It is said that “a picture is worth a thousand words,” a snapshot or an image of a complex reality, but it can never tell the whole story. Likewise, protein crystal structures divulge important molecular features, but a comprehensive understanding requires an arsenal of techniques and expertise from a variety of fields. The interplay between and importance of structural and functional studies are nicely illustrated by recent reports on the membrane-embedded protease rhomboid. For several years, molecular and biochemical findings provided strong evidence that rhomboids were a new type of serine protease with an active site within the boundaries of the lipid bilayer (1). In the past several months, studies of five crystal structures (2–6) have confirmed this theory but have offered conflicting ideas regarding substrate access to the active site. In this issue of PNAS, Baker et al. (7) bring us full circle, presenting compelling functional support for one of these hypotheses.
Rhomboids are found in virtually all organisms and were first discovered as proteases in 2001 (8). Evidence for proteolytic function included conserved serine and histidine residues that were essential for activity in cells, sensitivity to certain serine protease inhibitors, and a cleavage site within the transmembrane (TM) region of the substrate. Bacterial expression and purification (9, 10) further demonstrated that rhomboids work alone, without the need for other protein factors. The recent crystal structures (all of bacterial rhomboid orthologs) have confirmed the six-TM helix topology of the protein (Fig. 1a) and removed any doubt about their specific identity as serine proteases: the key Ser-201 and His-254 residues, located on TM helices 4 and 6, respectively, are indeed adjacent to each other and oriented in essentially the same way as found in classical soluble serine proteases. Two unusual features of the structure include a large loop (L1) between TM1 and TM2 that dips into the membrane, and TM4, which begins with the catalytic serine 10 Å below the membrane surface. The atypical TM4 helps form a water-lined active-site cavity, a feature that may also be true of at least one other membrane-embedded protease, the multicomponent γ-secretase complex (11, 12). The rhomboid structures have also generated hypotheses about the location and nature of the oxyanion hole, a pocket that stabilizes the tetrahedral intermediate in the reaction after nucleophilic attack by the catalytic serine. In silico docking of a peptide substrate into the active site (5) suggests that this stabilization is provided by the main-chain amide of the catalytic serine and a histidine side chain from TM2. Other researchers (6) propose that either this histidine or the adjacent Asn-154, also in TM2, contributes to the oxyanion hole. Cocrystallization with a transition-state analog inhibitor may provide more definitive identification of this pocket so critical to catalysis.
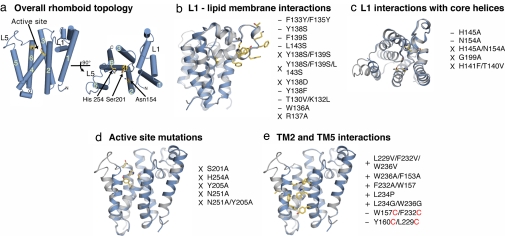
Fig. 1.
Escherichia coli rhomboid GlpG structure and effects of mutations on enzyme activity. Cartoon representations of GlpG monomers are from Protein Data Bank ID code 2NRF, with relevant residues highlighted in yellow ball-and-stick. Orientation is parallel to membrane normal (a Left, b, d, and e) and perpendicular (a Right and c). −, decreased activity; X, abolished activity; +, enhanced activity. (a) Overall rhomboid topology and location of the active site. (b) Mutations that perturb L1–lipid interactions impair activity. (c) Disruption of L1–core TM helix interactions decreases activity. (d) All mutations in the active site lead to abolished activity. (e) Enhancing access at TM2–TM5 interface increases activity, and closing access via disulfide linkages (red) decreases activity.
Although the crystal structures have clarified fundamental features of rhomboid, especially the overall protein fold and nature of the active site, the question of substrate accessibility has remained controversial. The catalytic Ser–His dyad lies within the interior of the protein, sequestered from the lipid bilayer, while the substrates are type I integral membrane proteins [which include the mitochondrial protein OPA1 in mammals (13), the growth factor Spitz in Drosophila (8), and the quorum-sensing protein TatA in Providencia stuartii (14)]. Presumably, lateral gating is necessary for substrate access to the active site, but where on rhomboid does it occur and how does it happen?
Two structure-based hypotheses have been proposed. In the first model (2), substrate enters after L1 swings out of the bilayer, revealing a V-shaped gap between TM1 and TM3. The unusual structure of L1, composed of two short amphipathic helices, suggests that it might play an important functional role. However, this domain is invariant in all rhomboid structures to date. Moreover, substrate entry from this direction does not place the scissile bond on the face of the catalytic dyad. A major conformational change of the protease structural core would be necessary to accommodate this model, one not immediately apparent from the crystal structures or previous functional work on rhomboid.
The second hypothesis (3, 4) is that substrate enters between TM2 and TM5, with the latter helix serving as the gate. Indeed, the position of TM5 varies among the reported crystal structures, and in one (3) appears to provide an opening for direct access to the hydrophilic catalytic center. Loop L5, which connects TM5 to TM6, can also move quite readily: it is disordered in some structures and can be perturbed by soaking with DMSO (6). Finally, several structures contain a lipid molecule bound at the TM2/TM5 interface (4, 5), and with the anionic phosphate head group interacting with the catalytic residues and putative oxyanion hole (4) (i.e., mimicking the tetrahedral intermediate), suggesting that this region may provide a conduit to the active site.
To test these models for substrate access, Baker et al. (7) generated and measured activity for some 40 rhomboid mutants, changing key residues in loops L1 and L5, in and around the active site, and in TM2 and TM5. The results were remarkably clear: All mutations to L1 disrupted activity, whereas mutations in TM2 and TM5 could cause increases in activity, up to 10-fold above that of wild type. L1 mutants were designed to allow this loop to move from its submerged position in the membrane to open the V-shaped gap to the interior. In particular, hydrophobic residues facing the bilayer were mutated to hydrophilic ones (Fig. 1b), and key points of interaction between L1 and core helices were mutated as well (Fig. 1c). Whereas the L1 gating model would predict that such changes might increase proteolytic activity, all of these mutations in L1 instead either decreased activity or abolished it altogether. Likewise, mutations around the active site (Fig. 1d), designed to increase access of the catalytic dyad to substrate entering from the L1 face, all destroyed activity.
In contrast, mutations in TM2 and TM5 (Fig. 1e), designed to disrupt interaction between these helices, resulted in increased activity. Swapping helix-breaking residues into TM5 (e.g., L234P) likewise increased activity, consistent with the idea that this helix must tilt away from the protein core to open the gate for substrate access. Conversely, closing this gate decreases activity. Cysteine mutants were designed to allow disulfide bond formation between residues close in space on TM2 and TM5 (e.g., W157C/F232C). These mutants exhibited reduced activity upon oxidation, but activity was partially restored upon chemical reduction of the disulfides back to free thiols. Together, this functional mutagenesis study clearly supports the TM5 gating model.
So what is the function of L1? All mutations in this domain reduced or eliminated activity, and this result combined with the observation that L1 remains invariant among the reported structures suggests that L1 serves as a scaffold. The large and unusual L1 domain is buttressed against the core of the protein, and some of the points of contact are highly conserved. Moving it away would make the interior accessible to lipids through the V-shaped gap, and such interactions, along with the lack of support from L1, might destabilize the core structure. Perhaps this is one means of regulating rhomboids: although L1 is positioned the same way in all of the structures, interaction with a regulator protein in the context of the cell might result in movement and enzyme inactivation. Exquisite control of this protease activity is very important: unlike other membrane-embedded proteins (γ-secretases, signal peptide peptidases, site 2 proteases) that require prior regulated proteolytic events (15), rhomboids cleave the full-length form of their substrates.
Although these rhomboid studies represent remarkable advances, we still have much to learn about the fundamentals of proteolysis within the lipid bilayer. Structure–function studies of other rhomboid orthologs, particularly those in which the substrate is known (e.g., Drosophila Rhomboid-1) may provide not only further backing of the TM5 gating model but additional insight into substrate specificity. The issue of specificity is especially important: rhomboids undoubtedly encounter many different TM helices that are potential substrates, so how does the enzyme distinguish among them? Are there functional roles for the diverse N- and C-terminal domains that are found among rhomboid orthologs? In addition, what are the kinetic details of catalysis, and how do they compare with soluble serine proteases? Beyond the basic research interest in intramembrane proteolysis, rhomboids are also emerging as potential drug targets [e.g., for parasitic infections (16–18)], underscoring the need for continued synergy between structural and functional studies of rhomboid.
Footnotes
The authors declare no conflict of interest.
See companion article on page 8257.
References
- 1.Urban S. Genes Dev. 2006;20:3054–3068. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Zhang Y, Ha Y. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Shem A, Fass D, Bibi E. Proc Natl Acad Sci USA. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. Proc Natl Acad Sci USA. 2007;104:750–754. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Ha Y. Proc Natl Acad Sci USA. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker RP, Young K, Feng L, Shi Y, Urban S. Proc Natl Acad Sci USA. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urban S, Lee JR, Freeman M. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 9.Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. EMBO J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban S, Wolfe MS. Proc Natl Acad Sci USA. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tolia A, Chavez-Gutierrez L, De Strooper B. J Biol Chem. 2006;281:27633–27642. doi: 10.1074/jbc.M604997200. [DOI] [PubMed] [Google Scholar]
- 12.Sato C, Morohushi Y, Tomita T, Iwatsubo T. J Neurosci. 2006;26:12081–12088. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D'Adamio L, et al. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson LG, Strisovsky K, Clemmer KM, Bhatt S, Freeman M, Rather PN. Proc Natl Acad Sci USA. 2007;104:1003–1008. doi: 10.1073/pnas.0608140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe MS, Kopan R. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 16.Brossier F, Jewett TJ, Sibley LD, Urban S. Proc Natl Acad Sci USA. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker RP, Wijetilaka R, Urban S. PLoS Pathog. 2006;2:e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger TW, Blackman MJ. J Cell Biol. 2006;174:1023–1033. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]