Abstract
Vesicular transport during exocytosis is regulated by Rab GTPase (Sec4p in yeast), which is activated by a guanine nucleotide exchange factor (GEF) called Sec2p. Here, we report the crystal structure of the Sec2p GEF domain in a complex with the nucleotide-free Sec4p at 2.7 Å resolution. Upon complex formation, the Sec2p helices approach each other, flipping the side chain of Phe-109 toward Leu-104 and Leu-108 of Sec2p. These three residues provide a hydrophobic platform to attract the side chains of Phe-49, Ile-53, and Ile-55 in the switch I region as well as Phe-57 and Trp-74 in the interswitch region of Sec4p. Consequently, the switch I and II regions are largely deformed, to create a flat hydrophobic interface that snugly fits the surface of the Sec2p coiled coil. These drastic conformational changes disrupt the interactions between switch I and the bound guanine nucleotide, which facilitates the GDP release. Unlike the recently reported 3.3 Å structure of the Sec4p·Sec2p complex, our structure contains a phosphate ion bound to the P-loop, which may represent an intermediate state of the nucleotide exchange reaction.
Keywords: exocytosis, guanine nucleotide exchange factor, GTPase, x-ray crystallography
Exocytosis is a fundamental biological process for the secretion of physiologically active substances, membrane recruitment, and membrane protein targeting to a specific domain of the cell surface. Secretory vesicles loaded with membrane components and cargo fuse with the plasma membrane in the last stage of exocytosis (1). Rab family proteins play a crucial role in the regulation of the membrane fusion process (2–4). Eleven Rab proteins have been identified in the budding yeast, Saccharomyces cerevisiae (5). Among the yeast Rab proteins, Sec4p is essential for the exocytic events in yeast (6). Sec4p is anchored onto the secretory vesicles through the C-terminal Cys residues modified with geranylgeranyl groups (7). The Rab protein oscillates between the active GTP form and the inactive GDP form, as do the other small GTPases, and functions as a molecular switch to ensure the accurate delivery of the transport vesicle to the target membrane (8). The conformational transition of two regions, termed “switch I” and “switch II,” substantiates the function of the Rab protein as a molecular switch (9). The downstream effectors for the Rab proteins recognize the configuration of the switch regions, and specifically interact with the GTP-bound form (10). The octameric protein complex, Exocyst, is one of the downstream effectors for Sec4p (11–13). The Sec15p subunit of Exocyst interacts with Sec4p in a GTP-dependent manner (13). This Sec4p–Sec15p interaction is crucial for Exocyst assembly, and reportedly tethers the secretory vesicle to the target membrane (14). Sec4p shuttles between the donor organelle, the trans-Golgi network, and the plasma membrane via the transport vesicle (15). In the present plausible model, the anterograde and retrograde transport events in exocytosis are coupled with GDP-GTP exchange and GTP hydrolysis, respectively, on Sec4p (16). The guanine nucleotide exchange reaction is catalyzed by the specific guanine nucleotide exchange factor (GEF), which facilitates the release of the bound GDP and the subsequent GTP rebinding (17).
Sec2p (759 aa, Mw 105 kDa) is the specific GEF for Sec4p in yeast (18). Sec2p is divided into N-terminal (residues 1–160) and C-terminal (residues 161–759) regions (19–21). The N-terminal region is solely sufficient for the GEF activity on Sec4p, whereas the C-terminal region does not affect the GEF activity (21). Instead, the C-terminal region binds with Sec15p and another Rab GTPase, Ypt32p, which recruits Sec2p to the secretory vesicle (21, 22). Thus, on the secretory vesicle, Sec2p activates Sec4p, and the activated Sec4p interacts with Sec15p (13). Sec15p in turn interacts with the C-terminal region of Sec2p, to displace Ypt32p from Sec2p. In a recent study, Sec2p, Sec4p, and Sec15p were reported to directly interact with each other, which is crucial for Sec2p localization and polarized cellular growth (22).
Two exocytic Rabs, Rab3A and Rab8, are known as mammalian orthologs of Sec4p: Rab8 is involved in the transport of Rab8-specific vesicles (23), whereas Rab3A regulates neurotransmitter release (24, 25). The GEFs for Rab8 and Rab3A were identified as Rabin8 (26) and GRAB (27), respectively. The N-terminal regions of these mammalian GEFs were predicted to share structural and sequence similarities with the yeast Sec2p GEF domain, suggesting that a common structural motif is used for the Rab GEF reactions from yeast to mammals.
We recently determined the crystal structure of S. cerevisiae Sec231–160p, which revealed that the Sec2p GEF domain folds into a parallel dimeric coiled coil extending >180 Å (28). Although the Sec4p·Sec2p complex structure was recently reported (29), the resolution and the Rfree value (3.3 Å and 0.325, respectively) were insufficient to elucidate the detailed interactions (e.g., hydrogen bonds) between Sec2p and Sec4p.
In this study, we determined the crystal structure of the Sec419–187p·Sec251–142p complex with a phosphate ion at 2.7 Å resolution. The interactions between Sec2p and the switch regions of Sec4p induce drastic conformational changes in both of the Sec4p switch regions as well as in Sec2p. The profound conformational changes disrupt the conserved interactions between switch I and the bound guanine nucleotide, which facilitates the GDP release. Due to the presence of a phosphate ion bound to the P-loop, our Sec4p·Sec2p·phosphate complex structure may represent the nucleotide exchanging intermediate state, which is distinct from that of the previous Sec4p·Sec2p complex (29). Based on the available structures of the GDP- and GTP-bound forms of Sec4p, Sec2p, the previous Sec4p·Sec2p complex, and the present intermediate state complex, we could delineate the entire nucleotide exchange mechanism of Sec4p by Sec2p.
Results and Discussion
Overall Structure.
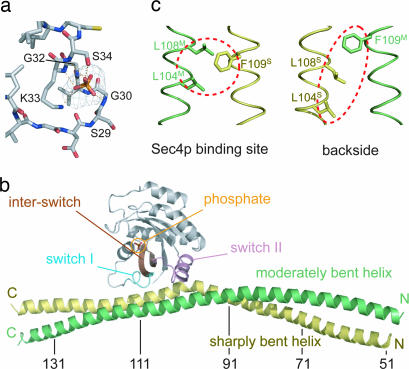
We prepared eight Sec2p truncated mutants (residues 1–160, 18–160, 31–160, 68–160, 1–142, 1–195, 68–195, and 51–142) and screened a wide variety of crystallization conditions to obtain Sec4p·Sec2p complex crystals suitable for a structure determination. Although all of these truncated mutants formed a stable complex with the nucleotide-free Sec419–187p, only Sec251–142p was successfully cocrystallized with Sec419–187p. The crystals of the Sec4p·Sec2p·phosphate complex belong to the space group I222, with unit cell parameters of a = 116.6 Å, b = 117.4 Å, and c = 123.3 Å, and contained one complex per asymmetric unit. We prepared SeMet-labeled Sec419–187p to form the complex crystals, and determined the phases by the multiwavelength anomalous dispersion method, using the selenium edge [supporting information (SI) Table 1]. Density modification with solvent flattening produced a clear electron density map. The atomic model of the Sec4p·Sec2p·phosphate complex was manually built and refined to a resolution of 2.7 Å, with an Rfree value of 0.298. The average temperature factor of Sec4p is rather high (96 Å2), probably due to the lack of crystal packing by Sec4p, which is fixed only by the interaction with Sec2p in the crystal. The average temperature factor in our complex structure is lower than that of the recently reported Sec4p·Sec2p complex structure, and the electron density is clearly observed in all regions of Sec2p and Sec4p, in contrast to the previous structure (29). The refined atomic model includes two phosphate ions, with one bound to the Arg-120 and Lys-123 residues of Sec2p, and the other tightly bound to the Sec4p P-loop (residues 28–34), which normally surrounds the β phosphate of the guanine nucleotide (30) (Fig. 1a). Either a phosphate or sulfate ion was previously observed to mimic the β phosphate of the bound guanine nucleotide, to stabilize the P-loop in the small GTPases complexed with their GEFs, such as in the Rac·Tiam (31), Ran·RCC1 (32), Rho·SopE (33), and Rho·LARG (34) complexes.
Fig. 1.
Structure of the Sec4p·Sec2p·phosphate complex. (a) Simulated-annealing Fo − Fc omit map, contoured at the 5.0 σ level, of the bound phosphate. Sec4p is colored gray. Oxygen, nitrogen, sulfur, and phosphorous atoms are colored red, blue, yellow, and orange, respectively. Hydrogen bonds are indicated by dotted orange lines. (b) Overall structure of the Sec4p·Sec2p·phosphate complex. The switch I, interswitch, switch II, and other regions of Sec4p are colored cyan, brown, pink, and gray, respectively. The sharply and moderately bent helices of Sec2p are colored yellow and green, respectively. The phosphate ion, which mimics the β phosphate of the guanine nucleotide, is shown by a stick model. Every 20 residues are labeled in Sec2p. (c) The asymmetric coiled-coil of the Sec4p binding site of Sec2p. (Left) The Sec4p binding site of Sec2p, highlighting Leu-104, Leu-108, and Phe-109 by red dotted ellipses. (Right) The backside view. The coloring scheme is the same as in b.
The overall structure of the Sec4p·Sec2p·phosphate complex is shown in Fig. 1b. One Sec419–187p molecule is bound with two Sec251–142p molecules, which form a dimeric parallel coiled coil, as in the Sec4p-free Sec231–160p structure (28). The Sec4p-binding site of Sec2p is located across the dimeric coiled coil of the Sec2p GEF domain, which well explained our recent mutational analysis (28). Around the Sec4p-binding site, the Sec2p coiled-coil helices are aligned in a “staggered” manner; one Sec251–142p monomer shifts by 0.5 pitch along the helical axis, relative to the other monomer, causing a configurational difference between the two faces of the Sec4p-binding region, despite their same amino acid composition (Fig. 1c). Due to this asymmetry around the Sec4p-binding site, Sec2p binds in an unusual 2:1 stoichiometry to Sec4p. One of the Sec2p helices bends sharply (≈30°) and another bends moderately (≈3°), in the upstream region of the “staggered” segment of Sec2p, to restore a regular coiled coil in the N-terminal portion. This molecular asymmetry is intrinsic, as observed in our recently reported structure of Sec231–160p alone (28). We referred to the two helices of the Sec231–160p dimer as a “sharply bent helix” and a “moderately bent helix” (28), based on their structural features. Hereafter, we include the superScript “S” and “M” with the residue number of the Sec2p molecules, to indicate the “sharply” and “moderately” bent helices, respectively.
Detailed Interactions Between Sec2p and Sec4p.
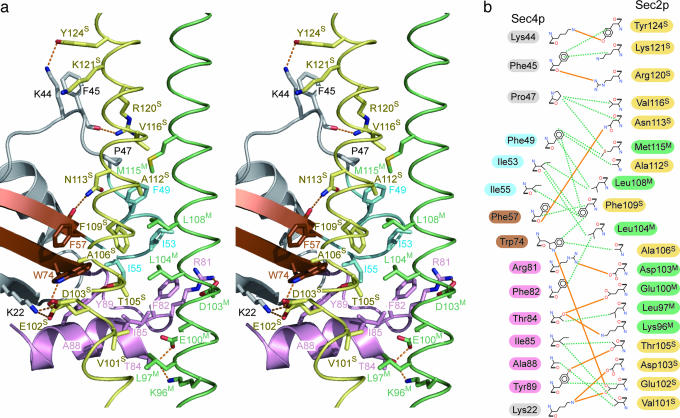
Sec419–187p is bound to the Sec251–142p homodimer with a buried surface area of 2,286 Å2, which is similar to that of the other Rab·RabGEF complex, Rab8·MSS4 (2,216 Å2) (35). The highly conserved region (residues 96–124) of Sec2p binds with Lys-22 and residues 44–89 of Sec4p, including the switch I (residues 48–56), interswitch (residues 57–75), and switch II (residues 76–93) regions. The switch I and II regions of Sec4p are relatively ordered in the Sec4p·Sec2p·phosphate complex (SI Fig. 6), where the average temperature factors for all atoms in the switch I and II regions (50 Å2 and 73 Å2, respectively) are lower than those for the other regions of Sec4p (100 Å2). In contrast, in the GDP-bound Sec4p structure, the average temperature factors for the switch I and switch II regions (50 and 40 Å2, respectively) are higher than those for the other regions (31 Å2) (9).
The interactions between Sec2p and the switch I region of Sec4p are mostly hydrophobic. The side chains of Leu-104M, Leu-108M, and Phe-109S in Sec2p form a hydrophobic core at the center of the molecular interface (Fig. 2). This hydrophobic core is surrounded by the side chains of Phe-49, Ile-53, and Ile-55 in the Sec4p switch I region (Fig. 2). The side chain of the Sec4p Phe-49 is lined up along with the side chains of the Sec4p Pro-47 and the Sec2p Val-116S and Met-115M, whereas that of Ile-55 are aligned with the side chains of Phe-57 and Trp-74 in the Sec4p interswitch region. The Sec4p Trp-74 is spatially stabilized by a hydrogen bond between its Nε atom and the Oδ atom of the Sec2p Glu-102S (Fig. 2). Furthermore, the main chain carbonyl oxygen of the Sec4p Phe-45 and the Nζ atom of the Sec4p Lys-44 hydrogen bond with the Nη atom of the Sec2p Arg-120S and the Oη atom of the Sec2p Tyr-124S, respectively, which may stabilize the structure just upstream of the switch I region of Sec4p. In contrast to the switch I region, the interaction between the switch II region and Sec2p is mediated by both hydrophobic and hydrogen bonding interactions. Val-101S of Sec2p hydrophobically interacts with Ile-85 and Ala-88 in the Sec4p switch II region. The Nη and Oη atoms of the Sec4p Arg-81 and Tyr-89 hydrogen bond with the Oε and Oγ atoms of the Sec2p Asp-103M and Thr-105S, respectively (Fig. 2). Furthermore, the Oγ atom of the Sec4p Thr-84 hydrogen bonds with the Oδ and Nζ atoms of the Sec2p Glu-100M and Lys-96M, respectively (Fig. 2).
Fig. 2.
Sec4p·Sec2p binding interface. (a) Stereoview of the detailed Sec4p·Sec2p binding interface. The coloring scheme is the same as in Fig. 1b. Hydrogen bonds are shown by dotted orange lines. (b) Schematic representation of the Sec4p·Sec2p interface. The hydrogen bonds (with a 3.4 Å cutoff) are displayed as solid orange lines. The hydrophobic contacts (with a 4.0 Å cutoff) are displayed as dotted green lines. The labels of the switch I, interswitch, switch II, and other regions of Sec4p are highlighted in cyan, brown, pink, and gray, respectively. The labels of the sharply and moderately bent helices of Sec2p are highlighted in yellow and green, respectively.
The molecular interactions in the present Sec4p·Sec2p·phosphate complex are consistent with our recent mutational analysis based on the Sec2p structure (28). Alanine substitutions for Leu-104, Phe-109 (responsible for the interaction with the switch I region), Glu-100, Glu-102, and Thr-105 (responsible for the interaction with the switch II region) impaired the GEF activity (28).
Reorganization of the Sec2p Dimeric Coiled Coil.
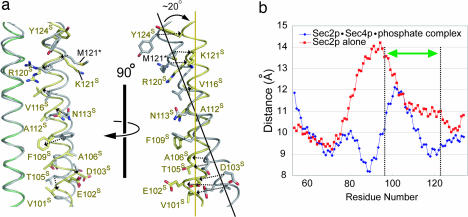
Our recent structural study on Sec231–160p alone revealed that the relative orientation of the two helices of the Sec231–160p dimer around the Sec4p-binding site (residues 100–121) is almost completely conserved among the 10 Sec231–160p dimers in the crystallographic asymmetric unit of the Sec231–160p crystals (PDB code 2E7S). In contrast, superposition of the moderately bent helix of Sec231–160p onto the equivalent helix of the Sec4p-bound Sec251–142p (rmsd 0.521 Å over the 16 Cα atoms of residues 100–115) clearly showed that the relative orientations of the Sec2p dimeric helices around the Sec4p-binding site are quite different between the two forms (Fig. 3a). In the Sec4p-free form, the two coiled-coil helices cross by 20°, whereas in the Sec4p-bound form, the two Sec2p helices are reorganized and are almost parallel around the Sec4p-binding site (Fig. 3a). In addition to the Sec4p binding site, the other regions (residues 77–95) of Sec2p also undergo structural changes. In the Sec4p-free Sec2p, the Arg-87 side chains are oriented toward the opposite helix, pushing the coiled-coil helices apart (28). The side chains of Ala-90 and Ala-94 are also oriented toward the opposite helix, in the absence of contact with any residues in the opposite helix (28). Due to this loose packing, the average inter-helical distance of residues 77–95 is 12.8 Å in the Sec4p-free Sec2p structure, which is larger than that of a canonical coiled coil (≈9.6 Å on average) (36) (Fig. 3b). In contrast, in the Sec4p·Sec2p·phosphate complex, the two helices of the Sec2p dimer approach each other to form a more tightly packed coiled coil. Arg-87 of Sec2p contacts the equivalent residue in the opposite helix in a “knobs-into-holes” manner with their Cβ atoms, forming an intermolecular salt bridge with Glu-83 (data not shown). Ala-90 and Ala-94 are also involved in the coiled-coil core formation. As a result, the average interhelical distance of residues 77–95 is largely reduced to 9.3 Å in the Sec4p·Sec2p·phosphate complex (Fig. 3b), and the buried surface area increases from 407 Å2 (Sec4p-free) to 875 Å2 (Sec4p-bound) in residues 77–95 of the Sec2p dimer interface. Although residues 77–95 do not directly interact with Sec4p, this tight interhelical packing is likely to facilitate the productive conformational change of the Sec4p-binding site in the present Sec4p·Sec2p·phosphate complex.
Fig. 3.
Structural comparison of the free and Sec4p-bound forms of Sec2p. (a) Comparison of the Sec4p binding region of Sec2p. The moderately bent helix of Sec2p in the Sec4p·Sec2p·phosphate complex and the equivalent helix in the free form are superposed over residues 100–115. The coloring scheme of Sec2p in the Sec4p·Sec2p·phosphate complex is the same as in Fig. 1c, whereas the Sec2p free form is colored gray. The labels of the mutated residues in the Sec2p free form (28) are denoted by asterisks. Dotted black arrows indicate positional changes of the amino acid residues. Yellow and black solid lines represent the helical axes of the Sec4p-bound Sec2p and the Sec2p free form, respectively. (b) The distance between the α helical axes of Sec2p alone (red) and the Sec4p·Sec2p ·phosphate complex (blue), plotted as a function of residue number. The two-headed green arrow shows the Sec4p binding site of Sec2p.
Furthermore, the side chain of Phe-109S flips toward the side chains of Leu-104M and Leu-108M in the opposite helix of Sec2p (Fig. 3a). As described above, these three residues provide a hydrophobic platform to attract the side chains of Phe-49, Ile-53 and Ile-55 in the switch I region, as well as Phe-57 and Trp-74 in the interswitch region of Sec4p. The functional role of the Sec2p Phe-109S in the GEF mechanism will be discussed below.
Conformational Transition of Sec4p.
The crystal structures of the GDP- and GppNHp-bound forms of Sec4p alone were previously reported (hereafter referred to as Sec4p·GDP and Sec4p·GTP, respectively) (9). The Sec4p structure in the present Sec4p·Sec2p·phosphate complex can be superposed well on the structures of both Sec4p·GDP and Sec4p·GTP (Cα atomic rmsd values of 0.727 Å and 0.732 Å, respectively), except for residues 42–47, the switch I region (residues 48–56) and the switch II region (residues 76–93). In contrast, the conformations of the switch I and switch II regions drastically changed upon complex formation (Fig. 4). In addition, the structure around the guanine binding motif, 133NKxD and 162SAK, also slightly changed (data not shown); however, this may be due to the absence of the bound nucleotide, because this region does not interact either directly or indirectly with Sec2p in the present Sec4p·Sec2p·phosphate complex. The drastic conformational change in the switch I region is induced by the aforementioned extensive hydrophobic interaction with Sec2p (Fig. 2). Concomitantly, Phe-45 and Pro-47 of Sec4p are pulled out to form a hydrophobic patch that snugly fits the binding surface of the Sec2p dimeric coiled coil. The distances of the shifts of Phe-45 and Pro-47 are estimated to be 14 and 16 Å, respectively, during the transition from Sec4p·GDP to the Sec2p-bound Sec4p (Fig. 4b).
Fig. 4.
Comparison of the Sec4p·GDP and Sec2p-bound Sec4p structures. (a) Comparison of the overall structures of Sec4p·GDP and Sec2p-bound Sec4p. Sec4p·GDP was superposed onto Sec4p in the Sec4p·Sec2p·phosphate complex, except for the residues 42–47, and the switch I (residues 48–56) and switch II (residues 76–93) regions. Sec4p·GDP is colored yellow, and Sec4p in the Sec4p·Sec2p·phosphate complex is colored as in Fig. 1b. The carbon atoms of the Sec4p-bound GDP are green, and the bound cobalt ion is yellow. (b) Comparison of the Sec2p binding region in the Sec4p·GDP and Sec2p-bound Sec4p structures. Coloring schemes are the same as in a. Dotted black arrows indicate positional changes of the amino acid residues.
Sec2p binding also induced a structural rearrangement of the switch II region in the present Sec4p·Sec2p·phosphate complex, because the switch II region directly interacts with the switch I region (SI Fig. 7). In the previous Sec4p·GTP structure, the two switch regions of Sec4p·GTP have two interaction sites: Ile-55 in the switch I region forms a hydrophobic core with Tyr-89 in the switch II region and Trp-74 in the interswitch region (referred to as “site 1”) (Sup. Fig. S2b). Ile-53 in the switch I region hydrophobically interacts with Phe-82 in the switch II region, whereas the Oγ atom of Thr-51 in the switch I region hydrogen bonds with the Nε atom of Gln-79 in the switch II region (referred to as “site 2”) (SI Fig. 7b). These switch I-switch II interactions stabilize the α helical conformation of the switch II region in the Sec4p·GTP structure, whereas the switch II region of Sec4p·GDP is unwound, due to the loss of both interactions (SI Fig. 7c). Intriguingly, although the present Sec4p·Sec2p·phosphate complex maintains the site 1 interaction, but lacks the site 2 interaction, the switch II region adopts the α helical conformation, which is stabilized by not only the site 1 interaction but also the aforementioned interactions with Sec2p (SI Fig. 7a and Fig. 2). These interactions attract the switch II region toward the Sec2p dimeric coiled coil.
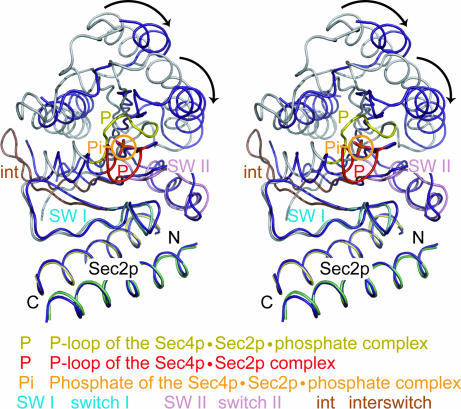
In the recently reported Sec4p·Sec2p complex structure, the P-loop is further reorganized upon complex formation to prevent GDP or GTP from rebinding (29). In contrast, in our structure, the P-loop conformation is stabilized by the tightly bound phosphate ion and resembles that in the nucleotide-bound form of Sec4p (Fig. 1a). Because the phosphate ion mimics the β phosphate of the bound guanine nucleotide, we propose that our structure represents the intermediate state before the GDP release in the nucleotide exchange reaction. As shown in Fig. 5, when we superposed the structure of the Sec4p binding site of Sec2p (residues 100–120) in the present Sec4p·Sec2p·phosphate complex onto that in the previously-reported Sec4p·Sec2p complex (Cα atomic rmsd values of 0.563 Å), the structures of Sec2p and the Sec2p binding region of Sec4p (switch I and switch II regions) were well superposed. In contrast, the P-loop of the phosphate-free Sec4p·Sec2p complex is restructured and moves toward the switch I region. In addition to the P-loop conformational change, the other regions of Sec4p, which do not interact with Sec2p, rotate relative to the Sec4p·Sec2p interface without conformational changes, to fill the space previously occupied by the P-loop. This structural reorganization may stabilize the nucleotide-free Sec4p structure.
Fig. 5.
Comparison of the present Sec4p·Sec2p·phosphate complex and the nucleotide-free Sec4p·Sec2p complex structures (29) (stereo). The Sec2p structures in the two complexes are superposed over residues 100–120. Sec4p in the nucleotide-free complex is colored purple, and its P-loop is colored red. Sec4p in the Sec4p·Sec2p·phosphate complex is colored as in Fig. 1b, and its P-loop is colored yellow. The black arrows indicate the structural reorganization in Sec4p upon nucleotide dissociation.
Guanine Nucleotide Exchange Mechanism.
We now have five crystal structures available to elucidate the nucleotide exchange mechanism of Sec4p by Sec2p: Sec4p·GDP, Sec4p·GTP (9), Sec2p GEF domain (28), the recently reported Sec4p·Sec2p complex (29), and the present Sec4p·Sec2p·phosphate complex representing the nucleotide exchanging intermediate state. Based on these structures, we can delineate the nucleotide exchange reaction on Sec4p by Sec2p as follows: the Sec2p dimer approaches Sec4p·GDP, and the side chain of the Sec2p Phe-109S flips toward the Sec2p Leu-104M and Leu-108M residues on the opposite helix, to avoid the steric hindrance with Phe-57 in the interswitch region of Sec4p (SI Fig. 8). The flipped Phe-109S interacts with Leu-104M and Leu-108M, forming a hydrophobic core to interact with Sec4p (Fig. 2 and SI Fig. 8). Upon the complex formation, the helices of Sec2p are reorganized and approach the switch II region of Sec4p to stabilize the complex, possibly providing sufficient binding energy for the subsequent nucleotide exchange catalysis (SI Fig. 8). Concomitantly, the Sec4p switch I region approaches the Sec2p hydrophobic core (Fig. 2 and SI Fig. 8), significantly changing the positions of Phe-45 and Pro-47 (Fig. 4b), which are essential to bind the base and ribose moieties of GDP (9). Consequently, the bound GDP loses its affinity to Sec4p, and dissociates. Finally, the dissociation of GDP leads to the P-loop conformational change and reorganization of the Sec4p structure, without changing the Sec4p·Sec2p interface (Fig. 5), to prevent the rebinding of GDP. It should be noted that we could not conclude here whether these conformational changes in Sec2p and Sec4p sequentially or simultaneously during the exchange reaction.
A previous study showed that the Ile50Ala mutant of Sec4p reduces the GDP-GTP exchange rate, and suggested that Ile-50 contributes to the dissociation of the magnesium ion during the guanine nucleotide exchange reaction (29). However, because the distance between Ile-50 and the magnesium ion is ≈3.7 Å in our Sec4p·Sec2p·phosphate structure, Ile-50 cannot dissociate the magnesium ion, whereas Ile-50 superposed well between both structures (data not shown). The Cδ atom of Ile-50 on the switch I region contacts with the switch II region by van der Waals interactions. Therefore, the replacement of Ile-50 by Ala might disturb the conformational transition of the switch I region, and reduce the GDP-GTP exchange rate of Sec4p. The opening up of switch I, which is promoted by the unusual coiled-coil GEF (i.e., Sec2p), is commonly observed in the complex structures of other small GTPases and their GEFs, including the nucleotide-free Rab8·MSS4 complex (35). On the other hand, the large conformational change of the P loop has been found only in the nucleotide-free Rab8·MSS4 complex (35), and thus, may be characteristic of the Rab family GTPases.
In living cells, Sec2p should facilitate not only the GDP release from Sec4p but also the subsequent GTP uptake by Sec4p. The conformational change of the switch I region alone succinctly explains the mechanism of the GDP release, but not that of the favorable uptake of GTP rather than GDP. We can speculate about the putative role of the switch II region to facilitate GTP uptake by Sec4p. In the present Sec4p·Sec2p·phosphate complex, the switch II region adopts an α helical conformation and retains some of the switch I-switch II interactions, as observed in Sec4p·GTP (SI Fig. 7). Therefore, we suggest that the α helical switch II conformation in the present complex might be advantageous for accepting the incoming GTP.
Methods
Protein Expression and Purification.
The genes encoding Sec251–142p and Sec419–187p were PCR amplified from S. cerevisiae genomic DNA. The amplified PCR products were cloned into the BamHI and XhoI sites in the pGEX-6P-1 expression vector (GE Healthcare), to create N-terminally GST-tagged proteins. E. coli strain Rosetta DE3) cells (Invitrogen) were transformed with the expression vector, and were cultured in LB containing 100 mg/liter ampicillin. The expression was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside at an A600 of 0.5, and then the Escherichia coli cells were incubated overnight at 20°C. The cells were collected by centrifugation at 8,000 × g for 15 min, and were disrupted by sonication in PBS containing 1 mM DTT and 1 mM phenylmethylsulfonyl fluoride (PMSF). The lysates were centrifuged at 12,000 × g for 40 min, and the supernatants were loaded onto a Glutathione Sepharose FF column (GE Healthcare), previously equilibrated with PBS containing 1 mM DTT. The GST-fused proteins were eluted with 50 mM Tris·HCl buffer (pH 8.0) containing 50 mM NaCl, 1 mM DTT and 15 mM reduced glutathione. The GST tags were cleaved by PreScission protease (GE Healthcare), and the reduced glutathione was removed by dialysis against 50 mM Tris·HCl buffer (pH 8.0), containing 50 mM NaCl and 1 mM DTT. The cleaved GST tags were then removed by chromatography on a Glutathione Sepharose FF column (GE Healthcare). The eluate was loaded onto a MonoQ anion exchange column (GE Healthcare), previously equilibrated with 50 mM Tris·HCl buffer (pH 8.0) containing 1 mM DTT. The proteins were eluted with a linear gradient of 0–1 M NaCl. The protein concentrations were estimated by the use of a Bio-Rad protein assay (Bio-Rad Laboratories). Sec251–142p was incubated with a 1.5-fold molar excess of Sec419–187p at 4°C, and was dialyzed overnight against 10 mM Tris·Cl buffer (pH 7.2), containing 10 mM EDTA, 50 mM NaCl and 5 mM 2-mercaptoethanol, to remove the guanine nucleotide and Mg2+. The Sec251–142p·Sec419–187p complex was loaded onto a Superdex 200 16/60 (prep grade) column (GE Healthcare), previously equilibrated with 10 mM Tris·Cl buffer (pH 7.2) containing 50 mM NaCl and 5 mM 2-mercaptoethanol, to remove the unbound Sec419–187p. The selenomethionine-labeled Sec419–187p was prepared in the same way as the native Sec419–187p, except that Sec419–187p was overproduced in the methionine-auxotroph E. coli strain, B834 (DE3), which was cultured in Core medium (Wako) with 30 μg/ml l-selenomethionine (Nakalai Tesque).
Crystallization and Data Collection.
The purified Sec251–142p·Sec419–187p complex was concentrated to 18 mg/ml by using an Amicon Ultra-15 5,000 MWCO filter (Millipore), following the manufacturer's instructions. Initial crystallization screening was performed by using the automatic crystallization system, Hydra96 plus One (Matrix). We tested more than 900 conditions, using crystallization reagent kits supplied by Hampton Research and Jena Bioscience. Preliminary crystals grew under several conditions, and among them we selected the best condition: 150 mM K2HPO4 containing 17% PEG 3350 and 500 mM dimethyl(2-hydroxyethyl)ammonium propane sulfonate, without buffer. Hanging drops were prepared by mixing 1 μl of reservoir with 1 μl of protein solution, and were equilibrated with 500 μl of reservoir solution at 20°C. The native crystals, which were used for the final structure refinement, belong to the space group I222, with the unit cell parameters, a = 116.6 Å, b = 117.4 Å, c = 123.3 Å. Diffraction data of the native and selenomethionine-labeled protein crystals were collected at the beamlines BL41XU in SPring 8 (Hyogo, Japan) and BL5A in the Photon Factory (Tsukuba, Japan), respectively, and were processed with the DENZO and SCALEPACK programs (37) and the CCP4 program suite (38).
Structural Determination and Refinement.
Phases were determined by a multiwavelength anomalous dispersion method using the selenium edge. Two of the three possible selenium sites were identified with the programs SHELXC and SHELXD (39). Refinement of the selenium sites and phase calculations were carried out using the MAD data set up to 3.0 Å, with the program SHARP (40). The calculated phases were improved by solvent-flattening and histogram matching with the program RESOLVE (41). The atomic model was built with the program O (42). The model was refined against the native data set up to 2.7 Å resolution by using the program CNS (43). Data collection, phasing, and refinement statistics are shown in Table 1. All molecular graphics were prepared with the program PyMOL (DeLano Scientific; www.pymol.org).
Supplementary Material
Acknowledgments
We thank the beam-line staffs at BL41XU of SPring-8 (Harima, Japan) and BL5A of PF (Tsukuba, Japan) for technical help during data collection. This work was supported by a SORST Program grant from JST (Japan Science and Technology) (to O.N.), a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) (to O.N. and S.F.), grants from MEXT (to O.N. and S.F.), and Kurata Memorial Hitachi Science and Technology Foundation grants (to S.F.).
Abbreviation
- GEF
guanine nucleotide exchange factor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2EQB).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701550104/DC1.
References
- 1.Farquhar MG. Annu Rev Cell Biol. 1985;1:447–488. doi: 10.1146/annurev.cb.01.110185.002311. [DOI] [PubMed] [Google Scholar]
- 2.Martinez O, Goud B. Biochim Biophys Acta. 1998;1404:101–112. doi: 10.1016/s0167-4889(98)00050-0. [DOI] [PubMed] [Google Scholar]
- 3.Tuvim MJ, Adachi R, Hoffenberg S, Dickey BF. News Physiol Sci. 2001;16:56–61. doi: 10.1152/physiologyonline.2001.16.2.56. [DOI] [PubMed] [Google Scholar]
- 4.Grosshans BL, Ortiz D, Novick P. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazar T, Gotte M, Gallwitz D. Trends Biochem Sci. 1997;22:468–472. doi: 10.1016/s0968-0004(97)01150-x. [DOI] [PubMed] [Google Scholar]
- 6.Salminen A, Novick PJ. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- 7.Newman CM, Magee AI. Biochim Biophys Acta. 1993;1155:79–96. doi: 10.1016/0304-419x(93)90023-6. [DOI] [PubMed] [Google Scholar]
- 8.Bourne HR, Sanders DA, McCormick F. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 9.Stroupe C, Brunger AT. J Mol Biol. 2000;304:585–598. doi: 10.1006/jmbi.2000.4236. [DOI] [PubMed] [Google Scholar]
- 10.Merithew E, Hatherly S, Dumas JJ, Lawe DC, Heller-Harrison R, Lambright DG. J Biol Chem. 2001;276:13982–13988. doi: 10.1074/jbc.M009771200. [DOI] [PubMed] [Google Scholar]
- 11.TerBush DR, Maurice T, Roth D, Novick P. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 12.Guo W, Grant A, Novick P. J Biol Chem. 1999;274:23558–23564. doi: 10.1074/jbc.274.33.23558. [DOI] [PubMed] [Google Scholar]
- 13.Guo W, Roth D, Walch-Solimena C, Novick P. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novick P, Medkova M, Dong G, Hutagalung A, Reinisch K, Grosshans B. Biochem Soc Trans. 2006;34:683–686. doi: 10.1042/BST0340683. [DOI] [PubMed] [Google Scholar]
- 15.Zajac A, Sun X, Zhang J, Guo W. Mol Biol Cell. 2005;16:1500–1512. doi: 10.1091/mbc.E04-10-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walworth NC, Goud B, Kabcenell AK, Novick PJ. EMBO J. 1989;8:1685–1693. doi: 10.1002/j.1460-2075.1989.tb03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quilliam LA, Khosravi-Far R, Huff SY, Der CJ. BioEssays. 1995;17:395–404. doi: 10.1002/bies.950170507. [DOI] [PubMed] [Google Scholar]
- 18.Walch-Solimena C, Collins RN, Novick PJ. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair J, Muller H, Peterson M, Novick P. J Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkind NB, Walch-Solimena C, Novick PJ. J Cell Biol. 2000;149:95–110. doi: 10.1083/jcb.149.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortiz D, Medkova M, Walch-Solimena C, Novick P. J Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medkova M, France YE, Coleman J, Novick P. Mol Biol Cell. 2006;17:2757–2769. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattula K, Peranen J. Curr Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- 24.Takai Y, Sasaki T, Matozaki T. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 25.Takai Y, Sasaki T, Shirataki H, Nakanishi H. Genes Cells. 1996;1:615–632. doi: 10.1046/j.1365-2443.1996.00257.x. [DOI] [PubMed] [Google Scholar]
- 26.Hattula K, Furuhjelm J, Arffman A, Peranen J. Mol Biol Cell. 2002;13:3268–3280. doi: 10.1091/mbc.E02-03-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo HR, Saiardi A, Nagata E, Ye K, Yu H, Jung TS, Luo X, Jain S, Sawa A, Snyder SH. Neuron. 2001;31:439–451. doi: 10.1016/s0896-6273(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 28.Sato Y, Shirakawa R, Horiuchi H, Dohmae N, Fukai S, Nureki O. Structure (London) 2007;15:245–252. doi: 10.1016/j.str.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Dong G, Medkova M, Novick P, Reinisch KM. Mol Cell. 2007;25:455–462. doi: 10.1016/j.molcel.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraste M, Sibbald PR, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 31.Worthylake DK, Rossman KL, Sondek J. Nature. 2000;408:682–688. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 32.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 33.Buchwald G, Friebel A, Galan JE, Hardt WD, Wittinghofer A, Scheffzek K. EMBO J. 2002;21:3286–3295. doi: 10.1093/emboj/cdf329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristelly R, Gao G, Tesmer JJ. J Biol Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 35.Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. EMBO J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Shea EK, Klemm JD, Kim PS, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project N. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 39.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 40.Fortelle Ed l. Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 41.Terwilliger TC. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones TA, Zou JY, Cowan SW, Kjeldgaard Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.