Abstract
Proteins are involved in various equilibria that play a major role in their activity or regulation. The design of molecules that shift such equilibria is of great therapeutic potential. This fact was demonstrated in the cases of allosteric inhibitors, which shift the equilibrium between active and inactive (R and T) states, and chemical chaperones, which shift folding equilibrium of proteins. Here, we expand these concepts and propose the shifting of oligomerization equilibrium of proteins as a general methodology for drug design. We present a strategy for inhibiting proteins by “shiftides”: ligands that specifically bind to an inactive oligomeric state of a disease-related protein and modulate its activity by shifting the oligomerization equilibrium of the protein toward it. We demonstrate the feasibility of our approach for the inhibition of the HIV-1 integrase (IN) protein by using peptides derived from its cellular-binding protein, LEDGF/p75, which specifically inhibit IN activity by a noncompetitive mechanism. The peptides inhibit the DNA-binding of IN by shifting the IN oligomerization equilibrium from the active dimer toward the inactive tetramer, which is unable to catalyze the first integration step of 3′ end processing. The LEDGF/p75-derived peptides inhibit the enzymatic activity of IN in vitro and consequently block HIV-1 replication in cells because of the lack of integration. These peptides are promising anti-HIV lead compounds that modulate oligomerization of IN via a previously uncharacterized mechanism, which bears advantages over the conventional interface dimerization inhibitors.
Keywords: allostery, protein equilibrium, shiftides, peptides, drug design
The design of molecules that shift protein equilibria is of great therapeutic potential, as demonstrated in the cases of allosteric inhibitors, which shift the equilibrium between active and inactive (R and T) states, and chemical chaperones, which shift folding equilibrium of proteins. Here, we propose the shifting of the oligomerization equilibrium of proteins as a powerful method to modulate their activity. Our approach is based on the allosteric model (1), according to which an allosteric protein is in equilibrium between R and T states, and ligand binding can shift this equilibrium toward one of the states. This approach was used therapeutically to develop molecules for the treatment of sickle cell anemia by heterotropic ligands that shift the R/T equilibrium of hemoglobin toward the high-affinity R state (2). A similar principle is applied for the development of drugs against protein-misfolding diseases (3): Chemical, or pharmacological, chaperones specifically bind the native states of misfolded proteins and shift the folding equilibrium toward the native state, leading to protein refolding and reactivation (4, 5). We have described a peptidic chemical chaperone that refolds and reactivates oncogenic mutants of the tumor suppressor p53 (6). In this article, we expand the application of shifting protein equilibrium as a potential therapeutic strategy and describe the development of “shiftides”: a class of peptides that inhibits proteins by shifting their oligomerization equilibrium. We apply this approach to design potent inhibitors of HIV-1 integrase (IN) protein.
Currently, the clinically approved anti-HIV drugs inhibit the viral enzymes reverse transcriptase and protease or prevent the penetration of HIV-1 into cells (7). The major problem with anti-HIV therapy is the high mutation rate in the viral genome, resulting in the emergence of drug-resistant virus strains. Thus, there is a constant need to identify new drug targets and to develop drugs directed against them. An attractive approach is to inhibit the oligomerization of the viral enzymes. Attempts were made to develop molecules that competitively bind at the dimerization interfaces (8, 9). However, such molecules were not developed into efficient inhibitors (8) because they usually bind relatively weakly to their large target proteins, and the high binding energy needed to disrupt the oligomerization interface cannot be supplied by a small molecule (10). Here, we propose the shifting of the oligomerization equilibrium as an alternative and more effective approach to disrupt protein oligomerization and demonstrate its application for inhibition of IN.
IN catalyzes integration of the reverse-transcribed viral DNA into the host genome. It is essential for HIV-1 replication, and mammalian cells do not harbor homologous enzymes. The integration proceeds by two steps (11): (i) 3′ end processing, in which IN creates the DNA template for integration by removing dinucleotides from the 3′ ends of both ends of the viral DNA LTRs after reverse transcription in the cytoplasm; and (ii) strand transfer, which is, after nuclear import, integration of the viral DNA template into the target host DNA. IN is in equilibrium among dimeric, tetrameric, and high-order oligomeric states (12–14). Dimeric IN binds at each end of the viral DNA during the 3′ end processing in the cytoplasm (15). After nuclear import, the two LTR DNA-bound dimers approach each other in the presence of the cellular protein lens epithelium-derived growth factor (LEDGF)/p75 and form a tetramer, and the integration proceeds to the strand-transfer step (16). The free IN tetramer does not bind DNA directly, and tetramerization occurs only by the interaction between two DNA-bound IN dimers (12). Incorrect oligomerization of IN in terms of time and localization may prevent the essential native assembly of its complexes with the viral DNA LTR ends (17). IN activity requires binding to the cellular protein LEDGF/p75 (18), which activates IN in vitro and in cells by tethering it to the host chromosomes (19–22). Although low concentrations of LEDGF/p75 stimulate IN to bind DNA (19) as well as its enzymatic activity (23), overexpression of the LEDGF/p75 IN-binding domain inhibits HIV-1 replication and blocks nuclear import of IN, suggesting that competitive inhibition of the LEDGF–IN interaction may be a target for anti-HIV drug design (24).
Despite the efforts invested, the only two IN inhibitors currently in phase II clinical trials (25) are the strand-transfer inhibitors designated MK-0518 (Merck & Co., Whitehouse Station, NJ) (7, 26, 27) and GS-9137 (Gilead, Foster City, CA) (7). Here, we demonstrate an alternative approach for the design of IN inhibitors, which block its catalytic activity at both integration steps in an allosteric mode by modulating its dimer/tetramer oligomerization equilibrium. We used LEDGF-derived peptides as shiftides that bind specifically to IN tetramer and shift the oligomeric state of IN to an inactive tetrameric form. The LEDGF-derived peptides inhibit the enzymatic activities of IN in vitro, penetrate cells, and block integration of viral DNA and HIV-1 propagation in cell culture. The anti-HIV activity of the peptides establishes the shiftide mechanism as a general approach for drug design.
Results
Design of LEDGF-Derived Peptides That Modulate the IN Oligomerization Equilibrium.
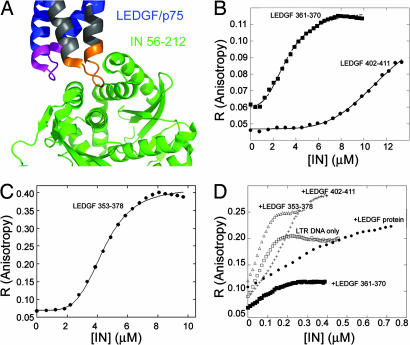
Any molecule that preferentially binds the tetrameric state of IN should shift the oligomerization equilibrium toward this tetrameric form. We sought molecules that specifically bind IN tetramers but not dimers. LEDGF/p75 binds specifically to the tetrameric form of IN (19) via two well defined loops (18). Based on the crystal structure of the IN–LEDGF/p75 complex (18), we designed and synthesized three fluorescein-labeled peptides derived from these IN-binding loops of LEDGF/p75 (Fig. 1A and Table 1).
Fig. 1.
Ligand binding of IN: fluorescence anisotropy studies. (A) Crystal structure of the LEDGF–IN complex. The LEDGF-derived peptides used in this study are LEDGF 353–378 (gray), LEDGF 361–370 (orange), and LEDGF 402–411 (magenta). Coordinates are taken from ref. 18. (B and C) Fluorescence anisotropy-binding studies. IN was titrated into the fluorescein-labeled LEDGF/p75 peptides (100 nM): LEDGF/p75 361–370 and LEDGF/p75 402–411 (B) and LEDGF/p75 353–378 (C). Data were fit to the Hill equation. (D) The effect of the LEDGF/p75 and peptides derived from it on the DNA binding of IN. IN was titrated into fluorescein-labeled HIV-1 LTR DNA (10 nM) alone and in the presence of 1 μM LEDGF 361–370, LEDGF 353–378, LEDGF 402–411, and full-length LEDGF. Binding affinities and Hill coefficients are given in Table 2.
Table 1.
The LEDGF/p75-derived peptides used in this study
| Peptide | Sequence |
|---|---|
| LEDGF/p75 353–378 | WIHAEIKNSLKIDNLDVNRCIEALD |
| LEDGF/p75 361–370 | WNSLKIDNLDV |
| LEDGF/p75 402–411 | WKKIRRFVSQVIM |
IN Binds to the LEDGF-Derived Peptides as a Tetramer and to the DNA as a Dimer.
Fluorescence anisotropy was used to determine the binding affinity of IN to the LEDGF/p75 peptides. IN bound LEDGF 353–378 and LEDGF 361–370 with a Kd of 4 μM and LEDGF 402–411 with a Kd of 12 μM. IN binding to all three peptides was strongly cooperative, with a Hill coefficient of ≈4 (Fig. 1 B and C and Table 2), confirming that IN binds the LEDGF peptides as a tetramer, similar to its binding to the LEDGF/p75 protein (12). IN binding to a fluorescein-labeled 36-bp double-stranded viral LTR DNA was in agreement with the previous reports (15, 28) and had a Kd of 37 nM and a Hill coefficient of 2 (Fig. 1D and Table 2). This finding suggests that IN binds the LTR DNA as a dimer.
Table 2.
Binding affinity of IN to the LTR DNA and the LEDGF/p75-derived peptides: Fluorescence anisotropy studies
| Peptide | Kd for IN binding, μM | Hill coefficient |
|---|---|---|
| LEDGF/p75 353–378 | 4.4 ± 0.2 | 4.4 |
| LEDGF/p75 361–370 | 3.7 ± 0.2 | 3.4 |
| LEDGF/p75 402–411 | 12 ± 0.6 | 4.5 |
| FL′ DNA LTR | 0.034 ± 0.001 | 2.1 |
| FL′ DNA LTR + LEDGF 361–370 | 0.099 ± 0.003 | 2.2 |
| FL′ DNA LTR + LEDGF 353–378 | 0.068 ± 0.003 | 2.4 |
| FL′ DNA LTR + LEDGF 402–411 | 0.20 ± 0.02 | 2.7 |
| FL′ DNA LTR + full-length LEDGF/p75 | 1.1 ± 0.1 | 1.5 |
Binding curves are shown in Fig. 1. FL′, fluorescein-labeled.
LEDGF-Derived Peptides Inhibit IN Binding to DNA by Shifting Its Oligomerization Equilibrium Toward the Tetramer.
We used fluorescence anisotropy to determine the effect of the peptides on the binding of IN to the fluorescein-labeled LTR DNA. All of the ligand binding and oligomerization studies were carried out at an ionic strength of 190 mM. All LEDGF peptides at molar ratio of 1:1 inhibited DNA binding of IN by 3- to 6-fold (Fig. 1D and Table 2). Full-length LEDGF inhibited DNA binding of IN by 30-fold, indicating a synergistic effect between the two peptides in the context of the full-length protein.
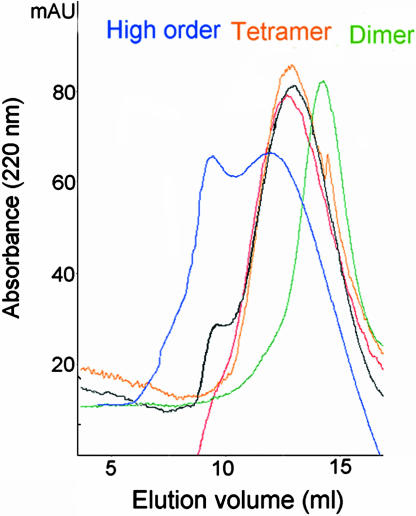
To reveal the mechanism of DNA-binding inhibition, we studied whether the peptides affect the oligomerization equilibrium of IN. Sedimentation equilibrium analytical ultracentrifugation (AUC) experiments showed that free IN is in equilibrium among high-order oligomers, tetramers, and dimers (Table 3). We used analytical gel filtration to study the effect of ligand binding on this equilibrium (Fig. 2 and Table 4). Unbound IN eluted as a high-order oligomer. IN was tetrameric in the presence of the LEDGF peptides and dimeric in the presence of LTR DNA, in agreement with our fluorescence anisotropy results. When incubated with both LTR DNA and the LEDGF peptides at a 1:1 ratio, IN was tetrameric as with the peptide only, indicating a shift of the oligomerization equilibrium caused by the peptide in the presence of DNA. The oligomeric state of the truncated mutant IN 52–288 was not affected by binding peptides or LTR DNA (data not shown), indicating that the effect is specific and that the N terminus of IN is involved in the binding process.
Table 3.
AUC results: IN oligomeric state
| [IN] | Mr, kDa | Oligomeric state |
|---|---|---|
| 80 μM | 180,677 ± 1,192 | High-order oligomer |
| 20 μM | 123,235 ± 805 | Tetramer |
| 2 μM | 68,240 ± 672 | Dimer |
Fig. 2.
Effect of ligand binding on the oligomeric state of IN. Oligomerization of IN in the presence of various ligands was studied by using analytical gel filtration. The samples were 14 μM IN 1–288 (blue), 14 μM IN 1–288 + 14 μM LEDGF 361–370 (black), 14 μM IN + 14 μM LEDGF 402–411 (red), 14 μM IN 1–288 + 14 μM DNA LTR (green), and 14 μM IN 1–288 + 14 μM DNA LTR + 14 μM LEDGF 361–370 (orange).
Table 4.
Gel filtration results: Effect of ligand binding on IN oligomeric state
| Sample | Mr, kDa | Oligomeric state | Elution volume, ml |
|---|---|---|---|
| IN | 200 | High-order oligomer | 9.5, 12 |
| IN + LEDGF 361–370 | 125 | Tetramer | 12.6 |
| IN + LEDGF 402–411 | 125 | Tetramer | 12.6 |
| IN + LTR DNA | 65 | Dimer | 13.6 |
| IN + DNA + LEDGF 361–370 | 130 | Tetramer | 12.8 |
The LEDGF Peptides Inhibit the Catalytic Activities of IN in Vitro.
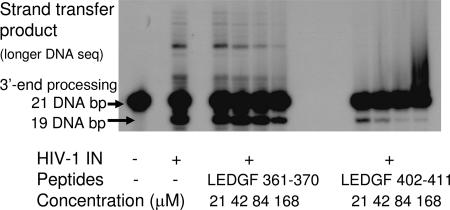
We tested whether the short LEDGF peptides inhibit the catalytic activities of IN in vitro. LEDGF 402–411 and LEDGF 361–370 strongly inhibited both the 3′ end processing and the consequent strand-transfer activities of IN. LEDGF 402–411 was more potent and showed significant inhibition at the lowest concentration tested, 21 μM (Fig. 3). The peptides also inhibited IN-mediated strand transfer when a processed LTR DNA served as a template (data not shown). Together, our results indicate that DNA binding and processing is inhibited by a shiftide mechanism (Fig. 4).
Fig. 3.
LEDGF peptides inhibit IN catalytic activities in vitro. IN was incubated with the LEDGF-derived peptides, and the 3′ end processing and strand-transfer enzymatic activities were analyzed as described in Materials and Methods.
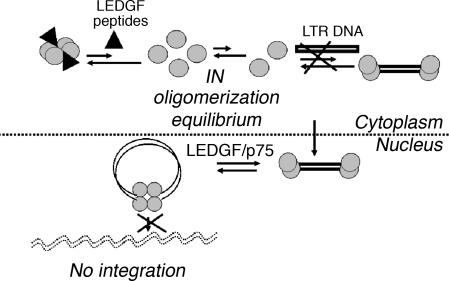
Fig. 4.
The shiftide model for IN inhibition. LEDGF-derived peptides are shiftides that shift the oligomerization state of IN toward the tetramer. The LEDGF peptides penetrate the cells and bind IN in the cytoplasm. They shift it to a tetrameric state, reducing its degree of binding to the unprocessed LTR DNA and preventing the 3′ end processing and consequently the strand-transfer catalytic activities.
The LEDGF Peptides Inhibit HIV-1 Replication in Cell Culture by Inhibiting Viral DNA Integration.
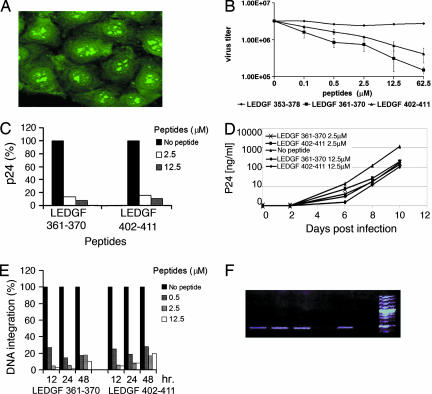
We determined whether the LEDGF peptides inhibit HIV-1 replication in cultured cells. Fluorescein-labeled LEDGF 361–370 and LEDGF 402–411, but not the longer LEDGF 353–378, penetrated HeLa CD4 cells (Fig. 5A and data not shown). These peptides were nontoxic to cells at the concentrations used, as measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) test (data not shown). The effect of LEDGF 361–370 and LEDGF 402–411 on HIV-1 propagation was studied by using TZM-bl multinuclear activation of a galactosidase indicator (MAGI) cells, which express the β-galactosidase gene under transcription activator region regulation (29). Both LEDGF 361–370 and LEDGF 402–411 significantly inhibited HIV-Tat-mediated expression of the reporter gene in a concentration-dependent manner (Fig. 5B), indicating that the transcription of viral genes was inhibited. The peptides inhibited HIV-1 replication in infected lymphoid cells, demonstrated by their ability to reduce the amounts of the viral p24 released by these cells (Fig. 5 C and D). We estimated the number of integrated proviruses in infected cells by using real-time PCR to verify that HIV-1 replication was blocked by preventing integration events. At concentrations <2.5 μM, both LEDGF peptides inhibited integration by >90% (Fig. 5E). To ascertain that the peptides did not affect earlier infection steps, such as the HIV-1 entry and/or RT activity, we estimated the total amount of reverse-transcribed viral cDNA in infected cells with PCR. Viral DNA was present in untreated cells and in cells treated with the LEDGF/p75-derived peptides but not in cells treated with the RT-inhibitor AZT (Fig. 5F), confirming that the observed reduction in viral gene expression and production of infectious viruses is attributable to inhibition of IN activity in the infected cells.
Fig. 5.
Inhibiting HIV-1 replication by the LEDGF/p75-derived peptides. (A) LEDGF 361–370 penetrates into HeLa cells, as visualized by using a confocal microscope. The peptide localizes both to the cytoplasm and the nucleus, especially to the nucleoli. (B) The LEDGF-derived peptides inhibit TAR-mediated transcription of HIV-1 genes in TZM-bl MAGI cells. (C) The LEDGF-derived peptides inhibit HIV-1 replication in cell culture. H9 T-lymphoid cells were incubated with the indicated peptides, and the total amount of the released virus was estimated based on the p24 protein content after 10 days. (D) Kinetics of the inhibition of p24 formation in T-lymphoid cells. (E) LEDGF-derived peptides inhibit the integration of HIV-1 to the genome. Real-time PCR studies after incubation with the peptides are shown. The results represent the percentage of integrated viral DNA. (F) Total viral DNA in HIV-1-infected cells. Cells were treated with 12.5 μM LEDGF 361–370 (lane 1), 12.5 μM LEDGF 402–411 (lane 2), 12.5 μM LEDGF 353–378 (lane 3), 2 μM AZT (lane 4), untreated cells (lane 5), and uninfected cells (lane 6).
Discussion
The Shiftide Concept: Expanding the Scope of Allosteric Inhibition and Oligomerization Inhibition.
We describe the shiftide concept, which utilizes peptides to modulate protein activity by specifically binding to an inactive oligomeric state of the target protein, resulting in a shift of the oligomerization equilibrium and an inhibition of activity. The shiftides act in a manner similar to the allosteric model (1), according to which ligand binding can shift an equilibrium toward R or T states, as in the case of hemoglobin (30–33): Heterotropic allosteric effectors lower the oxygen affinity of the T state upon binding to hemoglobin by binding to the T or R state and shifting the R/T equilibrium in favor of the bound state. Such effectors also affect the dimer–dimer affinity of hemoglobin and even lead to tetramer dissociation in extreme cases (34). The shiftides add an additional dimension to such allosteric inhibition because they modulate the equilibrium between various oligomeric states and not within a given oligomer.
Shiftides open new directions in the field of oligomerization inhibitors and are advantageous over conventional dimerization inhibitors (8, 9) or ligands that covalently attach several monomers together (35). There are intrinsic problems with competitive dimerization inhibitors because small molecules usually cannot supply enough binding energy for the large interfaces to be targeted, and the full-length protein will bind more tightly than a peptide derived from it (10). The shiftide approach targets oligomerization by binding at a different site of the protein, in an allosteric mode, which overcomes the drawbacks of targeting a protein–protein interaction interface and presents a way to modulate oligomerization in a noncompetitive allosteric mechanism.
The LEDGF/p75–IN Interaction as a Basis for Drug Design.
Our results show that peptides derived from LEDGF/p75 inhibit HIV-1 replication by blocking IN activity. However, they do not act by competitively inhibiting the LEDGF/p75–IN interaction, as was proposed for the LEDGF IN-binding domain (20, 24), but rather act as shiftides. The affinity of IN to DNA is three orders of magnitude stronger than its affinity to the peptides. Under a competitive situation at a 1:1 peptide:DNA ratio, IN dimers bind tightly to the DNA with nanomolar affinity. Weaker binding of the peptide to the unbound tetrameric fraction of IN follows later and shifts the equilibrium of free IN from the dimer toward the tetramer, which leads to a shift of the equilibrium from DNA-bound dimeric IN to free dimeric IN, resulting in dis-sociation of the IN–DNA complex (Fig. 1D). A higher pep-tide:DNA molar ratio would result in stronger inhibition of DNA binding.
According to our model, the peptides shift the oligomerization equilibrium of IN in the cytoplasm from a dimer, which binds the unprocessed LTR DNA and catalyses the 3′ end processing, to a tetramer that is unable to bind the unprocessed DNA and catalyze this reaction (Fig. 4). Thus, the viral DNA substrate is not ready for strand transfer, preventing the integration. Inhibition of IN is achieved before its binding to the full-length LEDGF/p75 and the tethering to the chromosomes, which takes place after nuclear import. Moreover, because the IN tetramer also is unable to bind directly to the processed DNA as shown by cross-linking experiments (12), shifting the oligomeric state of IN toward a tetramer inhibits the strand transfer of a processed DNA template. In summary, the shiftide approach results in inhibition of both integration steps, making it advantageous over strand-transfer inhibitors, which inhibit only the second integration step.
The LEDGF/p75 peptides inhibit the enzymatic activities of IN in vitro in the absence of the LEDGF/p75 protein, confirming that inhibition is not attributable to competition with the LEDGF–IN interaction. LEDGF 402–411 inhibited the enzymatic activity of IN more potently than LEDGF 361–370 did, although its affinity to IN was 3-fold weaker. This observation is because binding affinity is not the only factor that is responsible for the inhibitory activity of a molecule (Kd and Ki are different in many cases), as was shown for the RT inhibitors (36, 37). The peptides inhibit integration in cells, do not lower the amounts of reverse transcripts, but do reduce the number of proviral copies and HIV-1 Tat-mediated transcription, showing that the peptides do not affect the early, but do halt the late, events of HIV-1 replication. The peptides were active in cells in low micromolar concentrations, which sometimes are below KD possibly because of a high local concentration of the peptides in the cells, resulting in stronger interaction. Together, our results show that LEDGF-derived peptides inhibit HIV-1 replication by the mechanism of modifying the oligomerization state of IN in cells.
Our results suggest an additional possible explanation for the ability of LEDGF/p75 to stimulate IN activity in vivo: LEDGF/p75 protein may act in a mechanism similar to the peptides, as a natural heterotropic allosteric effector that mediates tetramerization of DNA-bound IN dimers. After 3′ end processing, the IN dimers bound to the two LTR DNAs penetrate the nucleus, bind LEDGF/p75, and tetramerize, and strand transfer occurs to complete the integration process. Further studies in cells are needed to prove this hypothetical model. Inhibition of IN by overexpression of LEDGF IN-binding domain also could be explained by a shiftide mechanism (24).
Implications for Drug Design.
The LEDGF peptides are only 10-aa long and efficiently penetrate cells. These properties make them potential lead anti-HIV compounds. To overcome the known problems with peptides as drugs, we are currently working on the conversion of these peptides into nonpeptidic lead compounds with improved activity, metabolic stability, and bioavailability. Administering such drugs in combination with existing therapy may improve the treatment of AIDS in the future. We propose that the shiftides approach, which furthers the classic approaches of allosteric inhibition and chemical chaperones, could be used as a general methodology to overcome the obstacles associated with classic dimerization inhibitors and to develop lead compounds against various diseases associated with oligomeric proteins.
Materials and Methods
Peptide Synthesis.
Peptides were synthesized on an ABI 433A peptide synthesizer (Applied Biosystems, Foster City, CA) and labeled with Trp at their N termini for UV spectroscopy. The peptides were labeled by using 5′ and 6′ carboxyfluorescein succinimidyl ester (Molecular Probes, Carlsbad, CA) at the N terminus. The peptides were purified on a Gilson (Middleton, WI) HPLC with a reverse-phase C8 semipreparative column (ACE) with a gradient from 5% to 60% acetonitrile in water [both containing 0.001% (vol/vol) trifluoroacetic acid]. Peptide concentrations were determined by using a UV spectrophoto-meter (Shimadzu, Kyoto, Japan) as described in ref. 38.
Protein Expression and Purification.
The His-tagged IN expression vector was a generous gift from A. Engelman (Harvard Medical School, Boston, MA), and its expression and purification were performed as described in ref. 39. His-tagged LEDGF/p75 was expressed and purified as described in ref. 40.
Fluorescence Anisotropy.
Measurements were performed at 10°C by using a PerkinElmer (Waltham, MA) LS-55 luminescence spectrofluorimeter equipped with a Hamilton microlab 500 dispenser (6, 41). The fluorescein-labeled peptide or DNA (1 ml, 0.05–0.1 μM in 20 mM Tris buffer, pH 7.4/185 mM NaCl) was placed in a cuvette, and the nonlabeled protein (200 μl, ≈100 μM) was added in 20 aliquots of 10 μl at 1-min intervals. The total fluorescence and anisotropy were measured after each addition by using an excitation wavelength of 480 nm and an emission wavelength of 530 nm. Data were fit to the Hill equation
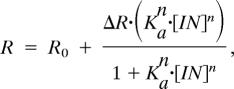 |
where R is measured anisotropy, ΔR is the amplitude of the anisotropy change from R0 (free peptide) to peptide in complex, [IN] is the added concentration of IN, and Ka is the association constant.
In the competition experiments, a mixture of LEDGF/p75 peptide (500 nM) and IN (4 μM) was incubated for 0.5 h and then titrated into fluorescein-labeled LTR DNA (10 nM). The LTR DNA sequence used was 5′-AGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGT-3′.
Analytical Gel Filtration.
Analytical gel filtration of IN (10 μM) was performed on an AKTA Explorer with a Superose 12 analytical column 30 × 1 cm (GE Healthcare–Amersham Pharmacia, Giles, U.K.) equilibrated with buffer (20 mM Tris, pH 7.4/1 M NaCl/10% glycerol). Proteins were eluted with a flow rate of 1 ml/min at 4°C, and the elution profile was monitored by UV absorbance at 220 nm. The column was calibrated with molecular weight standards (GE Healthcare–Amersham Pharmacia).
AUC.
The equilibrium sedimentation experiments were performed on a Beckman (Fullerton, CA) XL-I ultracentrifuge by using Ti-60 rotor and six-sector cells at 30,000 and 40,000 rpm, respectively, at 10°C. Sample volume was 50 μl. Samples were considered to be at equilibrium as judged by comparing several scans at each speed. Buffer conditions were 20 mM Tris (pH 7.4) and 10% glycerol. The ionic strength of the buffer was adjusted to 190 mM with a solution of 3 M NaCl in the same buffer. Data were processed and analyzed by using Ultra-Spin software (Centre for Protein Engineering; ultraspin.mrc-cpe.cam.ac.uk).
Cell Penetration Experiments.
The fluorescein-labeled peptides (10 μM in PBS) were incubated with HeLa cells for 2 h at 37°C. After three washes in PBS, the cells were visualized by confocal microscopy.
In Vitro Integration Assays.
The 3′ end processing and the strand-transfer activities of IN were performed as previously described (36, 37).
Cells.
Monolayer-adherent HeLa, HeLa MAGI (TZM-bl) (42), and HEK293T cells were grown in DMEM, whereas the T lymphocyte cell lines Sup T1 and H9 were cultivated in RPMI medium 1640. All media were supplemented with 10% (vol/vol) FCS, 0.3 g/liter l-glutamine, 100 units/ml penicillin, and 100 units/ml streptomycin (Biological Industries, Kibbutz Beit Haemek, Israel). Cells were incubated at 37°C in 5% CO2 atmosphere. The H9, Sup T1, and HeLa MAGI cells (TZM-bl) were provided by the National Institutes of Health Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD).
Viruses.
Wild-type HIV-1 and Δenv/VSV-G were generated by transfection (43) of HEK293T cells with pSVC21 plasmid containing the full-length HIV-1HXB2 viral DNA (44). Wild-type and Δenv/VSV-G viruses were harvested from HEK293T cells 48 and 72 h posttransfection with pSVC21 Δenv. The viruses were stored at −75°C.
Inhibiting HIV-1 Infectivity.
Cultured lymphocytes (1 × 105) were centrifuged for 3 min at 500 × g, the supernatant was aspirated, and the cells were resuspended in 0.2 to 0.5 ml of medium containing viruses at multiplicities of infection (MOIs) of 0.1 and 2. After absorption for 1 h at 37°C, the cells were washed and incubated in growth media for an additional 1 to 10 days.
H9 lymphoid cells were incubated with the indicated peptides for 2 h. After infection with wild-type HIV-1 at MOI 0.1, the cells were incubated for 10 days, and the amounts of p24 in the medium were determined by using the capture assay kit (SAIC, AIDS Vaccine Program, Frederick, MD), according to the manufacturer's instructions.
Titration of HIV-1 was carried out with the MAGI assay, as described by Kimpton and Emerman (29), by using TZM-b1 cells in 96-well plates at 10 × 103 cells per well. All cells were infected with the same MOI.
Estimation of the Amounts of Proviral DNA.
Real-time PCR experiments were performed as described in ref. 45. The second round of PCR was performed on 1/25 of the first-round PCR product in a mixture containing 300 nM each primer, 12.5 μl of 2× SYBR green master mix (Applied Biosystems) at a final volume of 25 μl, run on an ABI PRIZM 7700 (Applied Biosystems). The second round of PCR cycles began with a DNA-denaturing and polymerase-activation step (95°C for 10 min), followed by 50 cycles of amplification (95°C for 15 sec, 60°C for 60 sec). SVC21 plasmid containing full-length HIV-1HXB2 viral DNA was used to generate a standard linear curve in a range of 5 ng to 0.25 fg (R = 0.99). DNA samples were assayed with quadruplets of each sample.
Estimation of the Amount of Total Viral DNA.
After incubation of Sup T1 cells with the indicated peptides for 2 h, the cells were infected with a HIV-1 Δenv/VSV-G virus at MOI of 2 (as described above) for 6 h. Viral DNA sequence was amplified with the Gag-specific primers (Gag F, 5′-AGTGGGGGGACATCAAGCAGCCATG-3′; Gag R, 5′-TGCTATGTCAGTTCCCCTTGGTTCTC-3′). Gag fragment was amplified from 10 ng in a 25-μl reaction mixture containing 1× PCR buffer, 3.5 mM MgCl2, 200 μM dNTPs, 300 nM primers, and 0.025 units/μl Taq polymerase. The PCR conditions were as follows: a DNA denaturation and polymerase activation step of 5 min at 95°C and then 29 cycles of amplification (95°C for 45 sec, 60°C for 30 sec, and 72°C for 45 sec).
Acknowledgments
We thank Dr. Deborah E. Shalev, Prof. Chaim Gilon, and Dr. Michal Goldberg for their critical reading of the manuscript; and the Centre for Protein Engineering, Cambridge, U.K., for the use of the analytical ultracentrifuge facility. Part of this work was carried out in the Peter A. Krueger laboratory with the generous support of Nancy and Lawrence Glick and Pat and Marvin Weiss. This work was supported by a Bikura grant from the Israeli Science Foundation (to A.F.) and by an Israeli Science Foundation grant (to A. Loyter and M.K.).
Abbreviations
- IN
integrase
- LEDGF
lens epithelium-derived growth factor
- AUC
analytical ultracentrifugation
- MOI
multiplicity of infection
- MAGI
multinuclear activation of a galactosidase indicator
- LTR
long terminal repeat.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Monod J, Wyman J, Changeux JP. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 2.Beddell CR, Goodford PJ, Kneen G, White RD, Wilkinson S, Wootton R. Br J Pharmacol. 1984;82:397–407. doi: 10.1111/j.1476-5381.1984.tb10775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobson CM. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 4.Fan JQ. Trends Pharmacol Sci. 2003;24:355–360. doi: 10.1016/S0165-6147(03)00158-5. [DOI] [PubMed] [Google Scholar]
- 5.Ulloa-Aguirre A, Janovick JA, Brothers SP, Conn PM. Traffic. 2004;5:821–837. doi: 10.1111/j.1600-0854.2004.00232.x. [DOI] [PubMed] [Google Scholar]
- 6.Friedler A, Hansson LO, Veprintsev DB, Freund SM, Rippin TM, Nikolova PV, Proctor MR, Rudiger S, Fersht AR. Proc Natl Acad Sci USA. 2002;99:937–942. doi: 10.1073/pnas.241629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lataillade M, Kozal MJ. AIDS Patient Care STDS. 2006;20:489–501. doi: 10.1089/apc.2006.20.489. [DOI] [PubMed] [Google Scholar]
- 8.Camarasa MJ, Velazquez S, San-Felix A, Perez-Perez MJ, Gago F. Antiviral Res. 2006;71:260–267. doi: 10.1016/j.antiviral.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Divita G. J Biol Chem. 1994;269:13080–13083. [PubMed] [Google Scholar]
- 10.Arkin MR, Wells JA. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 11.Engelman A, Mizuuchi K, Craigie R. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 12.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deprez E, Tauc P, Leh H, Mouscadet JF, Auclair C, Hawkins ME, Brochon JC. Proc Natl Acad Sci USA. 2001;98:10090–10095. doi: 10.1073/pnas.181024498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deprez E, Tauc P, Leh H, Mouscadet JF, Auclair C, Brochon JC. Biochemistry. 2000;39:9275–9284. doi: 10.1021/bi000397j. [DOI] [PubMed] [Google Scholar]
- 15.Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. J Biol Chem. 2006;281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- 16.Chen A, Weber IT, Harrison RW, Leis J. J Biol Chem. 2006;281:4173–4182. doi: 10.1074/jbc.M510628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craigie R. J Biol Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 18.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Proc Natl Acad Sci USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. J Biol Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 20.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 21.Singh DP, Fatma N, Kimura A, Chylack LT, Jr, Shinohara T. Biochem Biophys Res Commun. 2001;283:943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 22.Rahman S, Lu R, Vandegraaff N, Cherepanov P, Engelman A. Virology. 2006;357:79–90. doi: 10.1016/j.virol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Cherepanov P, Devroe E, Silver PA, Engelman A. J Biol Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 24.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. J Virol. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pommier Y, Johnson AA, Marchand C. Nat Rev Drug Discov. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- 26.Hazuda DJ, Young SD, Guare JP, Anthony NJ, Gomez RP, Wai JS, Vacca JP, Handt L, Motzel SL, Klein HJ, et al. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz M, Morales-Ramirez JO, Nguyen BY, Kovacs CM, Steigbigel RT, Cooper DA, Liporace R, Schwartz R, Isaacs R, Gilde LR, et al. J Acquir Immune Defic Syndr. 2006;43:509–515. doi: 10.1097/QAI.0b013e31802b4956. [DOI] [PubMed] [Google Scholar]
- 28.Deprez E, Barbe S, Kolaski M, Leh H, Zouhiri F, Auclair C, Brochon JC, Le Bret M, Mouscadet JF. Mol Pharmacol. 2004;65:85–98. doi: 10.1124/mol.65.1.85. [DOI] [PubMed] [Google Scholar]
- 29.Kimpton J, Emerman M. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perutz MF. Nature. 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 31.Yonetani T, Park SI, Tsuneshige A, Imai K, Kanaori K. J Biol Chem. 2002;277:34508–34520. doi: 10.1074/jbc.M203135200. [DOI] [PubMed] [Google Scholar]
- 32.Yonetani T, Tsuneshige A. C R Biol. 2003;326:523–532. doi: 10.1016/s1631-0691(03)00150-1. [DOI] [PubMed] [Google Scholar]
- 33.Ackers GK, Doyle ML, Myers D, Daugherty MA. Science. 1992;255:54–63. doi: 10.1126/science.1553532. [DOI] [PubMed] [Google Scholar]
- 34.Schay G, Smeller L, Tsuneshige A, Yonetani T, Fidy J. J Biol Chem. 2006;281:25972–25983. doi: 10.1074/jbc.M604216200. [DOI] [PubMed] [Google Scholar]
- 35.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 36.Oz Gleenberg I, Avidan O, Goldgur Y, Herschhorn A, Hizi A. J Biol Chem. 2005;280:21987–21996. doi: 10.1074/jbc.M414679200. [DOI] [PubMed] [Google Scholar]
- 37.Oz I, Avidan O, Hizi A. Biochem J. 2002;361:557–566. doi: 10.1042/0264-6021:3610557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill SC, von Hippel PH. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 39.Jenkins TM, Engelman A, Ghirlando R, Craigie R. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 40.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. Nucleic Acids Res. 2006;34:1663–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedler A, Veprintsev DB, Rutherford T, von Glos KI, Fersht AR. J Biol Chem. 2005;280:8051–8059. doi: 10.1074/jbc.M411176200. [DOI] [PubMed] [Google Scholar]
- 42.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. J Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cullen BR. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 44.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, et al. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto N, Tanaka C, Wu Y, Chang MO, Inagaki Y, Saito Y, Naito T, Ogasawara H, Sekigawa I, Hayashida Y. Virus Genes. 2006;32:105–113. doi: 10.1007/s11262-005-5851-2. [DOI] [PubMed] [Google Scholar]