Abstract
Nitric oxide (NO) is used by mammalian immune systems to counter microbial invasions and is produced by bacteria during denitrification. As a defense, microorganisms possess a complex network to cope with NO. Here we report a combined transcriptomic, chemical, and phenotypic approach to identify direct NO targets and construct the biochemical response network. In particular, network component analysis was used to identify transcription factors that are perturbed by NO. Such information was screened with potential NO reaction mechanisms and phenotypic data from genetic knockouts to identify active chemistry and direct NO targets in Escherichia coli. This approach identified the comprehensive E. coli NO response network and evinced that NO halts bacterial growth via inhibition of the branched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase. Because mammals do not synthesize branched-chain amino acids, inhibition of dihydroxyacid dehydratase may have served to foster the role of NO in the immune arsenal.
Keywords: systems biology, chemoinformatics
Mammals possess complex immune systems that have evolved to prevent microbial invasion. Nitric oxide (NO) is one of the key chemicals used by mammalian cells to combat infections (1). Although NO is known to act in a bacteriostatic fashion (2), the mechanism underlying this action is not completely understood. NO interferes with biological processes either directly, by reacting with metal centers and free radicals, or indirectly, by promoting the formation of reactive nitrogen oxide species (RNOS), such as peroxynitrite and N2O3 (3). NO has been reported to react with various protein Fe–S clusters (4–7), and this reactivity has been implicated in the inhibition of tumor proliferation (8–10). NO also binds to the metal centers of respiratory enzymes, inhibiting bacterial respiration (11–13). Because NO is used to combat Escherichia coli in low oxygen environments, it is likely that respiration is not the only system targeted by NO.
E. coli contains NO-consuming proteins NO reductase NorV (14), NO oxidase HmpA (15, 16), and cytochrome c nitrite reductase NrfA (17). Expression of norV and hmpA is increased in response to NO by NO-specific transcription factors (TFs) NorR (18) and NsrR, respectively (19, 20). Our goal here was to identify the comprehensive NO response network, consisting of the direct NO targets leading to bacteriostasis, the response network resulting from bacteriostasis, bacterial NO sensors for self-defense, and the response network for self-defense.
Previous genome-scale studies have used DNA microarrays to identify the global genetic response to NO (21–24). Although transcriptome analysis provides substantial information regarding changes in gene expression, examination of individual genes cannot distinguish primary from secondary effects and does not directly identify the TFs and pathways leading to the transcriptional changes. Because many promoters are controlled by multiple TFs, the identity of the TF responsible for mediating transcriptional perturbations is not always clear in ad hoc analysis. The situation worsens when comparing samples with different growth rates, because growth rate differences introduce many changes in metabolic genes. Furthermore, the stochastic properties of microarray technology result in a significant amount of noise that can skew a “gene-level” perspective.
Instead of focusing on transcriptional perturbation of individual genes, we used a systems biology perspective to identify the TFs and pathways governing the NO response based on the transcriptome data as a whole and the known underlying transcription network structure. To attain this goal we used network component analysis (NCA), a mathematically based approach that reflects the underlying biological structure of regulatory networks (25–27). NCA is able to identify perturbed TFs and pathways from transcriptome data and separate the effects of multiple TFs. NCA accounts for the fact that many regulators require posttranslational modification, such as phosphorylation or ligand binding, to affect transcription. A change in TF activity (TFA) may not be accompanied by a change in TF transcript abundance. We were interested in identifying TFs that undergo a change in activity in response to NO. Because it was not feasible to directly measure the activity of all regulators, we used NCA as the first step to infer changes in TFA from the expression ratios obtained by transcriptome analysis.
Our approach was multipronged. (i) We used NCA of transcriptome measurements, which separates the effects of multiple TFs on the transcriptome (25, 26, 28), to identify NO-responsive TFs. (ii) Simultaneously, we created a putative reactome of RNOS targets by using known RNOS chemistry to scan the entire E. coli proteome and combined the results with the TFs responsive to NO to identify direct interactions between NO and the TFs. (iii) In addition, we used genetic knockouts and phenotypic experiments to validate the above screening results and determine key pathways responsible for NO-induced bacteriostasis. Combining these data allowed us to discriminate among the direct NO targets responsible for bacteriostasis, NO sensors for bacterial defense, and the underlying chemical and biological mechanisms. Finally, biochemical experiments were conducted to validate the direct NO target causing bacteriostasis, which is the Fe–S cluster of IlvD, a protein essential for branched-chain amino acid (BCAA) biosynthesis. NO is believed to alter the activity of Fe–S-containing proteins by reacting with the Fe–S center and forming a dinitrosyl–iron complex (DNIC) (3). Fe–S cluster properties, such as solvent accessibility, can impact NO's reactivity, which means that there will be variable sensitivity to NO among Fe–S-containing proteins (7). Finally, overexpression of IlvD alone, but not another Fe–S protein isopropylmalate isomerase (LeuCD), was able to rescue the NO-induced growth inhibition, further supporting IlvD as the crucial target for NO-induced bacteriostasis. In the process of deducing this NO target, a comprehensive network was mapped that included circuits for defense against NO and metabolic responses to NO challenge.
Results
E. coli Adaptively Responds to NO.
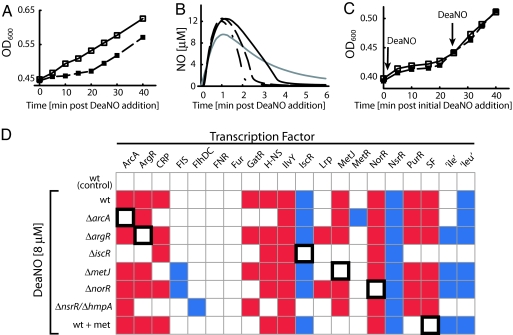
We used sodium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate (DeaNO), an NO donor with t½ = 2 min (37°C, pH 7.4), to deliver NO to the culture (29). Exposure of E. coli to 8 μM DeaNO induces bacteriostasis (Fig. 1A). Even though NO is below the detection limit within 5 min (Fig. 1B), growth inhibition lasts for 20 min. However, E. coli has an adaptive response to NO: the NO consumption rate increases after an initial DeaNO exposure (Fig. 1B), and subsequent DeaNO challenge after growth recovery does not impact growth (Fig. 1C). This adaptive behavior is indicative of a dynamic response network.
Fig. 1.
Phenotypic and regulatory response of E. coli to NO. (A) Exposure to 8 μM DeaNO (■) induces bacteriostasis in the WT relative to the control (□). (B) After an initial exposure to 8 μM DeaNO (——), the WT consumes NO more rapidly when exposed to additional doses of DeaNO at 10 min (— — —) and 20 min (— - — -). The gray line indicates NO release and degradation in the absence of E. coli. (C) Treatment of E. coli with 8 μM DeaNO induces resistance to subsequent DeaNO exposure. Treatment with a single dose of DeaNO induces bacteriostasis for ≈20 min (□). After recovery from bacteriostasis induced by 8 μM DeaNO, a subsequent dose of DeaNO does not induce an additional occurrence of bacteriostasis (■). (D) The regulatory network response to DeaNO is shown. NCA identifies TFs with activity significantly (P < 0.01 vs. random network) increased (red squares) or decreased (blue squares) when E. coli is exposed to 8 μM DeaNO for 5 min. The bolded box indicates expected results from TF deletion. For each condition, at least four biological replicate transcriptome measurements were taken.
Transcriptome Network Component Analysis Identified NO-Responsive Transcription Factors.
As a first step in unraveling the adaptive response network, we observed the global transcriptional response to DeaNO. Transcriptome analysis compared transcript levels immediately before and 5 min after addition of 8 μM DeaNO. The expression data were assigned 95% confidence intervals by using a MCMC (Markov Chain Monte Carlo) procedure (30); 709 genes were significantly perturbed [supporting information (SI) Table 1]. The MCMC procedure can identify statistically significant perturbations that do not meet the conventional ad hoc criterion of a 2-fold change, thus providing a better picture of network behavior. The complete MIAME-compliant dataset has been deposited in the GEO database (www.ncbi.nlm.nih.gov/geo).
Because ad hoc examination of individual genes cannot distinguish between primary and secondary effects, we used an integrated, multipronged approach to identify the NO response network. First, NCA identified perturbed TFs from transcriptome data. From this set of perturbed TFs, we can deduce the direct NO targets and relevant reaction chemistry. NCA uses an initial genome-wide transcriptional regulatory network, consisting of reported connections between genes and TFs [RegulonDB (31), Ecocyc (32), literature survey (20, 33)] to estimate TFA ratios (25–27). Because DeaNO induces growth retardation similar to the stringent response, a “virtual” stringent regulon, with a virtual regulator referred to as the stringent factor (SF), was constructed to account for growth-rate-associated effects (SI Table 2). The complete initial network is given in SI Table 3; genes with no known regulators were not included in the NCA.
NCA identified 13 significantly perturbed regulators (ArcA, ArgR, CRP, GatR, H-NS, IlvY, IscR, MetJ, NorR, NsrR, PurR, SF, and the leucine transcriptional attenuator, which is hereafter referred to as “leu”) in the DeaNO response transcriptome data (Fig. 1D). These results implicated potential NO sensors, NO targets, and secondary signaling pathways induced by NO. NorR and NsrR are known NO-specific TFs (18–20), and IscR has been recently recognized as involved in the NO response (24); the remaining 10 regulators have not been previously associated with the NO response. Some regulators, such as NorR, are easily identified from the raw transcriptome data. NorR has only two regulon members, norV and norW, and both are activated more than 10-fold in response to DeaNO (SI Table 1). However, other perturbations are not as obvious; only half of the 70 regulon members of ArcA are significantly perturbed, and the majority of these perturbations are <2-fold. The fact that many of the regulatory perturbations are not obvious in the transcriptome data demonstrates the utility of NCA relative to an ad hoc analysis.
To verify the NCA results, we deleted individual TFs, reexamined the transcriptome response to DeaNO, and again analyzed the data with NCA (Fig. 1D). Upon deletion of the TF in question (ArcA, ArgR, IscR, MetJ, NorR), NO-responsive transcriptional perturbations mediated by the TF were no longer observed, and NCA detected no change in regulator TFA, indicating that the apparent TFA changes are indeed caused by the TF and not by noise, artifacts in analysis, or other regulators.
For the ΔnsrR strain, very few transcriptional changes were observed during DeaNO challenge (SI Table 1), suggesting that a NO protective mechanism was derepressed. NsrR is a negative regulator of hmpA, an NO oxidase (19, 20). Deletion of hmpA in conjunction with nsrR restored most of the TFA changes observed in the WT strain (Fig. 1D), confirming that the major role of NsrR is repression of hmpA. To avoid the masking effect of derepressed HmpA expression, transcriptome analysis focused on the ΔnsrR/ΔhmpA strain instead of ΔnsrR only. In the ΔnsrR/ΔhmpA strain, the NsrR TFA appears to be perturbed (Fig. 1D); however, because the NsrR regulon (SI Table 3) is currently composed of four genes and only one (ytfE) is perturbed in this condition, this result is questionable.
Remarkably, deletion of iscR halves the number of perturbed TFs, suggesting that IscR regulates a protective mechanism against NO. IscR represses the general Fe–S cluster assembly and repair system (Isc), which contributes to the repair of NO-damaged Fe–S clusters (34). The basal expression of the Isc system, including IscS (SI Fig. 6) is increased in the iscR mutant. The decrease of regulator perturbations in the absence of iscR indicates that Fe–S cluster damage is a major component of the NO response network. Thus, our analysis of deletion mutants implicated the defensive roles of the NsrR and IscR regulons and highlights their importance. Because deletion of H-NS, IlvY, or CRP resulted in sickly strains, we excluded these mutants from this analysis.
Although previous ad hoc analyses have identified Fur (23, 24) and FNR (5, 24) as regulators responding to RNOS, the TFA of these regulators was not altered in our analysis (Fig. 1D). The transcriptomic response of fur and fnr deletion mutants resemble the WT response (data not shown), verifying that these TFs are not part of the NO response network in this condition.
Chemoinformatic Identification of Active NO Chemistry.
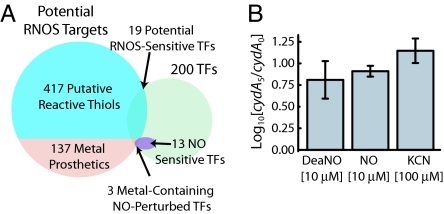
We reasoned that primary NO targets and sensors must react directly with NO, and, thus, we scanned the E. coli proteome for potential RNOS-reactive targets; proteins containing heme, copper, nonheme iron, and Fe–S prosthetics were flagged as NO-reactive (SI Table 4). Although NO is unlikely to directly react with thiols, NO-derived RNOS could possibly target thiol groups, and, therefore, we have included the putative reactive thiol motif (35) in the list of potential targets. Within the RNOS reactome, there are 19 potential NO-reactive TFs, three (IscR, NorR, NsrR) of which were significantly perturbed by 8 μM DeaNO (Fig. 2A). Of the known RNOS-sensitive TFs, only those containing nonheme iron (NorR) and Fe–S clusters (NsrR and IscR) were perturbed, suggesting that the pertinent NO chemistry in our study involves direct reactions of NO with metal groups. This conclusion narrowed the scope of our search for potential protein targets with the focus on Fe, Fe–S, or heme prosthetics.
Fig. 2.
Identification of the modes of RNOS chemistry that interact with genomic program of E. coli after exposure to DeaNO. (A) Chemoinformatic analysis of proteome of E. coli indicates that there are 554 potential RNOS-reactive sites. Of these sites, 417 are thiols and 137 are heme, nonheme iron, Fe–S clusters, or copper. Of the 13 NO-sensitive TFs, three contain NO-reactive metal prosthetics. (B) ArcA is activated in response to DeaNO exposure. Addition of cytochrome oxidase inhibitors (DeaNO, NO, KCN) increases the expression of cydA, a reporter gene for the ArcAB two-component system.
The list of potential NO targets includes the copper–heme and heme groups in cytochrome oxidases bo and bd, respectively. NO is known to inhibit bacterial respiration by direct reaction with these proteins (12, 13). Both cytochromes modulate the quinone pool, in turn modulating ArcAB activity (36). To assess whether the NO-mediated ArcA perturbation was an effect of the NO/cytochrome interaction, we exposed E. coli to KCN, which also targets the cytochromes, and measured the expression of an ArcA reporter gene, cydA, with real-time RT-PCR. KCN, like DeaNO and NO delivered in solution, increased cydA expression (Fig. 2B). In the absence of arcA or arcB, expression of cydA was not activated in response to NO, indicating that the activation signal was transduced by the ArcAB system (data not shown). Because ArcA responds similarly to KCN and NO, it is likely that ArcA activation is a consequence of cytochrome targeting by NO.
Phenotypic Analysis of Transcription Factor/Bacteriostasis Relationship.
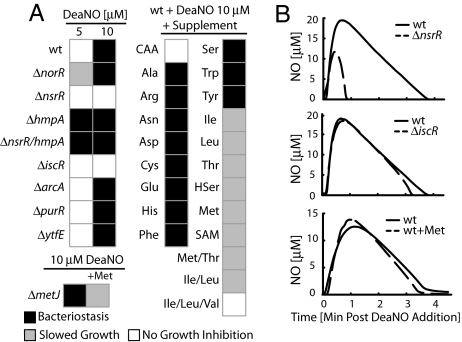
Next, we systematically deleted each NO-responsive TF and examined its role in the bacteriostatic response. Because we were interested in observing phenotypic differences relative to the WT, slow growing mutants (ArgR, H-NS, IlvY, CRP) were excluded from this analysis. arcA, purR, and metJ deletion mutants manifested no phenotype relative to the WT (Fig. 3A), indicating that they were not primary mediators of bacteriostasis in our condition. nsrR and iscR mutants exhibited increased NO resistance, and the norR mutation slightly increased sensitivity. NO measurements confirmed that NO consumption was increased in the nsrR mutant (Fig. 3B), consistent with the idea that hmpA expression is repressed by NsrR. In contrast, IscR deletion did not alter the NO consumption rate (Fig. 3B), suggesting that IscR deletion conferred NO resistance by derepression of Fe–S protein repair, rather than by increased NO consumption. YtfE, which has been linked to the anaerobic NO response (22) and Fe–S assembly (37), does not appear to play a role in protection from bacteriostasis (Fig. 3A) or NO consumption (data not shown). NorR deletion did not change the NO consumption rate appreciably (data not shown), given that this mechanism is mainly used under anaerobic conditions.
Fig. 3.
Phenotypic identification of the essential components of the bacteriostatic response and their impact on NO consumption. (A) The effect of NO-sensitive TF deletions and AA supplementation on NO-induced bacteriostasis. The change in the response to DeaNO mediated by the genetic deletion of the regulators IscR, NorR, and NsrR indicates that their regulons are involved in the response to NO. The alleviation of growth inhibition by BCAA, Met, and their precursors indicates that AA depletion is a key symptom of the offensive actions of NO. Deletion of hmpA in conjunction with nsrR indicates that NsrR combats bacteriostasis via regulation of hmpA expression. CAA, Casamino acids; HSer, homoserine; SAM, S-adenosylmethionine. (B) Deletion of NsrR strongly increases the ability of E. coli to consume NO, indicating that it plays a key role in regulating NO consumption. Deletion of iscR or supplementation with Met does not increase NO consumption, indicating that their roles may be related to secondary effects arising from the interaction of NO with E. coli.
Because the putative “stringent factor” cannot be deleted, we “short-circuited” this regulon by supplementation with casamino acids before NO challenge. Casamino acid supplementation conferred resistance to bacteriostasis (Fig. 3A). To determine whether there were any specific amino acids (AAs) that conferred resistance to DeaNO, we measured the effect of single AA supplements on growth (Fig. 3A). Met, Thr, and BCAAs reduced the impact of DeaNO on growth. When E. coli was grown in the presence of all three BCAAs, 8 μM DeaNO no longer affected growth (Fig. 3A and SI Fig. 7); however, when the cultures were supplemented with only two BCAAs (Ile and Leu) or two non-BCAAs (Met and Thr), 8 μM DeaNO still elicited a degree of growth inhibition (Fig. 3A). These results indicate that BCAAs are of key importance in NO-induced growth inhibition. Because homoserine, Met, SAM (S-adenosylmethionine), and Thr can reduce the extent of growth inhibition, but are not requisite for NO resistance, it is possible that these supplements confer protection by increasing metabolite flux toward BCAA biosynthesis. This hypothesis is supported by the observation that Met supplementation relieved growth inhibition (Fig. 3A) without dramatically altering NO consumption (Fig. 3B).
NO Induces Bacteriostasis by Damaging the Fe–S Cluster of IlvD.
Because IscR deletion and BCAA supplementation both confer resistance to NO without increasing NO consumption, it is plausible that they act through the same general mechanism. NO damages Fe–S clusters (7), and Fe–S cluster damage can lead to AA auxotrophies (38). Because the Fe–S cluster repair genes have increased expression in the ΔiscR mutant before NO treatment (SI Fig. 6), we hypothesized that stringent factor activation and bacteriostasis are a result of Fe–S cluster damage in BCAA biosynthesis pathways.
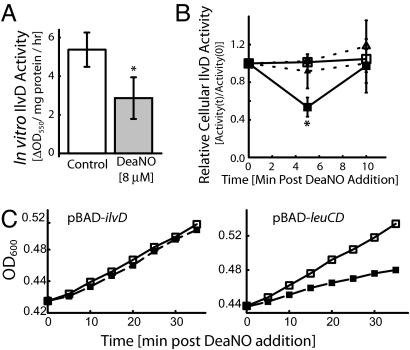
Of the 98 potential Fe–S NO targets in E. coli, only IlvD and LeuCD are involved in BCAA biosynthesis. Whereas IlvD is required for synthesis of all three BCAA, LeuCD is specific to Leu production. Because supplementation with all three BCAAs is required for complete growth protection (Fig. 3A and SI Fig. 7), it appears that IlvD, and not LeuCD, is the NO target leading to bacteriostasis. The Fe–S cluster of IlvD is vulnerable to attack from superoxide (39, 40); to determine whether IlvD is also vulnerable to NO, we measured the effect of DeaNO on enzyme activity. IlvD activity in crude extract was significantly (P < 0.01) decreased after exposure to DeaNO (Fig. 4A). Because we believe that deletion of iscR counters bacteriostasis by enhancing IlvD repair, we measured the impact of DeaNO on in vivo IlvD activity for the WT and ΔiscR strains. As expected, IlvD activity was transiently decreased by DeaNO in the WT, but not in the ΔiscR strain (Fig. 4B). Additionally, whereas increased expression of IlvD resulted in increased DeaNO resistance, overexpression of potential NO target LeuCD did not (Fig. 4C). These results show that NO damages the Fe–S cluster of IlvD and that this damage leads to bacteriostasis. However, these results do not imply that IlvD is the only Fe–S protein damaged by NO; it is simply the critical target for bacteriostasis. Because E. coli does not possess an IlvD-independent route to BCAA biosynthesis, inhibition of IlvD will result in the depletion of the BCAA pool and subsequent initiation of the stringent response and bacteriostasis.
Fig. 4.
DeaNO induces bacteriostasis by damaging the Fe–S cluster of IlvD. (A) DeaNO inhibits the activity of IlvD, a central enzyme required for BCAA biosynthesis, in crude cell extracts. The values are means ± SD (n = 7). ∗, Significantly (P < 0.01, Wilcoxon Mann–Whitney two-sided test) different from the control activity. (B) Deletion of iscR protects IlvD from inhibition by DeaNO. DeaNO (8 μM) temporally decreases WT IlvD activity (■) relative to the control (□). Deletion of iscR reduces the impact of DeaNO on IlvD activity (▴) relative to the control (▵). The values are means ± SD. (n = 3–4). ∗, Significantly (P < 0.05, Wilcoxon Mann–Whitney two-sided test) different from activity at 0 min. (C) Overexpression of IlvD confers resistance to DeaNO-induced bacteriostasis. Whereas overexpression of LeuCD, an Fe–S enzyme crucial for Leu biosynthesis, does not. Transformants containing the inducible vectors for ilvD (pBAD-ilvD) and leuCD (pBAD-leuCD) were grown in Mops/0.2% fructose with 0.1 mM arabinose to induce expression and were either exposed to cold buffer (□) or 20 μM DeaNO (■).
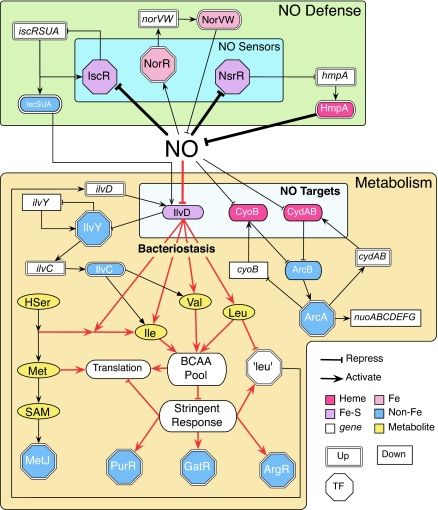
Taken together, E. coli responds to NO challenge at several levels (Fig. 5). First, NO induces the defense mechanisms of E. coli by direct interaction with NO sensors NsrR and NorR, which derepresses hmpA and induces norVW, respectively. These mechanisms destroy NO by oxidation and reduction. If these defense efforts are insufficient, NO begins to damage the Fe–S center of IscR, which in turn induces the isc operon for Fe–S repair. Meanwhile, damage to IlvD causes BCAA starvation and induces the stringent response and bacteriostasis. Disturbance of the BCAA pathway results in perturbation of other AA pathways. Meanwhile, NO directly inhibits respiratory enzymes, which in turn perturbs the ArcAB regulon. However, respiration inhibition may not induce bacteriostasis, because E. coli can grow without the aerobic electron transport system.
Fig. 5.
The essential NO response network of E. coli. The network can be divided into two key categories: the metabolic targets and the defense system. The metabolic targets include IlvD and cytochromes. NO inhibits bacterial growth by targeting IlvD, an essential BCAA biosynthesis enzyme. BCAA depletion results in a halt in translation and activates the stringent response. NO also induces a shift in the respiratory system by inhibiting cytochrome bo and bd oxidase. The defense system contains modules for NO detoxification and repair of critical damage; these modules are regulated by proteins that directly sense NO (IscR, NsrR, NorR). NsrR mediates the expression of the aerobic NO detoxification system, NorR controls anaerobic NO detoxification, and IscR controls expression of the Fe–S repair system.
Discussion
The guiding principle behind our approach was that chemical challenges to the cell would ultimately be manifested in a transcriptional response. Therefore, the transcriptome was a good starting point for attempting to reverse engineer the response network of an organism. Because the noise associated with global transcriptome analysis can confound a gene-level analysis, we approached this problem with a systems biology perspective that was backed by experimental validation. We used NCA to identify the regulatory interface between NO chemistry and transcriptomic output. Integration of the phenotypically relevant regulatory networks with the putative RNOS reactome revealed that NO primarily impacts E. coli through direct interaction with protein metal centers in a network that comprises NO sensors, NO defense, NO targets, and the ensuing metabolic response (Fig. 5). NsrR, NorR, and IscR are NO sensors and induce defensive effects. Previously recognized NO sensors NsrR and NorR mediate increased expression of NO detoxification systems. IscR regulates repair of NO-mediated Fe–S damage via the Isc system. Although other members of the IscR regulon possibly contribute to NO defense, the poor growth exhibited by iscS mutants (41) makes it difficult to characterize the NO response in the absence of the Isc system. NO targets are crucial proteins whose injury by NO induces a significant metabolic response. Cytochromes bo and bd have previously been recognized as NO targets (11–13), which was confirmed in our study; however, our approach further showed that IlvD damage was the underlying cause of bacteriostasis in our condition. Thus, by using this combined approach, we discovered a major component of the human–E. coli relationship that has relevance to treatment of infection.
In addition to causing bacteriostasis, IlvD damage results in a cascade of metabolic responses including the stringent response and perturbation of metabolic regulators PurR, MetJ, ArgR, GatR, and IlvY (Fig. 5). Decreased BCAA availability limits translation, which serves as a signal for activation of the SF and PurR TFs. Because the abrupt stoppage of translation removes a sink for the remaining AAs, activation of ArgR and MetJ may be the result of accumulation of Arg and Met. Additionally, the reduced Thr demand results in an increase in homoserine flux through the Met biosynthesis pathway and subsequent activation of MetJ. Furthermore, IlvD inhibition will result in a build-up of upstream intermediates which activate IlvY.
In support of the importance of BCAA biosynthesis to the struggle between the immune system and E. coli, a recent report by Roos and Klemm indicates that BCAA biosynthesis genes are up-regulated in E. coli growing in the urinary tract (42). Because humans do not possess the BCAA biosynthesis machinery, it makes sense that our immune system would target this system. Targeting IlvD will slow bacterial proliferation by inhibiting translation without inducing a similar effect in mammalian systems. Because IlvD is highly conserved (>97% protein sequence homology compared with the K12 strain) in the enterohemorrhagic O157:H7 and uropathogenic CFT073 strains, it is a prime candidate for therapy.
Materials and Methods
Cell Growth.
BW25113 was grown aerobically to mid-log (OD600, 0.4–0.5) from an initial OD600 of 0.05 at 37°C in Mops/0.2% glucose, as described previously (43), in baffled flasks at 1/10 to 1/5 total volume and shaken at 250 rpm. AA supplementation studies used the concentrations given in ref. 44. DeaNO (Cayman Chemical, Ann Arbor, MI) solutions were made on the day of use with cold Tris/saline buffer (pH > 10.5).
Cell Harvesting, RNA Purification, and Microarray.
Half of the cell culture was withdrawn for RNA purification as the reference time point; the remainder was harvested 5 min after treatment. Harvested cells were swirled in a dry ice/ethanol bath for 45 seconds, concentrated by centrifugation at 4°C and 8,000 × g for and 10 min, resuspended in 1 ml of RNA Later (Qiagen, Valencia, CA), and stored at −80°C until RNA purification. RNA purification, array design, hybridization, and analysis were performed as previously described (28), with the exception that the Array-Ready Oligo Set (Qiagen) was used for probes.
Network Component Analysis.
NCA is an algorithm that can identify changes in TFA when only a subset of a regulon is perturbed (25, 26), which is useful in deconvoluting the output of combinatorial regulation; however, caution should be taken when only a small number of perturbations are considered. NCA decomposed significant gene expression perturbations into TFA ratios and control strengths. The expression and connectivity matrices are, respectively, derived from SI Tables 1 and 3. To determine whether a TF was significantly perturbed in a given experiment, we built null distributions of TFAs and then performed z tests on individual data points of every TFA. The null TFA distributions were constructed by: (i) randomly selecting N genes from the genome, where N is the size of the original network; (ii) NCA decomposition of the expression data to obtain TFAs of the random network; and (iii) repeating steps i and ii 100 times.
Chemoinformatic Analysis.
Potential RNOS-reactive targets were identified by searching the proteome annotation of E. coli for iron, heme, or copper prosthetic groups. Reactive thiol candidates were identified by scanning the protein sequence of E. coli for the characteristic “acidic AA-Cys-basic AA” motif. All analyses were performed by using BioWarehouse version 3.5 (45); this database was queried for proteins with experimentally verified and computationally predicted cofactors known to be RNOS-reactive.
Real-Time RT-PCR.
Real-time PCR was performed in a Cepheid SmartCycler with QuantiTect RT-PCR SYBR mix (Qiagen). RT-PCR primer pairs were designed by using the MyProbes (Lars Rohlin, University of California, Los Angeles) software. Reverse transcription with SuperScript II (Invitrogen, Carlsbad, CA) used either 50 or 100 ng total RNA as template. RNA was checked for genomic DNA contamination by running a RT-PCR in the absence of reverse transcription. Transcript abundance was normalized by levels of chaA, which is not perturbed in any of the NO-related microarray data.
Gene Deletion.
Deletion mutants were generated by using the method described by Datsenko and Wanner (46). Deletions that could potentially disrupt expression of cotranscribed genes were designed to be nonpolar.
NO Concentration Measurements.
Extracellular NO concentration was measured by using a microchip NO electrode (ISO-NOPMC; World Precision Instruments, Sarasota, FL). Experiments were performed aerobically at 37°C in a total volume of 10 ml.
IlvD Assays.
IlvD in vitro activity was measured according to a previously described method (47) that which uses Val/Leu pathway substrate 2,3-dihydroxyisovalerate as substrate. Cold Tris buffer or 8 μM DeaNO was added to the crude extract/buffer mixture 10 min before substrate addition. All reagents were purchased from Sigma–Aldrich (St. Louis, MO), with the exception of 2,3-dihydroxyisovalerate, which was synthesized according to the method of Cioffi et al. (48). Whole-cell IlvD assays were similar to the in vitro assays, with modifications described by Wixom et al. (49). Cells were grown to mid-log phase, treated, washed twice with cold Tris buffer, pH 8.0, and concentrated 20-fold before measurement of IlvD activity.
Overexpression of IlvD and LeuCD.
ilvD and leuCD were placed under the control of the arabinose-inducible pBAD promoter in expression vectors pKJC1 and pKJC2 (Invitrogen). pJKC1 and pKJC2 relieve the AA auxotrophies of ilvD and leuCD deletion mutants, respectively, demonstrating functional expression of IlvD and LeuCD. Because of interference between glucose and the pBAD promoter, overexpression experiments were performed in Mops/0.2% fructose.
Statistical Analysis.
A two-tailed Wilcoxon Mann–Whitney test was used to assess the probability of statistically significant difference by using the R statistical computation environment (R Foundation for Statistical Computing, Vienna, Austria).
Supplementary Material
Acknowledgments
We thank Jon M. Fukuto for insightful discussions during the course of this work and assistance in preparing the 2,3-dihydroxyisovalerate and Alice Lee for assistance with experimental analysis. This work was supported by National Institutes of Health Grant 2R01HL065741, National Science Foundation Grant CCF-0326605, and the University of California, Los Angeles (UCLA)–Department of Energy Institute of Genomics and Proteomics. L.M.T. was supported in part by UCLA–National Science Foundation/Integrative Graduate Education and Research Traineeship Program Award DGE-9987641. L.R.J. was supported in part by the UCLA Chancellor's Dissertation Year Fellowship.
Abbreviations
- AA
amino acid
- BCAA
branched-chain AA
- DeaNO
Z-sodium (Z)-1-(N, N-diethylamino)diazen-1-ium-1, 2-diolate
- IlvD
dihydroxyacid dehydratase
- LeuCD
isopropylmalate isomerase
- NCA
network component analysis
- NO
nitric oxide
- RNOS
reactive nitrogen oxide species
- TF
transcription factor
- TFA
TF activity.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (GEO accession no. GSE7573).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610888104/DC1.
References
- 1.MacMicking J, Xie QW, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 2.Fang FC. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 3.Ignarro LJ. Nitric Oxide: Biology and Pathobiology. San Diego: Academic; 2000. [Google Scholar]
- 4.Soum E, Drapier JC. J Biol Inorg Chem. 2003;8:226–232. doi: 10.1007/s00775-002-0412-9. [DOI] [PubMed] [Google Scholar]
- 5.Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, Poole RK. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding H, Demple B. Proc Natl Acad Sci USA. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster MW, Cowan JA. J Am Chem Soc. 1999;121:4093–4100. [Google Scholar]
- 8.Drapier JC, Hibbs JB., Jr J Clin Invest. 1986;78:790–797. doi: 10.1172/JCI112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hibbs JB, Jr, Taintor RR, Vavrin Z, Rachlin EM. Biochem Biophys Res Commun. 1988;157:87–94. doi: 10.1016/s0006-291x(88)80015-9. [DOI] [PubMed] [Google Scholar]
- 10.Lancaster JR, Jr, Hibbs JB., Jr Proc Natl Acad Sci USA. 1990;87:1223–1227. doi: 10.1073/pnas.87.3.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevanin TM, Ioannidis N, Mills CE, Kim SO, Hughes MN, Poole RK. J Biol Chem. 2000;275:35868–35875. doi: 10.1074/jbc.M002471200. [DOI] [PubMed] [Google Scholar]
- 12.Butler C, Forte E, Maria Scandurra F, Arese M, Giuffre A, Greenwood C, Sarti P. Biochem Biophys Res Commun. 2002;296:1272–1278. doi: 10.1016/s0006-291x(02)02074-0. [DOI] [PubMed] [Google Scholar]
- 13.Borisov VB, Forte E, Konstantinov AA, Poole RK, Sarti P, Giuffre A. FEBS Lett. 2004;576:201–204. doi: 10.1016/j.febslet.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Gardner AM, Helmick RA, Gardner PR. J Biol Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 15.Gardner PR, Gardner AM. Free Radic Biol Med. 2001;31:S78–S78. doi: 10.1016/s0891-5849(01)00569-x. [DOI] [PubMed] [Google Scholar]
- 16.Poole RK. Biochem Soc Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 17.Poock SR, Leach ER, Moir JW, Cole JA, Richardson DJ. J Biol Chem. 2002;277:23664–23669. doi: 10.1074/jbc.M200731200. [DOI] [PubMed] [Google Scholar]
- 18.D'Autreaux B, Tucker NP, Dixon R, Spiro S. Nature. 2005;437:769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- 19.Bodenmiller DM, Spiro S. J Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. PLoS Comput Biol. 2005;1:415–431. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flatley J, Barrett J, Pullan ST, Hughes MN, Green J, Poole RK. J Biol Chem. 2005;280:10065–10072. doi: 10.1074/jbc.M410393200. [DOI] [PubMed] [Google Scholar]
- 22.Justino MC, Vicente JB, Teixeira M, Saraiva LM. J Biol Chem. 2005;280:2636–2643. doi: 10.1074/jbc.M411070200. [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. Proc Natl Acad Sci USA. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pullan ST, Gidley MD, Jones RA, Barrett J, Stevanin TM, Read RC, Green J, Poole RK. J Bacteriol. 2007;189:1845–1855. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Proc Natl Acad Sci USA. 2003;100:15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran LM, Brynildsen MP, Kao KC, Suen JK, Liao JC. Metab Eng. 2005;7:128–141. doi: 10.1016/j.ymben.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Yang YL, Liao JC. Metab Eng. 2005;7:280–290. doi: 10.1016/j.ymben.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Kao KC, Yang YL, Liao JC, Boscolo R, Sabatti C, Roychowdhury V. Abstr Pap Am Chem Soc. 2004;227:U216–U217. [Google Scholar]
- 29.Keefer LK, Nims RW, Davies KM, Wink DA. Methods Enzymol. 1996;268:281–293. doi: 10.1016/s0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- 30.Hyduke DR, Rohlin L, Kao KC, Liao JC. OMICS. 2003;7:227–234. doi: 10.1089/153623103322452369. [DOI] [PubMed] [Google Scholar]
- 31.Salgado H, Gama-Castro S, Martinez-Antonio A, Diaz-Peredo E, Sanchez-Solano F, Peralta-Gil M, Garcia-Alonso D, Jimenez-Jacinto V, Santos-Zavaleta A, Bonavides-Martinez C, et al. Nucleic Acids Res. 2004;32:D303–D306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gill M, Karp PD. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giel JL, Rodionov D, Liu MZ, Blattner FR, Kiley PJ. Mol Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 34.Yang W, Rogers PA, Ding H. J Biol Chem. 2002;277:12868–12873. doi: 10.1074/jbc.M109485200. [DOI] [PubMed] [Google Scholar]
- 35.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 36.Georgellis D, Kwon O, Lin ECC. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- 37.Justino MC, Almeida CC, Goncalves VL, Teixeira M, Saraiva LM. FEMS Microbiol Lett. 2006;257:278–284. doi: 10.1111/j.1574-6968.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- 38.Boehm DE, Vincent K, Brown OR. Nature. 1976;262:418–420. doi: 10.1038/262418a0. [DOI] [PubMed] [Google Scholar]
- 39.Flint DH, Tuminello JF, Emptage MH. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 40.Kuo CF, Mashino T, Fridovich I. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 41.Djaman O, Outten FW, Imlay JA. J Biol Chem. 2004;279:44590–44599. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 42.Roos V, Klemm P. Infect Immun. 2006;74:3565–3575. doi: 10.1128/IAI.01959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanner BL. In: Methods in Molecular Genetics. Adolph KW, editor. Orlando, FL: Academic; 1994. pp. 291–310. [Google Scholar]
- 44.Maloy SR. Experimental Techniques in Bacterial Genetics. Boston: Jones and Bartlett; 1990. [Google Scholar]
- 45.Lee TJ, Pouliot Y, Wagner V, Gupta P, Stringer-Calvert DW, Tenenbaum JD, Karp PD. BMC Bioinformatics. 2006;7:170. doi: 10.1186/1471-2105-7-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiritani K, Wagner RP. Methods Enzymol. 1970;17:745–750. [Google Scholar]
- 48.Cioffi EA, Shaw KJ, Bailey WF, Berg CM. Anal Biochem. 1980;104:485–488. doi: 10.1016/0003-2697(80)90104-9. [DOI] [PubMed] [Google Scholar]
- 49.Wixom RL, Garrett JL, Fetzek JP. Anal Biochem. 1971;42:262–274. doi: 10.1016/0003-2697(71)90034-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.