER-119 (Pharmingen) and streptavidin-coated beads (Dynabeads M-280 streptavidin; Brown Deer, WI). The resulting progenitor-enriched population was prestimulated for 48 hours in Iscove medium (Gibco, Carlsbad, CA), 20% fetal bovine serum (Hyclone, South Logan, UT), supplemented with interleukin-3 (IL-3; 20 ng/mL) (Preprotech, London, United Kingdom), interleukin-6 (IL-6; 30 ng/mL) (Peprotech), interleukin-7 (IL-7; 10 ng/mL) (Peprotech), and stem cell factor (SCF; 50 ng/mL) (R&D Systems, Minneapolis, MN) in nontissue culture grade plastic Petri dishes. After prestimulation the cells were plated onto Retronectin (Takara, Otsu, Japan)–coated plates and incubated with the retroviral supernatants twice daily for 2 days in the presence of cytokines.
Colony-forming unit assays
Transduced and normal Lin- cells were plated in semisolid methylcellulose medium (MC1) plus cytokines (10 μg/mL insulin, 200 ng/mL human transferrin, 50 ng/mL SCF, 10 ng/mL IL-3, 10 ng/mL IL-6, 3 U/mL erythropoietin [Epo], StemCell Technologies 03434, Victoria, BC, Canada) at a concentration of 1 × 103 cells per dish Erythroid colony-forming unit (CFU-E), granulocyte erythrocyte monocyte macrophage colony-forming unit (CFU-GEMM), and granulocyte macrophage colony-forming unit (CFU-GM) colonies were scored 10 to 14 days later, pooled, and reseeded at a density of 1 × 103 cells per dish into secondary methylcellulose (MC2) cultures. Part of the cells were used to determine the fraction of GFP-positive cells using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ). We continued MC replating until the cultures were exhausted (usually 3 to 4 consecutive rounds of plating) due to terminal differentiation of the cells.
Long-term culture-initiating cell (LTC-IC) assays
Transduced and normal BM cells (5 × 103 cells per dish) were plated onto irradiated M2-10B4 stromal layers37 in Myelocult media (StemCell Technologies M5300) to which freshly prepared 0.5 μg/mL hydrocortisone (hydrocortisone 21-hemisuccinate; Sigma Chemical) was added. The cells were cultured for 4 weeks, while replacing half the medium every 7 days. At the end of this period the cells were harvested, analyzed for GFP expression by fluorescence-activated cell sorting (FACS), and plated in methylcellulose media. We scored the number and type of colonies 2 weeks later. Serial MC replating and concomitant GFP analyses were performed for 6 to 8 weeks.
Bone marrow transplantation (BMT)
Female 6- to 12-week-old FVB mice were lethally irradiated (single dose of 950 cGy) and 24 hours later received transplants by tail vein injection with 300 μL PBS plus heparin (30 U/mL) containing 3 × 105 to 5 × 105 Lin- cells transduced with retrovirus or not. After BMT, the animals were evaluated daily for possible signs of disease. Peripheral blood was obtained monthly by retro-orbital phlebotomy and was analyzed by FACS to determine the percentage of white blood cells (WBCs), red blood cells (RBCs), and platelets expressing GFP. Complete blood counts (CBCs) were performed with a Hemavet 3700 (Drew Scientific, Cumbria, United Kingdom), and Giemsa-stained blood smears were analyzed to verify the presence of abnormal cells.
ENU mutagenesis
TEL2 BMT was performed exactly as described in “Bone marrow transplantation (BMT),” but 5 weeks after transplantation the recipients received intraperitoneal injections twice, at a 48-hour interval, with 100 mg/kg ENU. The health status of the animals was followed as described in “Bone marrow transplantation (BMT).”
Analysis of diseased mice and tissue preparation
All the following animal procedures were conducted in accordance with the U.S. Public Health Service Policy on the Humane Care and Use of Laboratory Animals. Peripheral blood of moribund animals was isolated for GFP analysis, blood smears, and CBCs. The animals were killed using CO2 asphyxiation, and the weight of the spleen and liver was recorded. All organs were recovered, fixed in 10% neutral-buffered formalin, processed and embedded in paraffin, sectioned at 4 to 5 μm, and stained with hematoxylin and eosin (H&E) for routine histologic examination. Sternum and hind limbs were subjected to an additional decalcification step before tissue processing. Select tissues were also processed for immunohistochemical analysis with antibodies to the hematopoietic markers CD3 (Dako, Carpinteria, CA), CD45R/B220 (Pharmingen, San Diego CA), terminal deoxynucleotidyl transferase (TdT; Supertechs, Bethesda, MD), myeloperoxidase (MPO; Dako), TER-119 (Pharmingen, San Diego, CA), GATA1 (Santa Cruz Biotechnology, Santa Cruz, CA), and green fluorescent protein (GFP; Clontech, Palo Alto, CA).
Single-cell suspensions were prepared from bone marrow, spleen, and liver for analysis of GFP expression by FACS and for the preparation of cytospin slides (5 × 104 cells per slide) for morphologic examination after May-Grünwald-Giemsa (MGG) staining. After lysis of the red cells, the leukocytes were analyzed immediately for surface marker expression or frozen in fetal calf serum (FCS)/10% DMSO for later analysis. Images of tissue sections and cytospins were obtained using a BX51 microscope equipped with a Uplan FL 40 ×/0.75 numeric aperture (NA) or a 100 ×/1.30 NA objective (Olympus, Tokyo, Japan). Images were acquired using a SPOT camera and SPOT Advanced imaging software (Diagnostic Instruments, Sterling Heights, MI). Original magnification for tissue sections and cytospins was 400 × and 1000 ×, respectively.
Flow cytometric analysis
Single-cell suspensions of BM, spleen, and liver were washed and incubated for 30 minutes on ice in staining medium (SM: DMEM supplemented with 10% fetal bovine serum) containing human γ-globulin (100 mg/mL; Sigma Chemical) to block Fc receptors. After washing, cells were incubated with monoclonal antibodies (CD3c, CD4, CD8, CD11b/Mac1, CD19, CD34, B220, TER-119, Gr1, Sca1, c-Kit, Flt3, all from Pharmingen, San Jose, CA; anti–mouse IgM from Southern Biotechnology Associates, Birmingham, AL) on ice for 30 minutes. After a final washing step, cells were resuspended in SM and analyzed using a BD Biosciences FACSCalibur flow cytometer (BD Biosciences, San Jose, CA), selecting single cells by gating on forward versus side light scatter. Wild-type and TEL2-expressing bone marrow cells were cultured for 2 weeks, and apoptotic cells were identified by FACS after annexin V–FITC staining. Propidium iodine staining was used to exclude dead cells.
Western blotting
For Western blotting, protein extracts were prepared from spleen samples of diseased TEL2 and healthy control mice, killed at the same time, using TRI Reagent (Sigma Chemical), following the manufacturer's instructions. After quantification using the BCA Protein Assay Reagent (Pierce Chemical, Rockford, IL), 30 μg total protein was separated on 10% SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) gels under reducing conditions and transferred onto a polyvinylidine difluoride membrane (Millipore, Billerica, MA). The membranes were incubated with the antibodies specific for Bcl-x (Transduction Laboratories, Lexington, KY), recognizing both Bcl-xl and Bcl-xs, Bcl2 antibody (554218; Pharmingen), and TEL2 antibody.30
Secondary transplantations
Lethally irradiated (9.5 Gy [950 rad]) recipient mice received injections in the tail vein with 1 × 106 freshly isolated bone marrow or spleen cells from moribund primary recipients after red cell lysis. Mice receiving transplants were followed and killed as described for the primary recipients.
Affymetrix GeneChip analysis
BM from 5FU-treated mice was either mock transduced (Un-BM) or transduced with MSCV-IRES-GFP (GFP-BM) or MSCV-TEL2-IRES-GFP (TEL2-BM) retrovirus as described in “Bone marrow transplantation (BMT).” After transduction, cells were allowed to recover in culture for 48 hours and were sorted for GFP expression by FACS. RNA of the GFP-positive cells and mock-transduced cells was isolated using Trizol (Invitrogen, Carlsbad, CA) following the manufacturer's recommendations.
RNA quality was confirmed by UV spectrophotometry and by analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). Ten micrograms of total RNA was processed in the St Jude Hartwell Center Core Facility according to the standard Affymetrix (Santa Clara, CA) protocol38 and analyzed on the Affymetrix MOE-430A GeneChip array. Signal values, detection calls, and pairwise expression analyses were performed using the default parameters within the statistical algorithm of the Affymetrix GCOS software version 1.2. Signal values were scaled to a 2% trimmed mean target value of 500. Stringent selection criteria were applied to identify differential expression associated with TEL2. The following pairwise comparisons were performed: TEL2-BM versus GFP-BM, TEL2-BM versus Un-BM, and GFP-BM versus Un-BM. Initial selection was based on differential expression between TEL2-BM and GFP-BM. Probe sets with a log2 ratio greater than one (more than 2-fold change) plus a “change call” (P < .006, Wilcoxon signed rank test) were selected for further evaluation. To best identify TEL2-associated changes, the initial selection was filtered to exclude probe sets with less than 2-fold change in TEL2-BM versus Un-BM and those with more than 2-fold change in GFP-BM versus Un-BM. Probe set annotations were obtained from the Affymetrix website.39 Functional classification of genes was performed using the Affymetrix Gene Ontology browser. Biologic processes significantly enriched within the TEL2-associated gene list were identified using the χ2 test (P < .001).
Results
TEL2 expression affects the colony-formation potential of primitive myeloid progenitors in vitro
To test the effect of forced TEL2 expression in mouse hematopoietic cells, Lin- cells of 5-FU treated donor mice were transduced with MSCV-IRES-GFP or MSCV-TEL2-IRES-GFP retrovirus (Figure 1A). FACS analysis 4 days later showed that 10% to 50% of the cells expressed GFP (not shown), and were used for in vitro colony-forming assays and in vivo bone marrow reconstitution of lethally irradiated recipient mice.
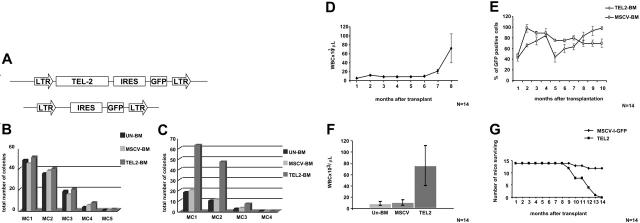
Figure 1.
BM-expressing TEL2 causes a myeloid proliferative disease in mice. (A) Schematic representation of the MSCV-IRES-GFP and MSCV-TEL2-IRES-GFP retroviral vectors used for the transduction of the Lin- BM cells. The TEL2 cDNA was cloned in the single EcoRI site and is followed by an IRES-GFP marker gene. LTR, long terminal repeat. (B) Myeloid clonogenic activity of BM cells transduced with TEL2 retrovirus (TEL2-BM) compared with BM cells transduced with vector (MSCV-BM) or untransduced BM cells (Un-BM). Bars indicate the number of colonies counted at each round of serial replating in MC1 to MC5. CFU (1000 cells per dish plated) counts show no difference among the 3 samples with regard to their successive colony-forming capacity. This graph depicts the result of 1 of 4 experiments that gave almost identical results. (C) Successive MC assays of TEL2-BM, MSCV-BM, and Un-BM cells after 4 weeks of LTC-IC culture on an M2-10B4 stromal layer showing an increased colony-forming capacity of the TEL2-BM cells during the first 2 rounds of MC culture. This graph depicts the result of 1 of 4 experiments that gave almost identical results. (D) Average monthly peripheral blood leukocyte counts of all mice receiving transplants with TEL2-BM, showing an increase of the WBCs starting at 6 months after transplantation. Error bars show the standard deviation of each data point. (E) Monthly percentage of GFP-positive cells in the peripheral blood of mice receiving transplants with MSCV-BM or TEL2-BM, showing a steady increase in GFP-positive cells in mice receiving transplants with TEL2-BM starting at 5 months after transplantation. Error bars show the standard deviation of each data point. (F) Comparison of the average leukocyte count—in peripheral blood of mice receiving transplants with UN-BM, MSCV-BM and TEL2-BM—at the moment of death of the TEL2-BM mice. (G) Combined Kaplan-Meier survival plot of 14 (2 × 7) lethally irradiated mice receiving transplants with MSCV-BM or TEL2-BM (n = 2).
To assess whether TEL2 expression conferred a proliferative advantage to hematopoietic cells in vitro, we compared the growth characteristics of Lin- cells transduced with empty retrovirus (MSCV-BM) or a TEL2 retrovirus (TEL2-BM) with that of uninfected Lin- cells (Un-BM). To assess the colony-forming capacity of committed progenitors we plated the cells into MC1 and 2 weeks later scored the cultures for the number and type of colony-forming units (CFU-E, CFU-GEMM, CFU-GM). After counting, 103 cells from the MC1 were replated into fresh MC and CFUs were scored, and this process was repeated until the CFU activity was exhausted (3 to 4 rounds). However, we observed no differences in the type, size, or number of the Un-BM, MSCV-BM, and TEL2-BM colonies (Figure 1B). We next tested whether TEL2 expression affected the growth of primitive progenitors and followed the clonogenic potential of TEL2-BM and MSCV-BM progenitors in myeloid MCs after long-term culture-initiating cell (LTC-IC) cultures. This experiment was repeated twice with both types of bone marrow containing 10% GFP-positive cells. After 4 weeks of LTC-ICs, cells were collected and plated into serial MC assays. While the percentage of GFP-positive cells in MSCV-BM was similar before and after LTC-ICs, the percentage of GFP-positive cells in the MC1 of TEL2-BM LTC-ICs had increased to 60% and the number of colonies in the MC1 was 3-fold higher than that in the MC1 of MSCV-BM cells. Upon serial replating, the elevated percentage of GFP-positive cells in the TEL2 MCs remained, but the number of colonies rapidly dropped to a level similar to that of MSCV-BM cells (Figure 1C). Thus, TEL2-expressing primitive progenitors appeared to have a growth advantage in LTC-IC assays only. When transduced Lin- bone marrow cells used for the LTC-ICs were directly plated onto tissue culture plastic, they formed their own stromal layer. Under these conditions TEL2-BM cells grew faster than control MSCV-BM cells (not shown), and the percentage of GFP-positive cells again increased from 10% to 60% during 3 weeks of culture. This suggested that interaction with stromal cells, whether in LTC-IC cultures or in stroma-forming BM cultures, provided a growth advantage to TEL2-BM cells.
Bone marrow–expressing TEL2 induces a myeloproliferative disease in FVB recipient mice
We simultaneously tested whether TEL2 expression in reconstituted BM30 would affect normal hematopoiesis in vivo and transplanted Un-BM, MSCV-BM, and TEL2-BM cells into 7 lethally irradiated FVB recipients and assessed their peripheral blood parameters and the percentage of GFP-positive cells monthly. The same experiment was repeated 3 months later, and the combined results of these experiments are presented in Figure 1D-G. All animals receiving transplants engrafted, and the peripheral blood counts (PBCs) remained stable for 6 months, followed by an increasing leukocytosis (Figure 1D) until the mice became moribund 2 to 3 months later. At the time the animals were killed, they showed an average peripheral WBC count of 75 × 109/L (75 × 103/μL) (Figure 1E), with all cells expressing GFP (Figure 1F). All TEL2-BM mice died within 8 to 14 months after transplantation (Figure 1G). Instead, mice receiving transplants with Un-BM or MSCV-BM cells showed normal blood parameters and remained healthy, although 2 MSCV-BM animals died of causes not related to hematopoietic disease.
Average CBCs of mice (Un-BM, MSCV-BM, and TEL2-BM) were determined at the time of death of the TEL2-BM mice (Table 1).
Table 1.
Average complete blood count (CBC) in peripheral blood of mice receiving transplants with Un-BM, MSCV-BM, and TEL2-BM, at the moment of death of the TEL2-BM mice
| UN-BM | MSCV-BM | TEL2-BM* | |
|---|---|---|---|
| Complete WBC count, × 109/L | 8.6 ± 3.6 | 10.3 ± 4.9 | 76 ± 35.32 |
| Neutrophil count, × 109/L | 1.48 ± 0.52 | 2.36 ± 0.87 | 14.7 ± 13.94 |
| Lymphocyte count, × 109/L | 4.99 ± 1.68 | 4.64 ± 1.6 | 15.3 ± 14.05† |
| Monocyte count, × 109/L | 0.55 ± 0.28 | 0.57 ± 0.39 | 3.2 ± 2.8 |
| Eosinophil count, × 109/L | 0.019 ± 0.008 | 0.013 ± 0.005 | 1.14 ± 0.96 |
| Basophil count, × 109/L | 0 | 0 | 0.23 ± 0.19 |
| MCH, pg | 13.42 ± 1.45 | 15.19 ± 1.31 | 17.9 ± 4.4 |
| RBC count, × 1012/L | 7.46 ± 0.71 | 7.6 ± 0.811 | 4.66 ± 1.65 |
| MCV, fL | 44.84 ± 3.14 | 49.23 ± 3.32 | 62.68 ± 14.29 |
| Platelet count, × 109/L | 1176 ± 68.63 | 1406 ± 204 | 1094 ± 303 |
| Hemoglobin level, g/L | 127 ± 17 | 130 ± 17 | 84 ± 14 |
The results are expressed as average ± SD. The 2-tail unpaired Student t test was used to compare the data. P ≤.05 was considered significant.
MCH indicates mean corpuscular hemoglobin; MCV, mean corpuscular volume.
P values calculated between the TEL2-BM and the MSCV-BM values were < .05
The high number of lymphocytes in the Hemavet counts of the TEL2-BM mouse blood samples is a gross overestimate because the blast cells and partially differentiated myeloid cells have the same size as lymphocytes and are therefore scored as “lymphocytes.” This was confirmed by morphologic analysis of the blood smears (Figure 2A)
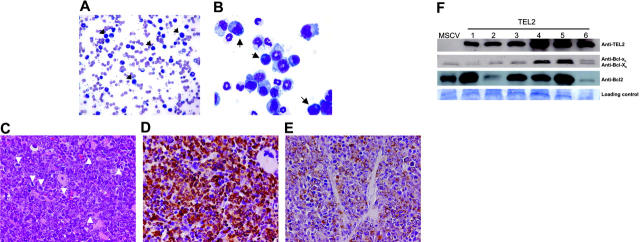
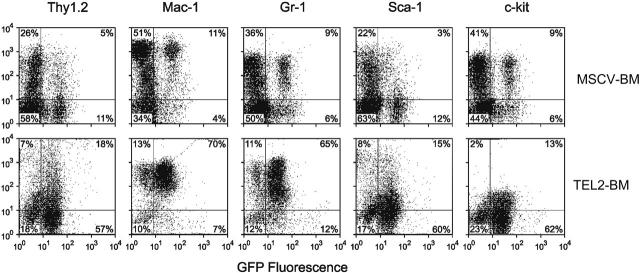
Assessment of cell morphology of peripheral blood smears (Figure 2A) showed anemia, large numbers of mature and immature neutrophils, and the presence of some blastlike cells. BM cytospins (Figure 2B) of affected mice revealed the presence of mostly myeloid cells with a similar abundance of mature and immature neutrophils, but also blastlike cells, monocytes, and promyelocytes. All TEL2-BM animals showed splenomegaly and hepatomegaly, due to extensive infiltration of a population of cells with a high mitotic index, which expressed GFP (Figure 2C-D). The spleen had completely lost its normal architecture (not shown), and the cell population in the red pulp consisted of erythroid precursors, megakaryocytes, and a vast excess of mature granulocytes and immature myeloid cells. Unlike the spleen, the liver infiltrate consisted of mature granulocytes and immature myeloid cells. The immature cells were the dominant constituent in both tissues, and most of them expressed MPO consistent with a granulocytic lineage (Figure 2E). Western blot analysis confirmed expression of TEL2 in the spleen of the diseased mice (Figure 2F). Given that the antiapoptotic effect of TEL2 on B-cell progenitors is associated with increased expression of Bcl2 but not Bcl-xl,27 we also tested the expression of these 2 antiapoptotic proteins in the same spleen samples. Compared with MSCV-BM spleen cells, 2 TEL2-BM tumor samples expressed an increased amount of Bcl-xl and Bcl-xs, whereas Bcl2 expression was increased in 4 of 6 tumor samples. Two tumors samples showed no up-regulation of Bcl2 or Bcl-xl, indicating that the antiapoptotic proteins were not consistently up-regulated in all tumors. FACS analysis of BM (Figure 3) showed that the malignant cells were Mac1+ and Gr1+. BM also showed an increase in Sca1+, c-Kit+, and Thy1.1+ progenitors. Together these features are consistent with a diagnosis of a chronic myelomonocytic leukemia (CMML)–like disease,40 suggesting that TEL2 enlarges an early myeloid progenitor population.
Figure 2.
Mice receiving transplants with TEL2-BM develop a chronic myelomonocytic-like disease. (A) Peripheral blood smear stained with May-Grünwald-Giemsa (MGG) of a diseased mouse that underwent TEL2-BM transplantation. Arrows indicate blastlike cells. (B) Cytospin preparation after MGG staining of bone marrow of a diseased mouse that underwent TEL2-BM transplantation, showing an excess of myeloid cells in early stages of differentiation. Arrows indicate blast cells. (C) Spleen—of a diseased mouse that underwent TEL2-BM transplantation—with an extensive red pulp infiltrate of myeloid cells with a high mitotic index (H&E, magnification × 40). Mitotic figures are indicated by white arrowheads. (D) The myeloid cells expressed GFP (brown stain), confirming their transplant derivation (anti-GFP, magnification × 40). (E) The myeloid population was primarily composed of immature cells that expressed MPO (brown stain), consistent with their granulocytic lineage (anti-MPO with cytoplasm localization, magnification × 40). (F) Top row shows a Western blot analysis of TEL2 expression in spleen samples of 1 MSCV-BM (MSCV, 10 months after transplantation) and 6 diseased TEL2-BM mice at the moment of death. The second row shows Bcl-xl and Bcl-xs expression in the same samples. Bcl-xl and Bcl-xs expression in leukemic spleen samples is not altered compared with that in the control spleen sample. The third row shows that 4 of 6 leukemic TEL2 spleens show increased Bcl2 expression (samples 1, 3, 4, and 5). The bottom row shows protein loading of the blot after staining with Coomassie blue.
Figure 3.
Flow cytometric cell surface marker analysis of BM cells of a diseased mouse that received TEL2-BM transplants and a healthy control mouse that received MSCV-BM transplants. (A) Top row shows expression of the indicated cell surface markers (y-axis) versus GFP expression (x-axis) in MSCV-BM cells. The bottom row shows expression of these markers and GFP in TEL2-BM cells. TEL2-BM cells show increased percentages of cells expressing all of the markers, but most cells are positive for Mac1 and Gr1. The malignant cells in the spleen, liver, and peripheral blood expressed the same complement of cell surface markers (not shown).
Transplantation of the diseased bone marrow into 12 sublethally irradiated secondary recipients failed to reproduce disease. The percentage of GFP-positive cells in the peripheral blood of the secondary recipients ranged between 5% and 35% during the first 3 months after transplantation but diminished to almost undetectable levels thereafter. Only 1 of 12 secondary recipients showed a distinct myeloproliferation (93 × 109/L [93 × 103/μL]) associated with organ infiltration, but the percentage of GFP-positive cells in peripheral blood was 12% and the population of cells infiltrating the different organs was GFP negative. We concluded that this unique case was caused by a genetic event not related to overexpression of TEL2.
Annexin V staining of FVB Lin- BM cells
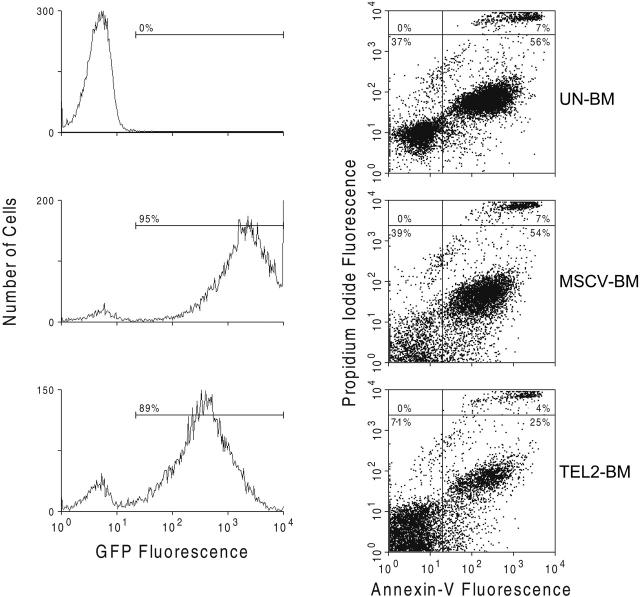
Because TEL2 expression in pre-B cells inhibits their rate of apoptosis,30 we assessed whether a similar effect of TEL2 in myeloid cells might provide a proliferative advantage in the mice receiving transplants. We analyzed the percentage of apoptotic cells by annexin V staining in Lin- Un-BM cells or in GFP-positive MSCV-BM and GFP-positive TEL2-BM cells after culture for 7 days in liquid media promoting myeloid proliferation. Compared with both Un-BM and GFP-positive MSCV-BM cells, GFP-positive TEL2-BM cells showed a consistent 50% decrease in the number of apoptotic cells (Figure 4). This suggested that the myeloproliferative disease caused by TEL2-BM cells in mice might in part be attributed to a reduced level of apoptosis of myeloid progenitors in vivo.
Figure 4.
TEL2 expression reduces the rate of apoptosis of BM cells cultured in vitro. UN-BM cells (top 2 panels) and retrovirus-transduced MSCV-BM cells (middle 2 panels) and TEL2-BM cells (bottom 2 panels) were inoculated in liquid culture in presence of myeloid growth factors. After 4 days of culture and staining with propidium iodide, cells were analyzed for GFP expression (left panels) and for annexin V expression (right panels) by FACS. The number of apoptotic cells in the TEL2-BM sample (bottom right quadrant of the dot plots) was half of that in the UN-BM and MSCV-BM samples. The same was true for the percentage of dead cells (top right quadrant of the dot plots).
To obtain information about TEL2-induced changes in gene expression, we transduced FVB Lin- cells with MSCV-IRES-GFP (MSCV-Lin- cells) or MSCV-TEL2-IRES-GFP retrovirus (TEL2-Lin- cells) and sorted GFP-positive cells 72 hours later by FACS. Relative RNA abundance in these 2 GFP-positive cell populations and in untransduced Lin- cells was determined by Affymetrix microarray analysis (MOE-430A). We then compared gene expression differences between Un-Lin- and MSCV-Lin- and MSCV-Lin- and TEL2-Lin- cells (Figure 5). This 3-way comparison allowed elimination of genes whose expression was changed due to GFP virus infection rather than TEL2 expression. A total of 195 genes was at least 2-fold up- or down-regulated (for the full list, see Table S1, available on the Blood website; see the Supplemental Table link at the top of the online article). Gene ontology analysis of this data set identified changes in expression of genes involved in apoptosis and differentiation/proliferation. This revealed changes in expression of numerous genes involved in apoptosis, including the proapoptotic genes Trp53inp1, Stk17b (DRAK2),41 Dapk2,42 and Bmf.43 We also noticed down-regulation of antiproliferative genes including Btg1,44 Btg2,45,46 Cdkn1B (p27Kip1),47,48 and Rb1cc1 49-51 and up-regulation of cKit, whose activation is associated with acute myelogenous leukemia (AML).52 There was also a general down-regulation of differentiation-associated genes, including cFes,53,54 Btg1, and Btg2 and of the tumor suppressor gene Tgfbr255 (Figure 4; Table S1). Notably, there was no up-regulation of Bcl2 or Bcl-x mRNA levels in TEL2-Lin– cells.
Figure 5.
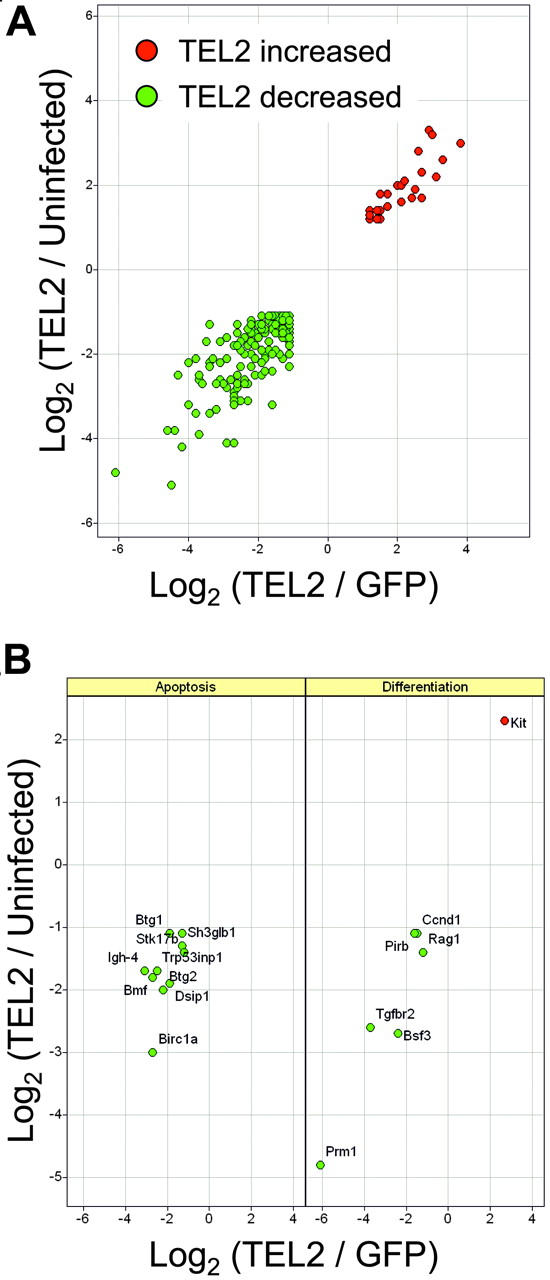
Microarray analysis of gene expression changes associated with TEL2 expression in mouse bone marrow. RNA from mouse bone marrow cells, either mock transduced (uninfected) or transduced with MSCV-TEL2-IRES-GFP (TEL2) or MSCV-IRES-GFP (GFP), were analyzed using the MOE-430A Affymetrix GeneChip microarrays. Log2 ratio values from pairwise comparisons are plotted. (A) Relative expression of 195 probe sets identified as differentially expressed (see “Materials and methods”) in both TEL2 versus GFP (x-axis) and TEL2 versus uninfected (y-axis) bone marrow. (B) Relative expression of differentially expressed probe sets found enriched (P < .001) in the Gene Ontology categories of apoptosis (left) or differentiation (right). Gene symbols are indicated in the panels.
ENU treatment of mice receiving transplants with TEL2 induces myeloid disease and T-cell lymphoblastic leukemia
The long latency of disease suggested that enforced TEL2 expression alone was insufficient to cause the myeloproliferative phenotype and that disease development depended at least on one or possibly more secondary mutations. To test this scenario we performed random mutagenesis by injecting 5 mice receiving transplants with vector-transduced BM and 10 mice receiving transplants with TEL2-transduced BM (30% GFP positive) with ENU 5 weeks after transplantation. The TEL2-BM-ENU animals started to die 2 months earlier than the MSCV-BM-ENU control group, and the average survival time was 2 months shorter (5 months) than that of MSCV-BM-ENU animals (7 months) (Figure 6A). Moribund mice had a large spleen and liver with cells staining positive for CD3 (Figure 6B) and TdT (Figure 6C), which were GFP negative (Figure 6D). Thus, the mice suffered from T-cell lymphoma not expressing TEL2. In addition, 3 of the mice had a significantly elevated WBC count (30 × 109/L to 50 × 109/L [30 × 103/μL to 50 × 103/μL]) in the peripheral blood. These mice had an abundance of poorly differentiated dysplastic GFP-positive (50% to 70%) cells in the peripheral blood (Figure 6E), which were not T cells because only a few of them stained positive for CD3 (Figure 6F). The BM of these mice as well as of mice that had normal peripheral WBCs did show a high percentage of Mac1+ (31% to 72%), Sca1+ (25% to 30%), and GFP-positive (50% to 70%) cells, but no cells were positive for CD3, CD4, or CD8 T-cell markers. Therefore, the mice also suffered from a myeloid disease that in 30% of the mice gave elevated WBCs. Two of the 5 MSCV-BM-ENU control mice died of solid tumors, whereas the remaining 3 died of unknown causes. However, no controls died of a hematopoietic malignancy because their peripheral blood counts remained normal and the percentage of GFP-positive cells remained constant.
Figure 6.
ENU treatment of mice receiving TEL2-BM transplants accelerates hematopoietic disease and causes T-cell lymphoma. (A) Kaplan-Meier survival curve of mice receiving transplants with bone marrow transduced with MSCV-IRES-GFP (MSCV-BM) or MSCV-TEL2-IRES-GFP (TEL2-BM) and treated with ENU 5 weeks after transplantation The mice receiving TEL2-BM transplants died 1.5 to 2 months earlier than the MSCV-BM mice, suggesting cooperation between TEL2 expression and ENU mutagenesis. (B) Spleen of a mouse with T-lymphoblastic lymphoma that underwent ENU treatment and TEL2-BM transplantation. The malignant lymphocytes in the red pulp express CD3 (brown color) while most of the lymphocytes associated with the follicular marginal zone do not express CD3 (anti-CD3 with cytoplasm/membrane localization, magnification × 40, marginal zone is on the left and the red pulp cells with CD3 expression are on the right). (C) The CD3+ malignant lymphocytes also expressed TdT (brown color), which is consistent with a lymphoblastic lymphoma (anti-TdT with nuclear localization, magnification × 40). (D) The T lymphocytes in the spleen do not stain with a GFP antibody; the only cells that are positive for GFP (bottom right corner) are B lymphocytes, which also stain positive for B220 (not shown). (E) Peripheral blood smear of a TEL2-BM/ENU mouse with increased WBCs (30 × 109/L [30 × 103/μL]) showing numerous dysplastic white cells (magnification × 100). (F) Blood smear of the same mouse stained with a CD3 antibody. Only a small percentage of cells (white arrowhead, red color) expressed this T-cell marker, indicating that the dysplastic cells are not of T-cell origin. The nucleated cells were counterstained with DAPI (blue color).
Discussion
TEL2, a new member of the ETS TFs, was identified by several groups a few years ago.9,19,28 The protein was coined TEL2 because of its high homology with TEL (38.2% identity). However, despite this homology, its function appears to be quite distinct from that of TEL. TEL has an antiproliferative effect26,27,56 whereas TEL2 has a mostly proliferative effect,27,29 although a direct assessment of the tumorigenic activity of TEL2 alone has not been determined. Our in vitro LTC-IC studies strongly suggested that TEL2 stimulated the growth of primitive progenitors, provided they are in contact with a feeder layer, a condition that mimics the bone marrow environment. This effect on myeloid cells is different from TEL2's effect on B-cell progenitors, whose growth is stimulated in the absence of feeder cells.27
TEL2 is specifically expressed in the human hematopoietic system, and its expression is up-regulated in some adult human leukemias29 and more than 30% of pediatric B-ALL.30 TEL2 also accelerated B lymphomagenesis in Eμ-Myc mice, a model for Burkitt lymphoma, and possibly cooperates with N-MYC/C-MYC in pediatric B-cell lymphoma.30 Therefore, we tested whether enforced TEL2 expression alone in mouse bone marrow predisposed the animals to hematopoietic malignancy. Our experiments indeed verified the oncogenicity of TEL2 in this setting, but the long latency of disease indicated that secondary mutations are necessary for disease development.
The disease was not transplantable in secondary recipients, although the proliferation of the myeloid cells appeared strictly cell autonomous given the tight correlation between GFP expression and the number of leukemic cells (Figure 1E). This indicated that progenitor cells expressing TEL2 were not immortalized and that the conditions in the reconstituted bone marrow were unable to support reconstitution by the MPD. Similar observations have been reported for MPD caused by NUP98-Hox A9.57 Annexin V analysis of TEL2-expressing bone marrow after a short period (1 week) of in vitro culture showed that TEL2 suppressed the apoptotic index of these cells as it does inhibit apoptosis in Myc-overexpressing or wild-type B-cell progenitors.30 Given the large excess of myeloid progenitors in the BM and the outcome of the Affimetrix array analysis of Lin- cells 72 hours after transduction with TEL2 or vector retrovirus suggested that TEL2 also inhibits apoptosis of myeloid progenitors. Several proapoptotic genes were repressed, such as Trp53inp1, Stk17b, Dapk2, and Bmf, which might mediate this effect. In addition, 4 of 6 mice analyzed for Bcl2 or Bcl-xl expression in myeloid cells, which invaded the spleen, showed up-regulation of Bcl2, Bcl-xl and Bcl-xs. Similar to the effect of TEL2 in B cells,30 up-regulation of Bcl2 and Bcl-x is probably not at the transcriptional level, because Bcl2 and Bclx mRNA was not increased in the Affimetrix arrays of TEL2-transduced bone marrow. We believe that Bcl2 and Bcl-x overexpression contributes to expansion of the myeloid cells, but currently we do not know which events substitute for Bcl2 and Bcl-x overexpression in the 2 tumor samples that did not overexpress these proteins. However, changes in expression of genes inhibiting (Btg1, Btg2, Cdkn1B, Rb1cc1) or stimulating (cKit) proliferation could have an additive effect on the enlargement of the myeloid progenitor pool. It remains to be determined whether any of these genes are direct transcriptional targets of TEL2, an issue that we will address in future studies.
It is reasonable to speculate that myeloid progenitors in the bone marrow of mice receiving transplants have a reduced apoptotic rate, but this does not lead to increased numbers of myeloid cells in the peripheral blood during the first 6 months after transplantation, a feature also apparent in mice expressing MLL fusion oncogenes and AML1-ETO.58,59 It is possible that compensatory mechanisms control the number of myeloid cells entering the periphery and that additional genetic changes, possibly those that affect cell proliferation, may override this check, resulting in accumulation of differentiating myeloid cells in the peripheral blood. In addition, expansion of the myeloid compartment during the first 6 months after transplantation would increase the frequency of secondary mutations and thereby promote development of disease. The extensive differentiation of the cells is the likely cause of the chronic character of the disease, which after initial detection took 2 to 3 months to kill the animal. The differentiation of the malignant cells in affected animals seems in conflict with the observation that TEL2 inhibits differentiation of HL60 and U937 myeloid cells.29 The reason for this is unknown but, unlike bone marrow, cell lines harbor many mutations, which might account for this difference in behavior.
Given the development of myeloproliferative disease in mice receiving transplants with TEL2, we expected that ENU mutagenesis would induce myeloid leukemia. This occurs in mice expressing RUNX1-ETO or CBFβ-MYH11,59,60 indicating that these fusion transcription factors specifically affect myeloid cells despite the fact that conditional RUNX1-ETO knock-in mice59 also express the fusion protein in their T cells.61 Our experiments indicate that in combination with ENU all TEL2-BM mice developed distinct myeloid aberrations in the bone marrow within 3 to 6 months after ENU treatment, which is much more rapid than TEL2-BM mice only (Figure 1G). However, only 3 mice showed elevated numbers of dysplastic cells in the peripheral blood, indicating a possible development of myeloid leukemia. Surprisingly, all mice also developed GFP-negative T-cell lymphoma. We must conclude that changes caused by TEL2 expression promoted T-cell lymphomagenesis, because none of the control ENU-treated mice developed T-cell lymphoma. It is possible that T-cell proliferation is stimulated indirectly by TEL2-expressing hematopoietic cells, which could contribute to increased genetic instability, making these cells more susceptible to ENU-mediated leukemic transformation. The lethal lymphoid disease might have prevented the myeloid preleukemic state of these mice to develop into AML.
Together our results suggest that TEL2 is a hematopoietic oncogene in the mouse, and its overexpression in some adult and 30% of pediatric ALL samples29,30 also argues for a causative role of TEL2 in human leukemogenesis.
Supplementary Material
Acknowledgments
We thank Blake McGourty for the supply of C57BL/6/129svJ-mixed background mice and technical assistance. We gratefully acknowledge Ann-Marie Hamilton Easton and Richard Ashmun for expert FACS analysis, Dorothy Bush for technical help, and Charlette Hill for editing this manuscript. We are especially grateful to Derek Persons for analyzing blood smears of TEL2/ENU mice.
Prepublished online as Blood First Edition Paper, October 18, 2005; DOI 10.1182/blood-2005-03-1196.
Supported by National Cancer Institute (NCI) grant CA72999-08 and the cancer center (CORE) support grant CA217G and by the American Lebanese Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital.
C.C. performed research, analyzed data, and wrote the paper; M.P. and J.B. performed research; J.E.R. performed histopathology and immunocytochemistry analyses; G.N. analyzed data; and G.C.G. designed research, analyzed data, and wrote the paper.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Degnan BM, Degnan SM, Naganuma T, Morse DE. The ets multigene family is conserved throughout the Metazoa. Nucleic Acids Res. 1993;21: 3479-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laudet V, Niel C, Duterque-Coquillaud M, Leprince D, Stehelin D. Evolution of the ets gene family. Biochem Biophys Res Commun. 1993;190: 8-14. [DOI] [PubMed] [Google Scholar]
- 3.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303: 11-34. [DOI] [PubMed] [Google Scholar]
- 4.Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75: 1-55. [DOI] [PubMed] [Google Scholar]
- 5.Janknecht R, Nordheim A. Gene regulation by Ets proteins. Biochim Biophys Acta. 1993;1155: 346-356. [DOI] [PubMed] [Google Scholar]
- 6.Kim CA, Phillips ML, Kim W, et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20: 4173-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacronique V, Boureux A, Valle VD, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278: 1309-1312. [DOI] [PubMed] [Google Scholar]
- 8.Baker DA, Mille-Baker B, Wainwright SM, Ish-Horowicz D, Dibb NJ. Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature. 2001;411: 330-334. [DOI] [PubMed] [Google Scholar]
- 9.Potter MD, Buijs A, Kreider B, van Rompaey L, Grosveld GC. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood. 2000;95: 3341-3348. [PubMed] [Google Scholar]
- 10.Fenrick R, Amann JM, Lutterbach B, et al. Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol Cell Biol. 1999;19: 6566-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassuk AG, Leiden JM. The role of Ets transcription factors in the development and function of the mammalian immune system. Adv Immunol. 1997;64: 65-104. [DOI] [PubMed] [Google Scholar]
- 12.Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19: 6432-6442. [DOI] [PubMed] [Google Scholar]
- 13.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288: 1439-1441. [DOI] [PubMed] [Google Scholar]
- 14.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16: 297-309. [DOI] [PubMed] [Google Scholar]
- 15.Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997;16: 4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golub TR. TEL gene rearrangements in myeloid malignancy. Hematol Oncol Clin North Am. 1997;11: 1207-1220. [DOI] [PubMed] [Google Scholar]
- 17.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2: 367-376. [DOI] [PubMed] [Google Scholar]
- 18.Tosi S, Giudici G, Mosna G, et al. Identification of new partner chromosomes involved in fusions with the ETV6 (TEL) gene in hematologic malignancies. Genes Chromosomes Cancer. 1998;21: 223-229. [PubMed] [Google Scholar]
- 19.Poirel H, Lacronique V, Mauchauffe M, et al. Analysis of TEL proteins in human leukemias. Oncogene. 1998;16: 2895-2903. [DOI] [PubMed] [Google Scholar]
- 20.Rubnitz JE, Downing JR, Pui CH. Significance of the TEL-AML fusion gene in childhood AML. Leukemia. 1999;13: 1470-1471. [DOI] [PubMed] [Google Scholar]
- 21.Rubnitz JE, Pui CH, Downing JR. The role of TEL fusion genes in pediatric leukemias. Leukemia. 1999;13: 6-13. [DOI] [PubMed] [Google Scholar]
- 22.Cools J, Mentens N, Odero MD, et al. Evidence for position effects as a variant ETV6-mediated leukemogenic mechanism in myeloid leukemias with a t(4;12)(q11-q12;p13) or t(5;12)(q31;p13). Blood. 2002;99: 1776-1784. [DOI] [PubMed] [Google Scholar]
- 23.Suto Y, Sato Y, Smith SD, Rowley JD, Bohlander SK. A t(6;12)(q23;p13) results in the fusion of ETV6 to a novel gene, STL, in a B-cell ALL cell line. Genes Chromosomes Cancer. 1997;18: 254-268. [DOI] [PubMed] [Google Scholar]
- 24.Yagasaki F, Jinnai I, Yoshida S, et al. Fusion of TEL/ETV6 to a novel ACS2 in myelodysplastic syndrome and acute myelogenous leukemia with t(5;12)(q31;p13). Genes Chromosomes Cancer. 1999;26: 192-202. [DOI] [PubMed] [Google Scholar]
- 25.Golub TR, Barker GF, Stegmaier K, Gilliland DG. The TEL gene contributes to the pathogenesis of myeloid and lymphoid leukemias by diverse molecular genetic mechanisms. Curr Top Microbiol Immunol. 1997;220: 67-79. [DOI] [PubMed] [Google Scholar]
- 26.Rompaey LV, Potter M, Adams C, Grosveld G. Tel induces a G1 arrest and suppresses Ras-induced transformation. Oncogene. 2000;19: 5244-5250. [DOI] [PubMed] [Google Scholar]
- 27.Fenrick R, Wang L, Nip J, et al. TEL, a putative tumor suppressor, modulates cell growth and cell morphology of ras-transformed cells while repressing the transcription of stromelysin-1. Mol Cell Biol. 2000;20: 5828-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu X, Shin BH, Akbarali Y, et al. Tel-2 is a novel transcriptional repressor related to the Ets factor Tel/ETV-6. J Biol Chem. 2001;276: 9421-9436. [DOI] [PubMed] [Google Scholar]
- 29.Kawagoe H, Potter M, Ellis J, Grosveld GC. TEL2, an ETS factor expressed in human leukemia, regulates monocytic differentiation of U937 cells and blocks the inhibitory effect of TEL1 on ras-induced cellular transformation. Cancer Res. 2004;64: 6091-6100. [DOI] [PubMed] [Google Scholar]
- 30.Cardone M, Kandilci A, Carella C, et al. The novel ETS factor TEL2 cooperates with Myc in B lymphomagenesis. Mol Cell Biol. 2005;25: 2395-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Center for Biotechnology Information. Collaborating on public cancer data. http://www.ncbi.nlm.nih.gov/CGAP. Accessed March 14, 2005.
- 32.Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20: 7419-7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawley RG. High-titer retroviral vectors for efficient transduction of functional genes into murine hematopoietic stem cells. Ann N Y Acad Sci. 1994;716: 327-330. [DOI] [PubMed] [Google Scholar]
- 34.Hawley RG, Fong AZ, Reis MD, Zhang N, Lu M, Hawley TS. Transforming function of the HOX11/TCL3 homeobox gene. Cancer Res. 1997;57: 337-345. [PubMed] [Google Scholar]
- 35.Persons DA, Allay JA, Allay ER, et al. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood. 1997;90: 1777-1786. [PubMed] [Google Scholar]
- 36.Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, eds. Current Protocols in Immunology Unit 10.17L. New York, NY: Wiley; 1999. [DOI] [PubMed]
- 37.Burroughs J, Gupta P, Blazar BR, Verfaillie CM. Diffusible factors from the murine cell line M2–10B4 support human in vitro hematopoiesis. Exp Hematol. 1994;22: 1095-1101. [PubMed] [Google Scholar]
- 38.Affymetrix. GeneChip Expression analysis technical manual. http://www.affymetrix.com/support/technical/manual/expression_manual.affx. Accessed March 14, 2005.
- 39.Affymetrix. NetAffx analysis center. http://www.affymetrix.com/analysis/index.affx. Accessed March 14, 2005.
- 40.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100: 238-245. [DOI] [PubMed] [Google Scholar]
- 41.Shao Y, Raiford KL, Wolpert CM, et al. Phenotypic homogeneity provides increased support for linkage on chromosome 2 in autistic disorder. Am J Hum Genet. 2002;70: 1058-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai T, Nomura F, Hoshino K, et al. Death-associated protein kinase 2 is a new calcium/cal-modulin-dependent protein kinase that signals apoptosis through its catalytic activity. Oncogene. 1999;18: 3471-3480. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Adachi M, Kawamura R, Imai K. Bmf is a possible mediator in histone deacetylase inhibitors FK228 and CBHA-induced apoptosis. Cell Death Differ. 2006;1: 129-140. [DOI] [PubMed] [Google Scholar]
- 44.Iwai K, Hirata K, Ishida T, et al. An anti-proliferative gene BTG1 regulates angiogenesis in vitro. Biochem Biophys Res Commun. 2004;316: 628-635. [DOI] [PubMed] [Google Scholar]
- 45.Duriez C, Moyret-Lalle C, Falette N, El-Ghissassi F, Puisieux A. BTG2, its family and its tutor. Bull Cancer. 2004;91: E242-E253. [PubMed] [Google Scholar]
- 46.Kuo ML, Duncavage EJ, Mathew R, et al. Arf induces p53-dependent and -independent antiproliferative genes. Cancer Res. 2003;63: 1046-1053. [PubMed] [Google Scholar]
- 47.Payne SR, Kemp CJ. p27(Kip1) (Cdkn1b)-deficient mice are susceptible to chemical carcinogenesis and may be a useful model for carcinogen screening. Toxicol Pathol. 2003;31: 355-363. [DOI] [PubMed] [Google Scholar]
- 48.Andreu EJ, Lledo E, Poch E, et al. BCR-ABL induces the expression of Skp2 through the PI3K pathway to promote p27Kip1 degradation and proliferation of chronic myelogenous leukemia cells. Cancer Res. 2005;65: 3264-3272. [DOI] [PubMed] [Google Scholar]
- 49.Kontani K, Chano T, Ozaki Y, et al. RB1CC1 suppresses cell cycle progression through RB1 expression in human neoplastic cells. Int J Mol Med. 2003;12: 767-769. [PubMed] [Google Scholar]
- 50.Chano T, Ikegawa S, Kontani K, Okabe H, Baldini N, Saeki Y. Identification of RB1CC1, a novel human gene that can induce RB1 in various human cells. Oncogene. 2002;21: 1295-1298. [DOI] [PubMed] [Google Scholar]
- 51.Chano T, Kontani K, Teramoto K, Okabe H, Ikegawa S. Truncating mutations of RB1CC1 in human breast cancer. Nat Genet. 2002;31: 285-288. [DOI] [PubMed] [Google Scholar]
- 52.Gilliland DG. Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002;39: 6-11. [DOI] [PubMed] [Google Scholar]
- 53.Carlson A, Berkowitz JM, Browning D, Slamon DJ, Gasson JC, Yates KE. Expression of c-Fes protein isoforms correlates with differentiation in myeloid leukemias. DNA Cell Biol. 2005;24: 311-316. [DOI] [PubMed] [Google Scholar]
- 54.Scheijen B, Griffin JD. Tyrosine kinase oncogenes in normal hematopoiesis and hematological disease. Oncogene. 2002;21: 3314-3333. [DOI] [PubMed] [Google Scholar]
- 55.Westbrook TF, Martin ES, Schlabach MR, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121: 837-848. [DOI] [PubMed] [Google Scholar]
- 56.Waga K, Nakamura Y, Maki K, et al. Leukemia-related transcription factor TEL accelerates differentiation of Friend erythroleukemia cells. Oncogene. 2003;22: 59-68. [DOI] [PubMed] [Google Scholar]
- 57.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001;20: 350-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17: 2298-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1: 63-74. [DOI] [PubMed] [Google Scholar]
- 60.Castilla LH, Garrett L, Adya N, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23: 144-146. [DOI] [PubMed] [Google Scholar]
- 61.Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103: 2522-2529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.