Abstract
Potentiation is the transition from higher-order, transcriptionally silent chromatin to a less condensed state requisite to accommodating the molecular elements required for transcription. To examine the underlying mechanism of potentiation an ≈13.7-kb mouse protamine domain of increased nuclease sensitivity flanked by 5′ and 3′ nuclear matrix attachment regions was defined. The potentiated DNase I-sensitive region is formed at the pachytene spermatocyte stage with the recruitment to the nuclear matrix of a large ≈9.6-kb region just upstream of the domain. Attachment is then specified in the transcribing round spermatid, recapitulating the organization of the human cluster. In comparison to other modifiers that have no effect, i.e., histone methylation, HP1, and SATB1, topoisomerase engages nuclear matrix binding as minor marks of histone acetylation appear. Reorganization is marked by specific sites of topoisomerase II activity that are initially detected in leptotene–zygotene spermatocytes just preceding the formation of the DNase I-sensitive domain. This has provided a likely model of the events initiating potentiation, i.e., the opening of a chromatin domain.
Keywords: chromatin, nuclear matrix, spermatogenesis, topoisomerase, potentiation
Chromatin loop domains typically range in size from 50 to 200 kb in length in somatic cells and partition the genome into discrete loci of coordinately regulated genes. This organization is a function of interactions with structural elements of a dynamic network of proteins termed the nuclear matrix (reviewed in ref. 1). As a cell differentiates, the demands on its genome change, requiring a mechanism that can selectively “potentiate,” i.e., open, regions of the genome as needed through development. Potentiation is the transition from higher-order structures to those more amenable to transcription, providing access to loci that are simultaneously active like the protamine cluster (2) or to genes that function in succession through development like the various members of the Hoxb cluster (3, 4). These transitions were first experimentally observed as an increased susceptibility to nuclease digestion in the vicinity of coordinately expressed genes (5–7). Potentiative changes in chromatin domains must occur before transcription of constituent elements can begin (8–10). Indeed, although a more relaxed chromatin conformation provides the requisite access for the transcriptional machinery, it will not drive transcription by itself. This is exemplified by the human α-globin cluster whose expression is constrained by histone modifications and several transcription factor complexes (11) as it shares a constitutively potentiated domain with MPG, a housekeeping gene that is active in DNA-damage repair (12). To date, the molecular components of potentiation are not well understood.
The nuclear matrix is rich in factors that organize and recruit complexes of proteins that modify chromatin structure (13–15). One of the best characterized examples of a structural nuclear matrix protein is special AT-rich binding protein 1. SATB1 is a phosphorylation-dependent (16) transcriptional regulator that has been associated with T cell (17) and erythroid (18) differentiation. This factor targets histone deacetylases and ATP-dependent nucleosome remodeling complexes (14) to specific base-unpairing regions containing matrix attachment regions (MARs), thereby regulating a number of factors involved in T cell differentiation.
Nuclear matrix proteins (19, 20) that have also been implicated in modifying DNA topology in chromatin are the topoisomerases. Type II topoisomerases, Top2, are ATP-dependent and catalyze transient single- or double-stranded breaks in DNA. This activity can pass one duplex of DNA through another, catenate and decatenate DNA, and is essential for the relief of torsional stress due to supercoiling (21, 22). Well studied examples of Top2 function include chromosome condensation and segregation during mitosis (23–26) as well as recombination and segregation in meiosis (27–31). Mammalian Top2 has two isoforms (α, Top2a; β, Top2b) that are somewhat functionally redundant (32), sharing a large degree of conservation in their N-terminal (ATPase) and catalytic domains.
Top2a is the predominant form in proliferating tissues, exhibiting peak expression in the late S phase and G2/M transition (24, 33) and its association with metaphase chromosomes (34, 35). Top2b activity has recently been shown to play a key role in transcription by creating transient double-stranded DNA breaks in the promoter regions of a number of genes (36). For example, estrogen-bound ERα directs an activating complex that includes Top2b, polyADP-ribose polymerase 1, and other factors typically associated with DNA repair to a promoter containing an estrogen response element, the pS2 promoter. Moreover, Top2b-null mouse embryos show a marked change in the expression of almost one-third of its developmentally regulated genes (37). Given its association with the nuclear matrix, catalytic activity (21, 22), association with several chromatin-modifying elements (15, 38, 39), and a new direct link to transcriptional regulation, Top2b presents an excellent chromatin potentiation candidate.
In humans, the three constituent genes, Protamine 1 (PRM1), Protamine 2 (PRM2), and Transition protein 2 (TNP2), reside in a single 28.6-kb DNase I-sensitive domain (2) on chromosome 16p13.2 (40). They are transcribed at the postmeiotic round spermatid stage of spermatogenesis and translated in elongating spermatids. These messages are bound as cytoplasmic messenger ribonuclear protein particles until histone replacement is initiated with the transition proteins in late elongating spermatids. At this time the mRNPs are activated and then translated into the peptides that will repackage and compact the male genome in terminally differentiated spermatozoa. In humans and mice the genes first acquire a DNase I-sensitive conformation in pachytene spermatocytes (41) that is even maintained in human spermatozoa. The analogous region in mice has thus far not been identified. Nuclear matrix associations have been observed at the human cluster as the two sperm-specific MARs that flank the DNase I-sensitive domain. The genes are subject to position effect when the arrangement and exclusion of the MARs flanking the human protamine domain were altered in transgenic mice, suggesting that these MARs confer an insulative role on the domain (42). Thus far, nuclear matrix association at the mouse cluster has not been investigated. It could provide the mechanism or staging ground for potentiation because it is involved in the recruitment of factors that mediate alterations in chromatin structure.
To address these issues, the endogenous mouse Prm1→Prm2→Tnp2 DNase I-sensitive domain was mapped and the developmental role of MARs throughout this region was assessed. ChIP revealed predictable patterns of histone acetylation among a high steady-state level of histone methylation. Neither SATB1 nor heterochromatin protein 1 (HP1) appeared to be associated with the domain throughout spermatogenesis. However, examination of Top2a and Top2b association throughout the region showed that they are a key factor in a novel topoisomerase II-based potentiative mechanism. The results of these studies are described here.
Results and Discussion
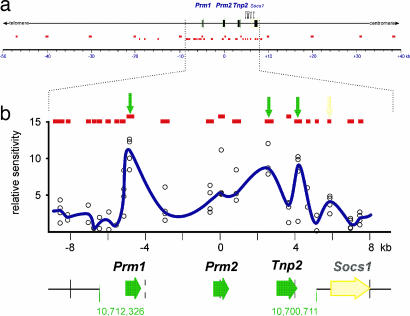
Whereas the 28.6-kb DNase I-sensitive human PRM1→PRM2→TNP2 domain is well characterized (2, 43) the analogous mouse domain thus far has not been delineated, although the mouse protamine cluster resides in a syntenic region of chromosome 16. To fill this void, nuclei from round spermatids were subjected to DNase I-sensitivity mapping by real-time PCR. Initially 12 amplicons within an ≈90-kb region (National Center for Biotechnology Information mouse genomic Build v.36.1; coordinates, 10,664,900–10,755,440) that encompasses the mouse protamine cluster were assayed as summarized in Fig. 1a and SI Table 1. This highlighted a central ≈16-kb region surrounding the mouse protamine cluster that was subjected to higher-resolution 21-amplicon analysis as shown in Fig. 1b and SI Table 1. High-resolution mapping identified an ≈11.6-kb DNase I-sensitive region extending from ≈1.2 kb upstream of Prm1 to ≈1.4 kb downstream of Tnp2, encompassing coordinates 10,700,711–10,712,326. Three sites of hypersensitivity within the protamine domain were also identified. The first was within the Prm1 promoter (≈10,711,006–10,711,161), the second just upstream of Tnp2 (≈10,703,079–10,703,454), and a third downstream of the Tnp2 gene (≈10,701,955–10,702,263) (Fig. 1b). A fourth hypersensitive site was also identified within a CpG island between Tnp2 and the neighboring Socs1 gene (≈10,700,311–10,700,559). This had been previously identified in somatic cells (44).
Fig. 1.
Definition of the DNase I-sensitive region encompassing the mouse protamine gene cluster. (a) DNase I sensitivity was initially assayed in round spermatid nuclei at 12 sites throughout a 90-kb region of mouse chromosome 16qA1 encompassing the protamine cluster using real-time PCR (upper row of small red boxes; see supporting information (SI) Table 1 for oligonucleotide sequences). A second, higher-resolution analysis of a subregion included an additional 21 amplicons (lower row of small red boxes). (b) Data from 21 amplicons (red boxes) immediately encompassing the gene cluster from four independent experiments is shown as a function of genomic position (open circles). A running average of the median of the four points is plotted in blue, identifying an ≈11.6-kb DNase I-sensitive domain encompassing coordinates 10,700,711–10,712,326 (National Center for Biotechnology Information public mouse genome assembly Build 36.1). DNase I-hypersensitive sites within the region assayed are shown as peaks and are marked by arrows.
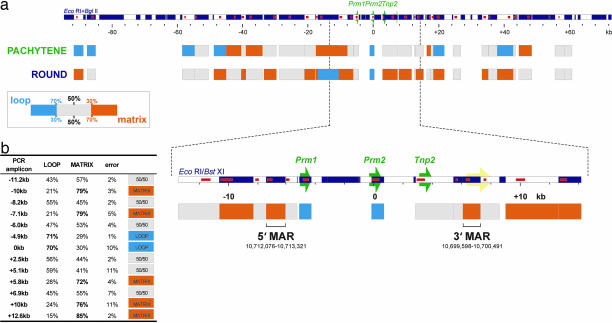
Nuclear matrix associations have been observed at the human cluster as the two sperm-specific MARs that flank the DNase I-sensitive domain. When the arrangement of the flanking MARs was experimentally altered in transgenic mice, the genes were subject to position effect. This has suggested that the MARs confer an insulative role on the domain (42, 45). To date, nuclear matrix association and the epigenetic modification of the mouse cluster have not been resolved. Accordingly, the nuclear matrix binding potential was assessed for 32 restriction fragments across an ≈142-kb region of mouse chromosome 16 encompassing the protamine domain as summarized in Fig. 2. Enriched populations of spermatogenic cells were treated with 2 M NaCl in the presence of DTT to extract nonnuclear matrix proteins. The resulting DNA loops were then cleaved with EcoRI and BglII, and then the matrix and loop DNAs were purified. The veracity of fractionation was confirmed by real-time PCR at four sites throughout the genome (SI Tables 1 and 2). An initial survey of earlier meiotic cell types did not show any significant nuclear matrix associations within this region (SI Table 3). However, seven restriction fragment-defined regions were attached to the pachytene spermatocyte nuclear matrix. Three were closely spaced indicating a large, ≈9.7-kb attachment, ≈2.6 kb upstream of Prm1 (Fig. 2a). Interestingly, a significant recruitment to the round spermatid nuclear matrix, both upstream and downstream of the protamine domain was observed, with 14 regions in total binding. The protamine domain itself remains within a small DNA loop between the bound regions. Mammalian sperm DNA loop sizes are in the range of 20–50 kb (46), and the data presented in Fig. 2 may suggest a periodicity. To address this issue, the periodicity of the observed strength of nuclear matrix association was modeled. Although a periodicity could be defined, the relatively small region of chromosome 16 examined (0.006%), when compared with whole-genome and its poor statistical confidence (r2 = 0.36), cautions our interpretation. This issue will likely be resolved upon whole chromosome analysis.
Fig. 2.
Nuclear matrix associations encompassing the mouse protamine domain. (a) Long-range nuclear matrix association of an ≈142-kb region encompassing the mouse protamine domain was assayed in pachytene primary spermatocytes (potentiated domain) and round spermatids (potentiated and transcribed domain). Nuclei were treated with 2 M NaCl, then restricted with EcoRI and BglII to release nonmatrix-associated, i.e., loop DNA. DNA was fractionated and then purified, and degree of nuclear matrix association for 32 restriction fragments was assayed by real-time PCR. Nuclear matrix binding potentials were calculated for the interrogated regions. Fragments that were highly enriched in the nuclear matrix-associated fraction (≥70% matrix/≤30% loop), termed matrix-attached, are shown in orange. Fragments that were highly enriched in the loop fraction (≥70% loop/≤30% matrix) are shown in cyan. All other regions, termed matrix-associated, are shown as gray boxes. A large ≈9.6-kb MAR upstream of the domain binds in pachytene spermatocytes. A considerable increase in the number of MARs was observed in round spermatids. (b) EcoRI and BstXI were used to resolve the round spermatid nuclear MARs. Nuclear matrix binding potentials for the 13 restriction fragments interrogated about the protamine domain are shown in the table (error displayed is one standard deviation of the mean for two independent experiments). Two restriction fragments, one upstream of Prm1 (coordinates 10,712,076–10,713,321) and another downstream of Tnp2 (10,699,598–10,700,491) demarcated the 5′ MAR and 3′ MAR, respectively. Genes of the protamine domain are indicated by green arrows, the neighboring Socs1 gene is indicated in yellow, restriction fragments are shown as alternating white and dark blue boxes, and PCR amplicons are indicated as red boxes within restriction maps.
To map the protamine domain nuclear matrix boundaries, DNA loops from round spermatid nuclear matrix preparations were cleaved with EcoRI and BstXI. Thirteen restriction fragments were interrogated across an ≈26-kb segment, spanning the protamine domain (SI Table 1), revealing four strong sites of nuclear matrix interaction. An ≈1.3-kb MAR (≈10,712,076–10,713,321) adjacent to the 5′ end of the DNase I-sensitive domain was identified as a single BstXI–BstXI fragment as shown in Fig. 2b. Similarly, a second BstXI–BstXI fragment ≈900 bp in size was present immediately 3′ of the domain (≈10,699,598–10,700,491). Thus, as defined by MAR and DNase I mapping, the mouse protamine domain exists as an ≈13.7-kb region of chromosome 16, spanning coordinates 10,699,598–10,713,321. Although more compact, the 5′-MAR→Prm1→Prm2→Tnp2→3′-MAR organization of the domain is identical to its human counterpart. Thus, recruitment of a large region several kilobases upstream of the domain that is detected in the pachytene spermatocytes is coincident with, but does not precede, potentiation. This supports the view that potentiation, i.e., the formation of an open chromatin structure that is requisite for expression, may engage but does not explicitly require nuclear matrix association.
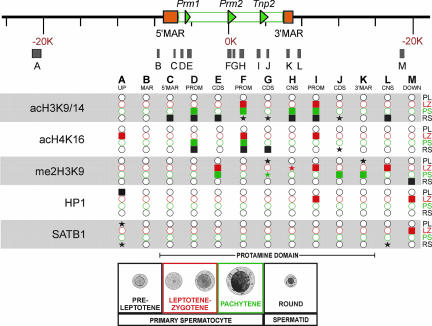
Epigenetic changes, together with key protein associations that mark both active and inactive chromatin, were assessed throughout meiotic and early spermiogenic stages of differentiation by using ChIP. ChIP-enriched templates from populations of preleptotene, leptotene–zygotene, and pachytene primary spermatocytes, as well as round spermatids, from two independent ChIP assays were isolated. A total of 13 regions encompassing the protamine domain (SI Table 1) were assessed in triplicate by real-time PCR (Fig. 3). These included two regions outside the domain (Fig. 3, regions A and M), the upstream pachytene MAR and the round spermatid 5′ MAR (regions B and C), the Prm1, Prm2, and Tnp2 genes (promoter and coding sequences, regions D and E, F and G, and I and J, respectively), as well as two conserved noncoding sequences (regions H and L) and the 3′ MAR (region K).
Fig. 3.
Epigenetic modification of the mouse protamine domain during spermatogenesis. Histone acetylation (acH3K9/14 and acH4K16), association of HP1 and SATB1, and histone methylation (me2H3K9) of the protamine domain were assessed by ChIP at 13 sites (A–M) in preleptotene (black; PL), leptotene–zygotene (red; LZ), and pachytene (green; PS) primary spermatocytes, as well as round spermatids (blue; RS). These included gene promoters (PROM: regions D, F, and I), coding sequences (CDS: regions E, G, and J), experimentally confirmed MARs (regions B, C, and K), CNS (regions H, and L). Regions A and M are upstream and downstream of the domain, respectively. Shaded boxes are those regions yielding a signal that was significantly greater than the preceding cell type in the spermatogenic pathway. Shaded stars represent a >4-fold increase compared with the preceding cell type. Open circles indicate regions with no significant association.
Acetylated histones H3 and H4 are epigenetic marks associated with active chromatin (47) that inhibit the formation of higher-order chromatin structures (48). Acetylation status of H3 at K9 and K14 and H4 at K16 were assessed throughout the protamine domain as summarized in Fig. 3. Low levels of H3 acetylation were established in the promoter regions at various meiotic stages (Fig. 3, regions D, F, and I). Coincident with transcription, levels of H3 acetylation increased in the promoter and coding regions in round spermatids (Fig. 3, regions D–G, I, and J). Similarly, several other regions, including the 5′ MAR (region C) as well as two conserved noncoding regions (region H, just downstream of Prm2, and region L, immediately downstream of the 3′ MAR), show a slight increase in H3 acetylation, indicating that, with transcription, the entire domain contains acetylated H3. In contrast, only marginal induction of H4 acetylation was noted at the promoter regions at various meiotic stages (Fig. 3, regions D, F, and I). The coding regions of Prm2 and Tnp2 showed a higher level of induction of H4 acetylation by the round spermatid stage (Fig. 3, regions G and J).
Coincident with transcription, acetylation of H3 and H4 increased in postmeiotic round spermatids at both promoter and coding sequences. This is consistent with many reports of hyperacetylated H3 localizing to transcriptionally active chromatin (reviewed in ref. 1). Curiously, one of the sites within the 5′ MAR showed H3 acetylation in round spermatids (Fig. 3), concomitant with nuclear matrix binding and transcription. The decondensed state marked by hyperacetylated chromatin may facilitate the interaction with the nuclear matrix that is known to harbor factors capable of modifying histones (49, 50). With the major changes in acetylation occurring in round spermatids, it follows that these modifications do not play a role in potentiating the protamine domain in pachytene spermatocytes.
The above modifications are positively associated with open, i.e., potentiated, chromatin. It is possible that the potentiation may be mediated by the termination of a negative signal (51, 52), as exemplified by the HP1 pathway, of which dimethyl (me2) H3K9 is an intermediate. Curiously, me2H3K9 was present, although highly variable, throughout most of this region of chromosome 16 during both expressing and nonexpressing stages of spermatogenic differentiation (Fig. 3). Regions G, J, and K within the domain, which were methylated in the meiotic stages, showed a decrease by the round spermatid stage. However, as shown in Fig. 3, the domain was not bound by HP1 α, β, or γ. Only region A and the promoter region of Tnp2 (region I), together with region M, exhibited transient associations with HP1, in the preleptotene and leptotene–zygotene spermatocyte stages, respectively.
Two well studied examples of MAR-binding proteins are SATB1 and Top2. SATB1 is a phosphorylation state-dependent regulator (16) of T cell and erythrocyte differentiation. It associates with MARs of several loci active in those pathways and then targets chromatin modifiers to them (14). Interestingly, Satb1-null mice exhibit spermatogenic arrest. As shown in Fig. 3, SATB1 was not associated with the protamine domain in any cell type. However, region A, ≈13 kb upstream of the domain, and region L, a conserved noncoding sequence (CNS), immediately downstream of the 3′ MAR, were associated in round spermatids. It is unlikely that it regulates the protamine domain because SAT proteins are more closely associated with the genes they regulate (16, 17, 53, 54). Perhaps SATB1 plays a role in suppressing germ cell expression of Socs1.
Another group of nuclear matrix proteins (19, 55) that have been implicated in modifying DNA topology in chromatin are the topoisomerases. Examples of topoisomerase II function include their involvement in histone-to-protamine exchange during spermiogenesis (56, 57), postfertilization genomic restructuring (58, 59), and chemotherapeutically induced DNA-damage repair mechanisms (60, 61). Two recent reports have implicated Top2b in transcription. The first shows direct involvement of Top2b in creating transient double-stranded DNA breaks in the promoter regions of several hormone-responsive genes (36). Top2b becomes part of an activating complex that includes polyADP-ribose polymerase 1 and other factors typically associated with DNA repair that targets hormone response elements within gene promoters. Ultimately this complex exchanges histone H1 for a high-mobility group protein on the nucleosome associating with that promoter, thus permitting transcription. In the second, Top2b-null mouse embryos show a marked change in the expression of almost one-third of developmentally regulated genes, mediated through the promoter (37). This suggested that Top2b regulation is involved in the alteration of the local chromatin structure during the initiation or elongation phases of transcription. Given its association to the nuclear matrix (19, 55), catalytic activity (21, 22), association with several chromatin-modifying elements (15), and a new direct link to transcriptional regulation, Top2b presents itself as an excellent chromatin potentiation candidate.
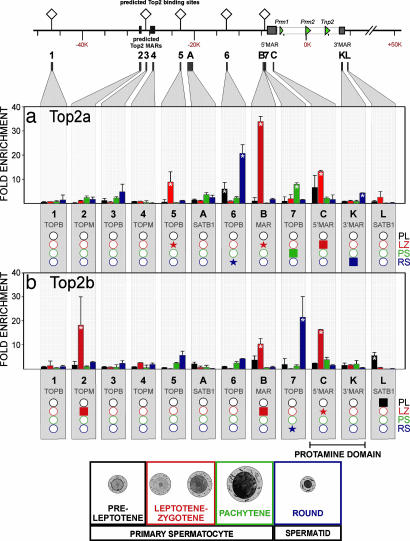
Five Top2–DNA consensus binding sites were identified in silico within a 100-kb region centered on the mouse protamine cluster as summarized in Fig. 4. Two additional clusters of Top2–DNA binding sites were identified by MAR-WIZ. The association of Top2 with the protamine domain throughout spermatogenesis was assessed by using ChIP. These sites were labeled sequentially 1–7 (Fig. 4 and SI Table 1). Site 1 is located ≈39 kb upstream of the domain, and site 7 is within the proximal end of the 5′ MAR. In addition, MARs (regions B, C, and K) and SATB1-associated regions (A and L) were examined. As shown in Fig. 4, Top2 was not associated with in silico-predicted sites 1, 3, and 4, as well as region A, in any cell type. In contrast, Top2a was strongly associated with site 6 in preleptotene spermatocytes, site 5 and regions B and C in leptotene–zygotene spermatocytes, site 7 in pachytene spermatocytes, and site 6 and region K in round spermatids. Similarly, Top2b was strongly associated with site L in preleptotene spermatocytes, site 2 and regions B and C in leptotene–zygotene spermatocytes, and site 7 in round spermatids. Interestingly, site 7 and regions B and C all interrogate the 5′ MAR. Together, these sites represent the highest concentration of topoisomerase II activity within the protamine domain.
Fig. 4.
Association of topoisomerase II with the mouse protamine domain during spermatogenesis. The binding of topoisomerase II α (Top2a) and β (Top2b) to the mouse protamine domain was assessed by ChIP at 12 sites (1–7, regions A, B, K, and L) in preleptotene, leptotene–zygotene, and pachytene primary spermatocytes as well as round spermatids. Sites included predicted Top2 binding sites (TOPB: 1, 3, and 5–7), predicted Top2 MARs (TOPM: 2 and 4), and experimentally confirmed MARs (regions B, C, and K), as well as sites flanking the domain that showed association with SATB1 in round spermatids (regions A and L). The region encompassing the 5′ MAR exhibits differential Top2a (a) and Top2b (b) associations throughout spermatogenesis. Values presented are means, and error bars represent the 95% confidence intervals. Asterisks denote a significant increase (P < 0.05) in that modification compared with the preceding cell types in the spermatogenic pathway. Below each panel, the data are summarized with shaded boxes representing those regions yielding a signal that was significantly greater than the preceding cell type in the spermatogenic pathway. Shaded stars represent a >4-fold increase compared with the preceding cell type. Open circles indicate regions with no significant association.
Acting as the more proficient “relaxase” of nucleosomal DNA (62), Top2b has been shown to directly interact with numerous gene promoters (37, 63). Of the various predicted topoisomerase II binding sites, MARs, and CNSs interrogated, the 5′ MAR showed the greatest level of association. Beginning with strong and pervasive interactions as early as leptotene–zygotene spermatocytes, transient interactions persist through the pachytene stage to culminate in a marked increase in round spermatids (Fig. 4). The significant associations detected in prepachytene meiotic stages suggest that modifiers of DNA topology are part of a potentiative mechanism marking the locus for decondensation to a DNase I-sensitive, open chromatin conformation.
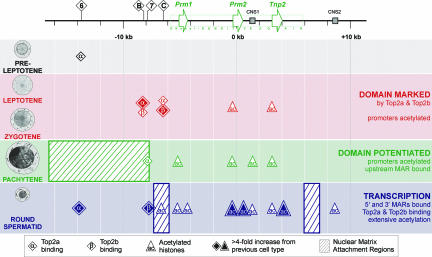
The results reported in this article underscore the elegant molecular event, potentiation, i.e., the opening of a chromatin domain, that underlies transcription. As summarized and presented in the model illustrated in Fig. 5, this is clearly referenced by a dynamic nuclear matrix network. Transient Top2a activity is first detected at site 6, ≈10 kb from the protamine domain in preleptotene spermatocytes. The function of this mark, if it is associated with this mechanism, is not clear. This is followed by a rather significant burst of Top2a and Top2b activity in leptotene–zygotene spermatocytes at regions B and C, marking region B for nuclear matrix attachment. Curiously, even though the domain still exists in a nuclease-insensitive state (41), the promoter regions of Prm2 and Tnp2 show low levels of histone H3K9/K14 acetylation. As the cell differentiates to the pachytene spermatocyte, when the potentiated conformation is adopted, a large region upstream of B is recruited to the nuclear matrix and its 3′ end is marked by Top2a. This is consistent with the view that cleavage by Top2a and Top2b marks the region for both matrix attachment and potentiation. Upon potentiation, acetylation of the constituent promoter elements and CNS (Fig. 5, CNS1) elements takes place and the preceding Top2a and Top2b marks at B and C are lost. Curiously, this mechanism repeats: Top2a activity is lost, and Top2b repositions the 5′ MAR upstream to demarcate the round spermatid domain along with the utilization of site 6 Top2a. This occurs coincidentally with the binding of the 3′ MAR in round spermatids immediately downstream of the DNase I-sensitive domain. Large inductions of acetylation throughout the domain are evident in the round spermatid. It is tempting to speculate that a mechanism similar to that reported for promoter cleavage by Top2b (36) is functional at the domain level. In this case the 5′ MAR region serves as a site of activity that mediates the decondensation of the adjacent chromatin domain.
Fig. 5.
The molecular events that precede transcription of the protamine domain. Before acquiring a potentiated conformation in the pachytene spermatocyte, the mouse protamine domain is marked at upstream sites B and C by Top2a and Top2b in leptotene and zygotene spermatocytes. The promoters of Prm2 and Tnp2 are marked by histone (H3K9/14) acetylation. Coincident with the potentiation of the domain, a large region immediately upstream of the Top2 cleavage sites of the gene cluster is recruited to the nuclear matrix. The promoters of all three genes and CNS1 within the domain are acetylated. A single site (site 7) of Top2a activity was detected toward the proximal end of this ≈9.6-kb MAR in pachytene spermatocytes. The genes of the protamine domain are transcribed postmeiotically in round spermatids. At this time, two MARs that flank the domain bind the nuclear matrix in conjunction with extensive histone acetylation throughout the domain and a single upstream CNS2. Significant induction of Top2a site 6 and Top2b site 7 activity were detected upstream of the domain. Top2 functions in concert with other nuclear matrix factors to mark this multigenic domain for potentiation and eventual transcription.
While the relationship among nuclear matrix association, Top2 activity, and potentiation is evolving, MAR deletions encompassing the Top2 sites significantly down-regulate protamine expression (42, 45). The data presented here suggest that midmeiotic associations of Top2a and Top2b in regions that localize near the mouse 5′ MAR constitute part of a topoisomerase–nuclear matrix association-dependent potentiative mechanism that results in a DNase I-sensitive protamine domain by the pachytene spermatocyte stage in preparation for postmeiotic transcription. It is tempting to analogize these interactions acting over a multigene domain to the topoisomerase-based rearrangements reported at a number of promoter regions (37, 63). The model outlined likely represents part of a larger mechanism that reorganizes chromatin in a manner that would guide cell fate.
Methods
Prediction of Top2–DNA consensus binding sites was carried out by using DS Gene (Accelrys, San Diego, CA) and MAR-WIZ (64). Pachytene stage primary spermatocytes and round and elongating spermatids were isolated from adult C57BL/6 males, whereas preleptotene, a mixed pool of leptotene and zygotene, and early pachytene spermatocytes were isolated from 18-day-old juveniles by unit gravity sedimentation (65, 66). After separation, cells were either immediately cross-linked for ChIP or stabilized in FSB (67) and then stored in liquid nitrogen.
Nuclease sensitivity was assayed as detailed in SI Text. PCR was carried out by using the HotStarTaq polymerase system (Qiagen, Valencia, CA) together with the Chromo4 real-time PCR detection system (Bio-Rad, Hercules, CA). Nuclear matrix association was assessed by using 2 M NaCl extraction (68) along with restriction digestion. ChIP was carried out with EZ-ChIP essentially as described by the manufacturer, using normal mouse IgG (background control), anti-acetyl-H3K9/K14, anti-acetyl-H4K16, anti-HP1 (recognizes α, β, and γ isoforms), anti-SatB1, anti-dimethyl H3K9, anti-Top2a, and anti-Top2b antibodies. Detailed methods and reagent information are available in SI Text.
Supplementary Material
Acknowledgments
The contributions of Adrian E. Platts to data analysis are gratefully acknowledged. This work was supported by National Institute of Child Health and Human Development Grant HD36512 (to S.A.K.) and the Department of Obstetrics and Gynecology of Wayne State University (S.A.K.).
Abbreviations
- MAR
matrix attachment region
- CNS
conserved noncoding sequence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700076104/DC1.
References
- 1.Martins RP, Krawetz SA. Gene Ther Mol Biol. 2005;9:229–246. [PMC free article] [PubMed] [Google Scholar]
- 2.Choudhary SK, Wykes SM, Kramer JA, Mohamed AN, Koppitch F, Nelson JE, Krawetz SA. J Biol Chem. 1995;270:8755–8762. doi: 10.1074/jbc.270.15.8755. [DOI] [PubMed] [Google Scholar]
- 3.Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Development (Cambridge, UK) 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 4.Chambeyron S, Bickmore WA. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keene MA, Corces V, Lowenhaupt K, Elgin SCR. Eur J Cell Biol. 1980;22:95–95. [Google Scholar]
- 6.Weintraub H, Groudine M. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 7.Lawson GM, Knoll BJ, March CJ, Woo SL, Tsai MJ, O'Malley BW. J Biol Chem. 1982;257:1501–1507. [PubMed] [Google Scholar]
- 8.Weintraub H. Cell. 1985;42:705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- 9.Rose SM, Garrard WT. J Biol Chem. 1984;259:8534–8544. [PubMed] [Google Scholar]
- 10.Gross DS, Garrard WT. Trends Biol Sci. 1987;12:293–297. [Google Scholar]
- 11.Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vickers MA, Vyas P, Harris PC, Simmons DL, Higgs DR. Proc Natl Acad Sci USA. 1993;90:3437–3441. doi: 10.1073/pnas.90.8.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stauffer DR, Howard TL, Nyun T, Hollenberg SM. J Cell Sci. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- 14.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 15.Tsai SC, Valkov N, Yang WM, Gump J, Sullivan D, Seto E. Nat Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 16.Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Mol Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- 18.Wen J, Huang S, Rogers H, Dickinson LA, Kohwi-Shigematsu T, Noguchi CT. Blood. 2005;105:3330–3339. doi: 10.1182/blood-2004-08-2988. [DOI] [PubMed] [Google Scholar]
- 19.Berrios M, Osheroff N, Fisher PA. Proc Natl Acad Sci USA. 1985;82:4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulikas T. Int Rev Cytol. 1995;162A:279–388. doi: 10.1016/s0074-7696(08)61234-6. [DOI] [PubMed] [Google Scholar]
- 21.Austin CA, Marsh KL. BioEssays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell A, Costenaro L, Mitelheiser S, Bates AD. Biochem Soc Trans. 2005;33:1460–1464. doi: 10.1042/BST0331460. [DOI] [PubMed] [Google Scholar]
- 23.Clarke DJ, Vas AC, Andrews CA, Diaz-Martinez LA, Gimenez-Abian JF. Cell Cycle. 2006;5:1925–1928. doi: 10.4161/cc.5.17.3200. [DOI] [PubMed] [Google Scholar]
- 24.Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 25.Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 26.Wood ER, Earnshaw WC. J Cell Biol. 1990;111:2839–2850. doi: 10.1083/jcb.111.6.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Handel MA, Cobb J, Eaker S. J Exp Zool. 1999;285:243–250. [PubMed] [Google Scholar]
- 28.Attia SM, Schmid TE, Badary OA, Hamada FM, Adler ID. Mutat Res. 2002;520:1–13. doi: 10.1016/s1383-5718(02)00079-7. [DOI] [PubMed] [Google Scholar]
- 29.Paliulis LV, Nicklas RB. BioEssays. 2003;25:309–312. doi: 10.1002/bies.10265. [DOI] [PubMed] [Google Scholar]
- 30.Sukhacheva TV, Bogush TA, Kolomiets OL. Bull Exp Biol Med. 2003;135:464–469. doi: 10.1023/a:1024919510096. [DOI] [PubMed] [Google Scholar]
- 31.Iwabata K, Koshiyama A, Yamaguchi T, Sugawara H, Hamada FN, Namekawa SH, Ishii S, Ishizaki T, Chiku H, Nara T, et al. Nucleic Acids Res. 2005;33:5809–5818. doi: 10.1093/nar/gki883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakaguchi A, Kikuchi A. J Cell Sci. 2004;117:1047–1054. doi: 10.1242/jcs.00977. [DOI] [PubMed] [Google Scholar]
- 33.Heck MM, Hittelman WN, Earnshaw WC. Proc Natl Acad Sci USA. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gimenez-Abian JF, Clarke DJ, Mullinger AM, Downes CS, Johnson RT. J Cell Biol. 1995;131:7–17. doi: 10.1083/jcb.131.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhat MA, Philp AV, Glover DM, Bellen HJ. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- 36.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 37.Lyu YL, Lin CP, Azarova AM, Cai L, Wang JC, Liu LF. Mol Cell Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeRoy G, Loyola A, Lane WS, Reinberg D. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 39.Johnson CA, Padget K, Austin CA, Turner BM. J Biol Chem. 2001;276:4539–4542. doi: 10.1074/jbc.C000824200. [DOI] [PubMed] [Google Scholar]
- 40.Nelson JE, Krawetz SA. DNA Seq. 1995;5:163–168. doi: 10.3109/10425179509029356. [DOI] [PubMed] [Google Scholar]
- 41.Kramer JA, McCarrey JR, Djakiew D, Krawetz SA. Development (Cambridge, UK) 1998;125:4749–4755. doi: 10.1242/dev.125.23.4749. [DOI] [PubMed] [Google Scholar]
- 42.Martins RP, Ostermeier GC, Krawetz SA. J Biol Chem. 2004;279:51862–51868. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- 43.Kramer JA, McCarrey JR, Djakiew D, Krawetz SA. Mol Reprod Dev. 2000;56:254–258. doi: 10.1002/(SICI)1098-2795(200006)56:2+<254::AID-MRD9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 44.Choi YC, Aizawa A, Hecht NB. Mamm Genome. 1997;8:317–323. doi: 10.1007/s003359900431. [DOI] [PubMed] [Google Scholar]
- 45.Kramer JA, Zhang S, Yaron Y, Zhao Y, Krawetz SA. Genet Test. 1997;1:125–129. doi: 10.1089/gte.1997.1.125. [DOI] [PubMed] [Google Scholar]
- 46.Ward WS, Partin AW, Coffey DS. Chromosoma. 1989;98:153–159. doi: 10.1007/BF00329678. [DOI] [PubMed] [Google Scholar]
- 47.Khorasanizadeh S. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 48.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 49.Sun JM, Chen HY, Davie JR. J Biol Chem. 2001;276:49435–49442. doi: 10.1074/jbc.M108364200. [DOI] [PubMed] [Google Scholar]
- 50.Ishihara SL, Morohashi K. Biochem Biophys Res Commun. 2005;329:554–562. doi: 10.1016/j.bbrc.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. Nat Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 53.Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 54.Seo J, Lozano MM, Dudley JP. J Biol Chem. 2005;280:24600–24609. doi: 10.1074/jbc.M414076200. [DOI] [PubMed] [Google Scholar]
- 55.Cockerill PN, Garrard WT. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 56.McPherson S, Longo FJ. Eur J Histochem. 1993;37:109–128. [PubMed] [Google Scholar]
- 57.Marcon L, Boissonneault G. Biol Reprod. 2004;70:910–918. doi: 10.1095/biolreprod.103.022541. [DOI] [PubMed] [Google Scholar]
- 58.St Pierre J, Wright DJ, Rowe TC, Wright SJ. Mol Reprod Dev. 2002;61:347–357. doi: 10.1002/mrd.10015. [DOI] [PubMed] [Google Scholar]
- 59.Shaman JA, Prisztoka R, Ward WS. Biol Reprod. 2006;75:741–748. doi: 10.1095/biolreprod.106.055178. [DOI] [PubMed] [Google Scholar]
- 60.Kantidze OL, Iarovaia OV, Razin SV. J Cell Physiol. 2006;207:660–667. doi: 10.1002/jcp.20597. [DOI] [PubMed] [Google Scholar]
- 61.Emmons M, Boulware D, Sullivan DM, Hazlehurst LA. Biochem Pharmacol. 2006;72:11–18. doi: 10.1016/j.bcp.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 62.Salceda J, Fernandez X, Roca J. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lis JT, Kraus WL. Cell. 2006;125:1225–1227. doi: 10.1016/j.cell.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 64.Kramer JA, Singh GB, Krawetz SA. Genomics. 1996;33:305–308. doi: 10.1006/geno.1996.0198. [DOI] [PubMed] [Google Scholar]
- 65.Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wykes SM, Krawetz SA. Mol Biotechnol. 2003;25:131–138. doi: 10.1385/MB:25:2:131. [DOI] [PubMed] [Google Scholar]
- 67.Kramer JA, Krawetz SA. BioTechniques. 1997;22:826–828. doi: 10.2144/97225bm07. [DOI] [PubMed] [Google Scholar]
- 68.Ostermeier GC, Liu Z, Martins RP, Bharadwaj RR, Ellis J, Draghici S, Krawetz SA. Nucleic Acids Res. 2003;31:3257–3266. doi: 10.1093/nar/gkg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.