Abstract
Activation-induced cytidine deaminase (AID) is a B cell enzyme essential for Ig somatic hypermutation and class switch recombination. AID acts on ssDNA, and switch regions of Ig genes, a target of AID, form R-loops that contain ssDNA. Nevertheless, how AID action is specifically targeted to particular DNA sequences is not clear. Because mutations altering cotranscriptional messenger ribonucleoprotein (mRNP) formation such as those in THO/TREX in yeast promote R-loops, we investigated whether the cotranscriptional assembly of mRNPs could affect AID targeting. Here we show that AID action is transcription-dependent in yeast and that strong and transcription-dependent hypermutation and hyperrecombination are induced by AID if cells are deprived of THO. In these strains AID-induced mutations occurred preferentially at WRC motifs in the nontranscribed DNA strand. We propose that a suboptimal cotranscriptional mRNP assembly at particular DNA regions could play an important role in Ig diversification and genome dynamics.
Keywords: hyperrecombination, hypermutation, messenger ribonucleoprotein biogenesis, R-loops
Activation-induced cytidine deaminase (AID) is a specific B cell enzyme believed to be responsible for the initiation of somatic hypermutation (SHM) and class switch recombination (CSR) during B cell differentiation (1–3). Many studies have shown that AID acts directly on DNA (4), the natural target for AID action in the variable and switch (S) regions for SHM and CSR, respectively. The specific mechanisms of AID function are still unclear, but evidence suggests that the preferential target of AID may be ssDNA (5, 6). Thus, in vitro experiments have shown that AID deaminates ssDNA and dsDNA that is transcribed (5–7), and transcription has been shown to be required for both SHM and CSR in vivo (8, 9).
Studies have shown a strand preference for the action of AID during in vitro transcription with AID-induced mutations detected preferentially in the nontranscribed (NT) strand (6, 10–12). Analysis of the products of SHM in B cells, however, reveals that both DNA strands are mutated (13). Other in vitro studies have shown that AID can deaminate both strands within regions of supercoiled DNA (14) and that the ssDNA-binding replication protein A is required for deamination of SHM targets (15). All of these findings are consistent with the idea that transient formation of ssDNA during transcription facilitates AID action.
Along the same line, formation of R-loops during transcription of the S regions has been proposed (16). Such R-loops would possibly provide AID with ssDNA substrates at its target region because there the transcribed (T) strand hybridizes with the nascent RNA and the NT strand is present as ssDNA. Because of its high G content, the NT DNA strand at S regions could be stabilized by the formation of parallel four-stranded G quartets (17, 18). Indeed, AID appears to bind specifically to G-loops within transcribed S regions (19) and can deaminate the displaced strand of a transcription-induced R-loop in vitro (20).
Interestingly, cotranscriptional messenger ribonucleoprotein (mRNP) biogenesis is an essential step in gene expression that may influence genetic integrity (21, 22). In the yeast Saccharomyces cerevisiae, hyperrecombination can be triggered by transcription in mutants depleted of THO/TREX, a conserved protein complex that functions at the interface between transcription and mRNA export (23, 24). THO is a conserved complex first identified in yeast and formed by Tho2, Hpr1, Mft1, and Thp2 (24). It interacts with the Sub2 RNA-dependent ATPase and the Yra1 RNA-binding protein to form the TREX complex (23). THO has been shown to be required for the transcription of long genes and genes containing either high GC content or multiple internal repeats (25, 26). It is believed that THO controls the cotranscriptional formation of export-competent mRNP during transcription elongation by controlling the assembly of heterogeneous nuclear ribonucleoproteins onto the nascent mRNA (21). Hyperrecombination of THO mutants is particularly evident in long and GC-rich DNA sequences such as the bacterial lacZ gene (26). Because hyperrecombination depends on the formation of DNA:RNA hybrids (22), it has been proposed that one function of THO/TREX is to prevent the nascent mRNA from interacting with the template DNA to form R-loops. The observation that the ASF/SF2 splicing factor has a similar role in chicken DT40 and HeLa cells (27) suggests that a number of RNA processing enzymes have an additional role in preventing R-loop formation.
One important and unresolved question is why AID is targeted at specific DNA regions during B cell differentiation, and we wondered whether this could be because of suboptimal mRNP formation. To test this possibility, we confirmed that AID can induce hypermutation and hyperrecombination at low levels in yeast cells (28, 29) and demonstrated additionally that this instability occurs in a transcription-dependent manner. Using yeast THO mutants as a way to generate suboptimal mRNP formation, we found that both hypermutation and hyperrecombination were strongly and synergistically increased in a transcription-dependent manner, with mutations primarily occurring in the NT strand. We propose that a suboptimal cotranscriptional mRNP assembly at particular DNA regions can regulate AID action, and we suggest that the control of mRNP biogenesis may play an important role in Ig diversification and genome dynamics.
Results
Mutations in the THO Complex Confer a Slight Transcription-Dependent Hypermutation Phenotype.
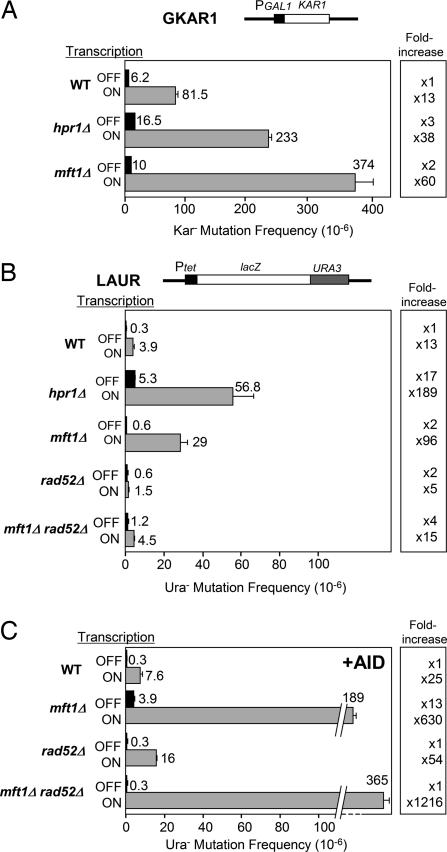
Because THO mutants share phenotypes of recombination-mediated genetic instability associated with transcription, it was important to know whether they showed, in addition to hyperrecombination, a transcription-dependent hypermutation phenotype. Two different plasmid-borne assays were developed for the analysis of forward mutations. The first assay is based on a fusion of the KAR1 ORF to the regulated GAL1 promoter (GKAR1 system). Because KAR1 overexpression is lethal in yeast (30), kar1 mutations can be selected on galactose medium. The second assay is based on a lacZ::URA3 translational fusion under control of the regulated Tet promoter (LAUR system) (31). In this assay, Ura− mutants are selected on synthetic complete (SC) medium supplemented with 5-fluoroorotic acid, whereas lacZ− mutants and lacZ+ are scored by color on SC plus X-Gal. As can be seen in Fig. 1 A and B, the mutation frequencies were increased 13-fold when transcription was high in both systems. Transcription-associated mutation (TAM) was also observed in rad52Δ cells although to a minor extent (Fig. 1B). The result is in agreement with previous reports indicating that transcription increases mutation in yeast cells (32) and Escherichia coli (reviewed in ref. 33).
Fig. 1.
Effect of transcription and AID on the frequency of mutation in S. cerevisiae wild-type and THO mutant strains. (A) Frequency of Kar− mutants obtained in the GKAR1 system. (B) Frequency of Ura− mutants obtained in the LAUR system and Rad52 dependency. (C) Effect of AID on the frequency of Ura− mutants in the LAUR system in wild-type, mft1Δ, rad52Δ, and mft1Δ rad52Δ strains. All experiments were performed under high (ON) and low (OFF) transcription. Diagrams of systems are shown above each graph. Mean mutation frequency and standard deviation of three to four different fluctuation tests are plotted.
Next, we used both the LAUR and GKAR1 assays to determine whether different THO mutations stimulated TAM. As seen in Fig. 1A, mft1Δ and hpr1Δ, which caused an elimination of the THO complex in yeast cells (34), increased the frequency of mutation 2- to 3-fold in GKAR1 at low transcription versus 38- to 60-fold at high transcription levels. Similarly, in the LAUR system, the increase in mutation frequency was significantly stronger for high transcription (96- to 189-fold) than for low transcription (2- to 17-fold) as assayed in both the mft1Δ and hpr1Δ backgrounds (Fig. 1B). Therefore, THO mutants share a transcription-dependent hypermutation phenotype that is not as strong as its hyperrecombination phenotype observed in several direct-repeat assays (26). Because all null mutations of the genes encoding the different THO subunits lead to deprivation of THO (34) and the same phenotypes (24), we decided to continue our analysis with mft1Δ, which, in contrast to hpr1Δ, has little impact on growth.
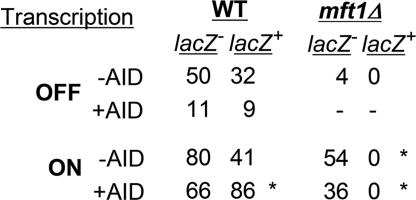
To gain insight into the molecular basis of mutations caused by THO mutations, we performed a genetic analysis of the pattern of mutations caused by mft1Δ. As expected from the fact that transcription through lacZ is the most sensitive step in THO mutants (26), every Ura− mutant was also lacZ− in all conditions tested, implying that all mutations mapped within lacZ (54 in high transcription and four in low transcription) (Fig. 2), whereas, in the wild type, 34–40% of the Ura− mutations were lacZ+ (lacZ−:lacZ+ ratios of 50:32 and 80:41 in low and high transcription, respectively) (Fig. 2); that is, they occurred within URA3. The mft1Δ mutations must, therefore, be either frameshifts or premature stop codons within lacZ or DNA rearrangements covering lacZ and URA3. Given the strong transcription-dependent hyperrecombination phenotype of THO mutants, it was important to determine whether lacZ− Ura− colonies in the mft1Δ background arose by recombination-dependent rearrangements, which are known to be Rad52-dependent. The spontaneous mutation frequency under high transcription in the rad52Δ strain was 2-fold lower than in the wild type (Fig. 1B), although it has been reported that TAM increases in rad52Δ cells (32, 35), indicating that some of the TAM events observed in the wild type may arise by a Rad52-dependent pathway in the LAUR construct. The rad52Δ mutation causes a decline in the mutation frequency of mft1Δ cells of 6.5-fold (from 2.9 × 10−5 to 4.5 × 10−6), indicating that most of TAM events in mft1Δ occur by RAD52-dependent rearrangements rather than by point mutations (Fig. 1B). In contrast to wild-type cells in which none of eight analyzed Ura− mutants contained detectable rearrangements, 62% of the mutants (23 of 37) contained rearrangements in mft1Δ [supporting information (SI) Fig. 5]. Thus, the absence of THO stimulates Rad52-independent mutations 2- to 3-fold (from 0.6 × 10−6 to 1.2 × 10−6 and from 1.5 × 10−6 to 4.5 × 10−6 in low and high transcription, respectively) (Fig. 1B). Indeed, 50% of Ura− mutations are lacZ+ in the mft1Δ rad52Δ mutant (7:7 lacZ−:lacZ+ ratio; data not shown), consistent with the conclusion that THO mutants also stimulate point mutations, although slightly, in a transcription-dependent manner.
Fig. 2.
Genetic analysis of spontaneous (−AID) and AID-induced (+AID) mutations. Distribution of lacZ−:lacZ+ mutations among Ura− mutants in wild-type and mft1Δ strains is shown. An asterisk indicates a statistically significant difference (P < 0.05) with respect to the wild-type value under the same transcription conditions, as determined by contingency χ2 analysis.
AID-Induced Mutation Is Transcription-Dependent in Yeast.
Taking advantage of the LAUR assay, we studied the ability of AID to induce mutation and whether this ability depended on transcription in wild-type cells. As can be seen in Fig. 1 B and C, AID expression in wild-type yeast did not lead to hypermutation at low transcription, but caused a 2-fold increase at high transcription (from 3.9 × 10−6 to 7.6 × 10−6). Consistent with a lack of an AID effect on mutation frequency under low transcription in the wild type, the ratio of lacZ−:lacZ+ among Ura− mutants was similar regardless of whether cells expressed AID (11:9 versus 50:32) (Fig. 2). However, a significant change (P < 0.05) in this ratio was observed under high transcription if AID was expressed (66:86 versus 80:41), indicating that the pattern of mutations was changed by AID in a transcription-dependent manner. Therefore, AID induces a weak transcriptional-dependent increase in the frequency of mutation and a change in the pattern of mutations.
AID-Induced Mutation Is Strongly Stimulated by THO Mutations.
THO mutations may create local regions that are susceptible to forming recombinogenic and mutagenic intermediates in a transcription-dependent manner. To determine whether such regions would be more susceptible to AID action, we tested the effect of AID on the frequency and pattern of mutations in mft1Δ cells. As can be seen in Fig. 1C, in mft1Δ cells the effect of AID on the frequency of mutations was greater than in the wild type, leading to a 13-fold increase in mutations at low transcription that went up to 630-fold at high transcription (Fig. 1C). Under low and high transcription, AID stimulated the mutation frequency 6.5 times (from 0.6 × 10−6 to 3.9 × 10−6 and from 2.9 × 10−5 to 1.9 × 10−4, respectively) in mft1Δ cells. Importantly, this hypermutator effect of AID was not dependent on Rad52. AID expression further increased Ura− mutations 81-fold (from 4.5 × 10−6 to 3.7 × 10−4) in mft1Δ rad52Δ cells under high transcription (Fig. 1 B and C), indicating that they are not due to rearrangements but most likely point mutations.
THO mutations enhance AID action presumably by creating a local structure in the transcribed DNA region that works as a target for AID. Therefore, we would expect that nonframeshift point mutations mapping at lacZ and conferring a lacZ− but not Ura− phenotype would occur at a high frequency in mft1Δ but not in wild-type cells expressing AID. As expected, we were not able to detect lacZ− Ura+ mutants from 5,922 colonies in wild-type cells expressing AID (frequency <10−3), whereas in mft1Δ expressing AID we counted 47 lacZ− Ura+ mutants of 4,692 colonies (frequency of 1 × 10−2) (SI Table 2). This result indicates that the frequency of Ura− AID-induced mutations in mft1Δ was underestimated in the LAUR system and indicates a strong effect of THO mutations on AID action. Therefore, transcription and AID result in a strong hypermutator phenotype in mft1Δ cells.
AID Mutates Preferentially the NT Strand in mft1Δ but Shows No Strand Preference in the Wild Type.
AID acts preferentially at WRC (GYW in the opposite strand) motifs (11) that are part of the WRCY motifs (RGYW in the opposite strand) found as hotspots of SHM (36) (where W is A or T, R is A or G, and Y is C or T). As R-loops in THO mutants (19) leave ssDNA opposite the DNA:RNA hybrid, one prediction is that most of the AID-induced mutations in mft1Δ cells should be at WRC motifs and in the NT strand. Therefore, we sequenced spontaneous and AID-induced mutations in wild-type and mft1Δ cells. As shown in Fig. 2, all mutants in mft1Δ cells were lacZ− Ura−, which can be explained if the Ura− phenotype results from frameshift mutations, stop codons, or rearrangements occurring at lacZ. This view is consistent with the fact that transcription through lacZ is the most sensitive step in THO mutants (26). Therefore, because point mutations at URA3 are not detected and lacZ− Ura− mutants represent a biased class of mutations at lacZ, we decided to sequence the lacZ sequence of the lacZ− Ura+ mutants that arose in mft1Δ cells expressing AID under high transcription conditions. The URA3 ORF from 19 and 53 independent lacZ+ Ura− mutants obtained from the wild type with and without AID overexpression, respectively, was also sequenced (SI Fig. 6 A and B). Results are summarized in Table 1 and SI Table 3. In wild-type cells not expressing AID <60% (11 of 19) were point mutations and only 9% of these (1 of 11) were within the WRC/GYW target motif of AID. In contrast, in the wild type and mft1Δ expressing AID, the percentage of point mutations increased to 86% (46 of 53) and 91% (22 of 24), respectively. Of these, 65% (30 of 46) and 54% (12 of 22), respectively, occurred at the WRC/GYW motif. Altogether, the data indicate that AID has a preferential function as a cytosine deaminase at the WRC/GYW motif, consistent with the preferential target of AID deamination observed in vitro (11).
Table 1.
Spontaneous (−AID) and AID-induced (+AID) base substitutions in the wild type (lacZ+ Ura−) and mft1Δ mutants (lacZ− Ura+) classified according to different sequence features
| Sequence features | WT | WT + AID | mft1Δ + AID |
|---|---|---|---|
| Mutations at C | 0 | 16 | 16 |
| Mutations at G | 7 | 21 | 5 |
| Mutations at WRC | 0 | 15 | 10 |
| Mutations at GYW | 1 | 15 | 2 |
| Transversions | 11 | 35 | 13 |
| Transitions | 0 | 11 | 9 |
| Point mutations | 11 | 46 | 22 |
| Total mutations | 19 | 53 | 24 |
DNA sequences are shown in SI Fig. 6 A and B.
As AID deaminates C within ssDNA, mutations occurring at C should reflect the action of AID directly on the NT strand, whereas mutations at G reflect the action of AID on C in the T strand. Therefore, we analyzed the NT:T ratio of AID-induced mutations in both strains. Whereas in the wild type expressing AID the NT:T ratio of mutations was 0.75:1 (16:21), this ratio increased significantly (P < 0.05) to 3:1 (16:5) in mft1Δ. Importantly, from these mutations, the WRC AID-target motif was mutated at an NT:T ratio of 1:1 in the wild type (15:15) and 5:1 in mft1Δ (10:2). The lack of strand preference of the mutation pattern produced by AID in the wild type was not observed previously (29) and is consistent with the transient opening of DNA strands by transcription presumably promoted by negative supercoiling. By contrast, the putative impairment of mRNP biogenesis caused by mft1Δ promotes a more stable formation of ssDNA on the NT strand and makes it an appropriate target for the action of human AID. The transversions:transitions ratio also decreased from 11:0 in the wild type to 35:11 and 13:9 in wild-type and mft1Δ cells expressing AID, respectively (Table 1). The increase in transitions is consistent with the direct action of AID in deaminating C to U, which leads to a C-to-T transition (G to A in the opposite strand). Consistently, the changes produced by AID were more evident for C (or G) with a strong bias for C in mft1Δ cells (Table 1).
AID-Induced Recombination Is Transcription-Dependent and Strongly Stimulated by THO Mutations in Yeast Cells.
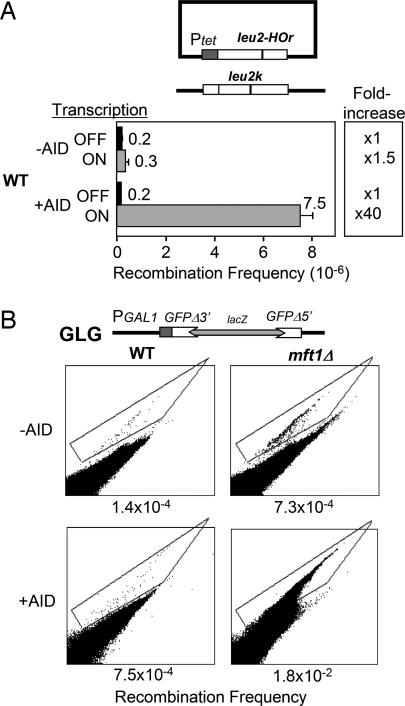
Finally, AID has also been shown to contribute to recombination in yeast (28, 29). This increase reflects the ability of AID to, in a direct or an indirect way, induce the formation of DNA double-strand breaks, which in yeast are preferentially repaired by recombination, whereas in human cells they are repaired by nonhomologous end-joining. Indeed nonhomologous end-joining is believed to be the major mechanism that leads to class switching in B cells (see ref. 37). We expected that AID should also increase recombination in a transcription-dependent manner in wild-type yeast and much more strongly in mft1Δ. For this purpose we used a plasmid–chromosome recombination assay between a CEN-plasmid leu2-r under control of the regulated Tet promoter and a chromosomal leu2-k mutation (38) in the wild type. As seen in Fig. 3A, AID did not change the frequency of gene conversion of leu2-r at low transcription. However, it caused a 40-fold increase at high transcription. Thus, like the mutagenic activity, the hyperrecombinogenic activity of AID is transcription-dependent in yeast. These findings are consistent with the view that both AID-induced mutation and recombination are the output products of a common intermediate.
Fig. 3.
Effect of AID on transcription-associated recombination. (A) Spontaneous (−AID) and AID-induced (+AID) frequency of gene conversion under high (ON) and low (OFF) transcription between a plasmid leu2-r and a chromosomal leu2-k allele in a wild-type strain. (B) Effect of AID expression on direct-repeat recombination in the GLG system. The frequency of GFP+ recombinants (signals inside the box) is indicated. y axis, green fluorescence (FL1H); x axis, unspecific fluorescence (FL2H). A diagram of each system is shown on the top of each panel.
Because direct-repeat recombinants are the type of event primarily increased by THO mutations (24), a direct-repeat recombination assay was developed for the analysis of deletions to assess whether THO mutations also strengthen AID-induced recombination. The assay is based on two truncated GFP repeats, which are under control of the GAL1 promoter and interrupted by a lacZ sequence. GFP+ recombinants are scored directly by FACS. As shown in Fig. 3B, mft1Δ and AID independently produced a 6-fold increase in GFP+ deletions, whereas the combination of mft1Δ and AID expression caused a strong synergistic 100-fold increase. Therefore, we conclude that impairment of mRNP formation caused by THO mutations enhances the action of AID in a transcription-dependent manner, resulting in a strong induction of both point mutations and direct-repeat recombination.
Discussion
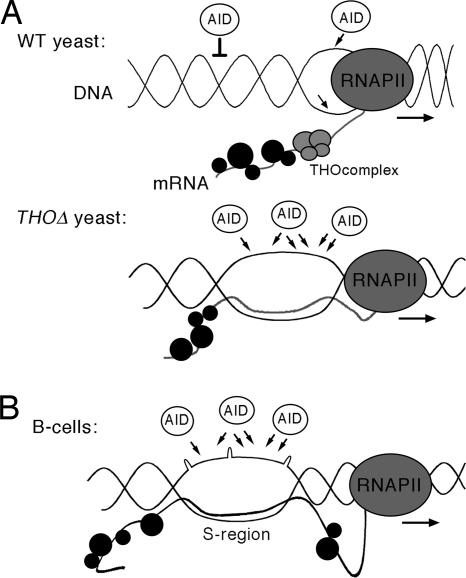
In this work we show that AID can function in yeast THO mutants similar to that in human B cells. In wild-type yeast not only can AID induce hypermutation and hyperrecombination, as it was previously shown, but, importantly, this induction depends on transcription. We show that such transcription-dependent hypermutation and hyperrecombination effects are weak in the wild type but strongly and synergistically enhanced in mutants of the THO complex. The synergistic effect of AID and THO mutations is also demonstrated at the molecular level by showing that most AID mutations occurred preferentially on the NT DNA strand. Because THO mutants are affected in mRNP biogenesis (21, 39), our results suggest that AID action can be enhanced by suboptimal mRNP formation. These results open the possibility that a suboptimal formation of the mRNP would enhance the probability of the nascent mRNA to form a DNA:RNA hybrid, leaving the NT strand as single-stranded. Such a single-stranded region would be stabilized by the G-loops in S regions, so that the final structure promotes AID targeting (Fig. 4).
Fig. 4.
A model to explain stimulation of genome instability by AID in yeast THO mutants compared with the S regions in B cells. (A) Transcription and mRNP formation mediated by THO in wild-type and THO-depleted yeast cells. In wild-type yeast, AID is able to act on the DNA when it is transcribed. Negative supercoiling behind the elongating RNA polymerase II (RNAPII) allows a weak AID action. In THO-depleted yeast, a cotranscriptional R-loop can be formed, allowing AID to act on the nontranscribed ssDNA, causing strong hypermutation and hyperrecombination. (B) A putative example of transcription and mRNP formation in the S region of Ig genes in human B cells. The G-rich S region could be refractory to a number of RNA-binding proteins, therefore forming a suboptimal mRNP at that particular region. Nascent RNA S regions could lead to local cotranscriptional R-loops or G-loops that would allow AID action.
For this study we first characterized new in vivo assays for the analysis of forward mutations under the control of transcription-regulated promoters in yeast, with which we have been able to show that transcription significantly enhances mutation, consistent with a previous result (32), in which this stimulation was accompanied by a change in the mutation spectrum (40). TAM has also been shown in bacteria with both spontaneous and damage-induced mutations (reviewed in ref. 33).
Mutants of the THO complex have a strong transcription-dependent hyperrecombination phenotype, as shown in direct-repeat systems (41, 42). Although recombination is also enhanced by transcription, the mechanisms underlying transcription-associated recombination may be different from those of TAM. Thus, impairment of replication progression by transcription may be a critical factor triggering transcription-associated recombination (43, 44), but this is not obvious for TAM. Our study reveals that THO mutations lead to a low hypermutator phenotype in the LAUR system, which is based on a highly expressed lacZ gene (Fig. 1). It is important to note that the gene expression defect and hyperrecombination phenotypes of THO mutants have been shown to be particularly strong at GC-rich DNA sequences such as lacZ (26). The mft1Δ-induced mutations have a clearly different pattern from those obtained in wild-type cells. In mft1Δ cells all mutations occurred within the lacZ gene of the lacZ::URA3 fusion, whereas in the wild type 40% of the mutations occurred at URA3. Interestingly, mutations originate at the same DNA region, lacZ, where hyperrecombination is strongly stimulated (41). Indeed, hypermutation associated with THO mutations is partially dependent on Rad52, and part of the hypermutation events observed in the LAUR assay are caused by Rad52-dependent DNA rearrangements (see Fig. 1B), therefore being a consequence of the strong hyperrecombination phenotype of these mutants. This result indicates that the recombinogenic structures generated in THO mutants may not be mutagenic by themselves.
Our study provides an appropriate system for the analysis of hypermutation caused by AID. In vitro and in vivo experiments in E. coli have shown the transcription dependency of AID deamination and hypermutation, respectively (5–8, 12). A hyperrecombination and hypermutation effect of AID was also observed in yeast (28), but whether this was transcription-dependent was not determined. In this work we show that hypermutation and hyperrecombination caused by AID in yeast are also transcription-dependent, confirming the hypothesis that the action of AID requires transcription of the target sequence (4, 6, 12). This is shown not only by an increase in the frequency of mutations but by the fact that most of these mutations occur at the AID-preferred DNA motif WRC (see Table 1). Importantly, the AID effect observed in yeast is still low, which suggests that there must be additional factors in human B cells responsible for the specific action of AID on its target DNA sequences. Indeed, our mutation spectrum analysis did not reveal a substantial number of A-T mutations (SI Table 3), which typically compose half of the mutational spectrum of SHM (13), consistent with the view that additional factors (e.g., mismatch repair, error-prone polymerases, chromatin structure, etc.) are regulated in B cells to mediate SHM (45).
The slight AID action observed in wild-type yeast may be explained by the prediction that the negatively supercoiled DNA putatively accumulated behind an elongating RNA polymerase can promote transient formation of ssDNA (14, 46), which is the preferential target of AID (5, 6). The results indicating that AID-induced mutations in wild-type yeast occurred at a 1:1 ratio in the NT:T strands would be consistent with this view, implying that both strands have equal probability of becoming single-stranded and, therefore, to be accessed by AID. Previous data in yeast have revealed a different NT:T strand ratio of mutations in the CAN1 gene (29). It is likely, therefore, that the pattern of mutations caused by AID in yeast may not be unique but dependent on a number of parameters that can include nucleotide sequence, chromatin structure, levels of expression, etc. Further investigations using different genes as mutation targets may be required to obtain a more complete view of AID action in yeast. Nevertheless, our results clearly show that a high impact of AID action in yeast is observed only when mRNP biogenesis is compromised by THO mutations. We detected a 5:1 bias in favor of the NT strand, a result that can be explained only if a more stable nontranscribed ssDNA structure is formed and simultaneously the T strand is protected from AID action. Because THO mutations have previously been shown to induce cotranscriptional R-loop formation, as determined in the same GC-rich lacZ gene used in this study (22), and S regions also form R-loops (16), formation of R-loops may be a natural way of stabilizing ssDNA for AID activity and of protecting the T strand in the form of the DNA:RNA hybrid. Our data in yeast show, therefore, that the strong AID effect on hypermutation in different THO-depleted yeast cells is consistent with the formation of R-loops during transcription (22, 34). Interestingly, recent reports have shown that AID interacts in vitro with the elongation complex (47). Because RNA polymerase II progression is reduced in THO mutants (23), we cannot discard that, in addition, AID action in THO mutants could be strengthened by a longer time in which AID could interact with the elongation complex, therefore increasing its time to act on DNA.
AID deamination seems to trigger the intermediates leading to class switch recombination in B cells, and some evidence suggests that class switching occurs by nonhomologous end-joining (37). Consequently, it is believed that AID deamination is the first event of a process ending in double-strand breaks. Whether this putative double-strand break is performed directly by AID or is formed after replication is a question yet to be addressed. Because double-strand breaks in yeast are preferentially repaired by recombination, it was expected that AID would affect recombination similarly to mutation. It was previously reported that AID induces recombination in yeast (28, 29). In this study we show that AID is able to increase the frequency of gene conversion only under high transcription (Fig. 3A), and, using a direct-repeat recombination assay, which allows us to detect hyperrecombination in THO mutants, we have been able to see a strong and synergistic effect of AID and THO mutations on recombination (Fig. 3B). Therefore, in yeast cells THO mutations enhance the transcription-dependent AID effect, whether detected as hypermutation or hyperrecombination.
In summary, our data clearly show that yeast THO mutants can be used as a model in vivo system for the study of AID action. Not only are hyperrecombination and hypermutation observed as a consequence of AID, but they occur in a transcription-dependent manner. More importantly, the pattern of mutations fits the previously shown pattern caused by AID in vitro (11), mutations occurring preferentially at the WRC motif. However, the pattern observed in vitro, in which mutations are preferentially found at the NT strand (6, 11, 29), is seen only in THO mutants. Therefore, our results open the possibility that mRNP biogenesis controlled AID action. It would be interesting to know whether the high G content of the S-region mRNA in B cells might limit its capacity to be assembled in an optimal mRNP, as THO mutations in general do in yeast, thereby increasing the reactivity of the nascent RNA with the DNA template (see Fig. 4). Yeast THO mutants can, therefore, be an excellent tool in deciphering the molecular mechanisms by which AID can induce both hypermutation and hyperrecombination.
Materials and Methods
Strains and Plasmids.
We used W303–1A isogenic strains WMK-2A (mft1Δ::KAN), U678-1C (hpr1Δ::HIS3), WRS52-4B (rad52Δ::KAN) [described previously (24)], WMR52-1D and WMR52-4D (rad52ΔKAN mft1Δ::KAN) (obtained in this study), and the BY derivative BER08-64A (his3::leu2-k) (38).
Centromeric plasmids pMR260 (30) carrying KAR1 under control of the GAL1 promoter, pCM184-LAUR (31) containing lacZ::URA3 under the tet promoter, and pCM184-L2HOr used for the plasmid–chromosome recombination assay (38) were described previously. Centromeric plasmids p414GALAID and p413GALAID carrying the human AID ORF under the GAL1 promoter were obtained by PCR amplification of AID from pGAID (6) using primers 5′-CTCTGGACGAAATTCCATGGACAGCCTCTTC-3′ and 5′-CCTGGAAGCTCGAGTCAAAGCTCCAAAGTA-3′ and cloning into the EcoRI-XhoI-digested pRS414GAL and pRS413GAL (38), respectively.
Centromeric plasmid pGLG containing the GFP direct-repeat construct was obtained by amplifying GFPΔ3′ and GFPΔ5′ fragments from pUG34 and pUG23 plasmids (48) by PCR using primers 5′-ACTAGTGCCATGATGTAAACATTG-3′, 5′-AATACAGGGTCGTCAGAT-3′, 5′-TTAAAGCCTTCGAGCGTCGGGCCCCCTTCT-3′, and 5′-ACTGGTCGA CTCCCAATTTTGGTTGAAT-3′ and subcloning into the SpeI-XbaI and ApaI-SalI sites of pRS413GAL, respectively. The lacZ ORF was subcloned at a BamHI between the GFP repeats.
Mutation and Recombination Analysis.
For the GKAR1 mutation assay, cells were cultured overnight in SC medium containing 2% glycerol-lactate as a carbon source. Afterward, the culture was split in two, one with 2% glucose (transcription OFF) and the other with 2% galactose (transcription ON), and cultured for another 9 h before mutant selection. For the LAUR mutation assay and the plasmid–chromosome recombination assay cells were cultured in SC plates with (transcription OFF) or without (transcription ON) 5 μg/ml doxycycline, from which independent colonies were obtained for the mutation or recombination analyses. Kar− and Ura− mutants were selected on SC containing 2% galactose and SC plus 5-fluoroorotic acid, respectively, and Leu+ recombinants on SC-Leu plates. lacZ+ and lacZ− were distinguished by color on SC plus X-gal medium. Median mutation and recombination frequencies were obtained by fluctuation tests as the median value of six independent colonies isolated from SC plates. The final frequency given for each strain and condition is the mean and standard deviation of three to four median values.
Miscellanea.
Yeast methodology, α-32P-labeled DNA probes, and Southern and Northern blots were performed by following standard procedures. GFP fluorescence was determined in a FACSCalibur (Becton Dickinson, San Jose, CA) from 106 cells grown in SC overnight and resuspended in 1 ml of H2O.
Supplementary Material
Acknowledgments
We thank A. S. Bhagwat (Wayne University, Detroit, MI) for kindly providing the hAID clone, J. Escalante for technical assistance, J. F. Ruiz for helpful discussions, F. Cortés-Ledesma and H. Gaillard for critical reading of the manuscript, and D. Haun for style supervision. This work was supported by Spanish Ministry of Science and Education Grants SAF2003-00204 and BFU2006-05276 and Junta de Andalucía Grants CVI102 and CVI624. B.G.-G. was recipient of a predoctoral training grant from the Spanish Ministry of Science and Education.
Abbreviations
- AID
activation-induced cytidine deaminase
- CSR
class switch recombination
- mRNP
messenger ribonucleoprotein
- NT
nontranscribed
- S
switch
- T
transcribed
- SC
synthetic complete
- SHM
somatic hypermutation
- TAM
transcription-associated mutation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702836104/DC1.
References
- 1.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 2.Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, Honjo T. Nature. 2002;416:340–345. doi: 10.1038/nature727. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Petersen-Mahrt SK, Harris RS, Neuberger MS. Nature. 2002;418:99–103. [PubMed] [Google Scholar]
- 5.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 6.Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Nucleic Acids Res. 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bransteitter R, Pham P, Scharff MD, Goodman MF. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters A, Storb U. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 9.Hein K, Lorenz MG, Siebenkotten G, Petry K, Christine R, Radbruch A. J Exp Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martomo SA, Fu D, Yang WW, Joshi NS, Gearhart PJ. J Immunol. 2005;174:7787–7791. doi: 10.4049/jimmunol.174.12.7787. [DOI] [PubMed] [Google Scholar]
- 11.Pham P, Bransteitter R, Petruska J, Goodman MF. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 12.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 13.Milstein C, Neuberger MS, Staden R. Proc Natl Acad Sci USA. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen HM, Storb U. Proc Natl Acad Sci USA. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhuri J, Khuong C, Alt FW. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 16.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 17.Dempsey LA, Sun H, Hanakahi LA, Maizels N. J Biol Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 18.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duquette ML, Pham P, Goodman MF, Maizels N. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 20.Yu K, Roy D, Bayramyan M, Haworth IS, Lieber MR. Mol Cell Biol. 2005;25:1730–1736. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilera A. Curr Opin Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Huertas P, Aguilera A. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 23.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 24.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voynov V, Verstrepen KJ, Jansen A, Runner VM, Buratowski S, Fink GR. Proc Natl Acad Sci USA. 2006;103:14423–14428. doi: 10.1073/pnas.0606546103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavez S, Garcia-Rubio M, Prado F, Aguilera A. Mol Cell Biol. 2001;21:7054–7064. doi: 10.1128/MCB.21.20.7054-7064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Manley JL. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Poltoratsky VP, Wilson SH, Kunkel TA, Pavlov YI. J Immunol. 2004;172:4308–4313. doi: 10.4049/jimmunol.172.7.4308. [DOI] [PubMed] [Google Scholar]
- 29.Mayorov VI, Rogozin IB, Adkison LR, Frahm C, Kunkel TA, Pavlov YI. BMC Immunol. 2005;6:10. doi: 10.1186/1471-2172-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose MD, Fink GR. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- 31.Jimeno S, Rondon AG, Luna R, Aguilera A. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta A, Jinks-Robertson S. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- 33.Aguilera A. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huertas P, Garcia-Rubio ML, Wellinger RE, Luna R, Aguilera A. Mol Cell Biol. 2006;26:7451–7465. doi: 10.1128/MCB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morey NJ, Greene CN, Jinks-Robertson S. Genetics. 2000;154:109–120. doi: 10.1093/genetics/154.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogozin IB, Kolchanov NA. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 37.Rooney S, Chaudhuri J, Alt FW. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Rubio M, Huertas P, Gonzalez-Barrera S, Aguilera A. Genetics. 2003;165:457–466. doi: 10.1093/genetics/165.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinciguerra P, Stutz F. Curr Opin Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Lippert MJ, Freedman JA, Barber MA, Jinks-Robertson S. Mol Cell Biol. 2004;24:4801–4809. doi: 10.1128/MCB.24.11.4801-4809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chavez S, Aguilera A. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prado F, Piruat JI, Aguilera A. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wellinger RE, Prado F, Aguilera A. Mol Cell Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prado F, Aguilera A. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo CJ, Martin A, Scharff MD. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 46.Drolet M, Bi X, Liu LF. J Biol Chem. 1994;269:2068–2074. [PubMed] [Google Scholar]
- 47.Besmer E, Market E, Papavasiliou FN. Mol Cell Biol. 2006;26:4378–4385. doi: 10.1128/MCB.02375-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niedenthal RK, Riles L, Johnston M, Hegemann JH. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.