Abstract
Helioxanthin is a natural product that inhibits the replication of a number of viruses. We found that a previously undescribed helioxanthin analogue, 8-1, exhibited potent anti-hepatitis B virus (HBV) activity with little cytotoxicity. 8-1 suppressed both HBV RNA and protein expression, as well as DNA replication of both wild-type and 3TC-resistant virus. Time-course analyses revealed that RNA expression was blocked first after treatment with 8-1, followed by viral proteins, and then DNA. 8-1 inhibited the activity of all HBV promoters by decreasing the binding of hepatocyte nuclear factor 4 (HNF-4), HNF-3, and fetoprotein factor to the precore/core promoter enhancer II region. The amount of HNF-4 and HNF-3 was decreased posttranscriptionally by 8-1 in HBV-producing cells, but not in HBV-negative cells. Therefore, 8-1 suppresses HBV replication by posttranscriptional down-regulation of critical transcription factors in HBV-producing cells, thus diminishing HBV promoter activity and blocking viral gene expression and replication. This mechanism is unique and different from other anti-HBV compounds previously described.
Keywords: hepatocyte nuclear factors, helioxanthin, HBV promoters
Hepatitis B virus (HBV) infection is still a major world health problem despite the availability of an effective vaccine. Approximately 350 million people are chronically infected with the virus worldwide, including 1.25 million in the U.S. and >200 million in China. HBV infection can persist for the life of the host, often leading to severe consequences such as liver failure, fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). Currently, there is no treatment that completely eliminates the infection in all chronically infected patients. The approved chemotherapeutic treatments (i.e., lamivudine, adefovir, entecavir, and telbivudine) inhibit virus replication by targeting the viral DNA polymerase, and after long-term treatment development of drug-resistant virus becomes problematic. IFN-α is also clinically useful for HBV infection, but it has substantial side effects as well. Some nonnucleoside inhibitors have also been reported to inhibit HBV replication by interfering with nucleocapsid assembly (1). Nevertheless, the search for compounds with novel antiviral targets and mechanisms is still needed.
A series of analogues of a nonnucleoside natural product, helioxanthin, were synthesized and exhibited a broad-spectrum of antiviral activity (2–4). Among those analogues, 5-4-2 and 8-1 (Fig. 1A), in addition to blocking hepatitis C virus [50% inhibitory concentration (IC50) = 1 and 10 μM respectively], HIV (4 and 15 μM), herpes simplex virus type 1 (0.29 and 1.2 μM), Epstein–Barr virus (11 and >25 μM), and human papilloma virus (0.2 and 5.8 μM), were most potent against HBV replication (0.08 and 0.1 μM). Because the cytotoxicity of 5-4-2 is comparatively higher than that of 8-1 (2), 8-1 was chosen for further investigation of its activity against HBV. We found that 8-1 potently inhibits HBV replication by an antiviral mechanism that has not been reported before. This class of compounds is therefore warranted for further development as new drug(s) for treatment of HBV infection and associated diseases.
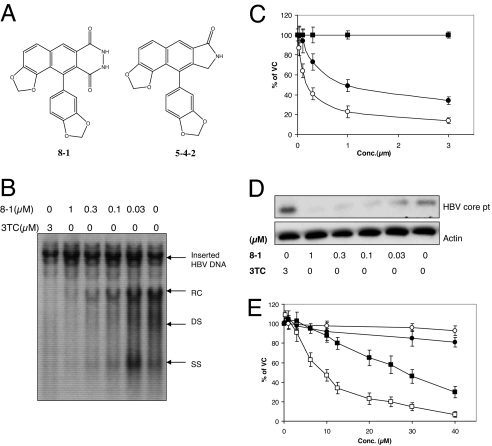
Fig. 1.
8-1 inhibits HBV DNA, RNA, and protein production with little cytotoxicity. (A) Chemical structure of the helioxanthin analogues 8-1 and 5-4-2. (B) Intracellular HBV relaxed circle (RC), double strand (DS), and single strand (SS) DNA synthesis was decreased dose-dependently after treatment with the indicated concentration of 8-1 or 3TC. (C) Inhibition of HBV RNA synthesis by 8-1 and 3TC in HepG2(2.2.15) cells. 8-1 inhibited HBV RNA production dose-dependently, but 3TC did not. Open circles, 8-1 (3.5 kb); filled circles, 8-1 (2.4/2.1 kb); filled squares, 3TC (3.5/2.4/2.1 kb). (D) Western blot analysis of HBV core protein synthesis in HepG2(2.2.15) cells treated by 8-1. 8-1 inhibited HBV core protein synthesis, but 3TC did not. (E) Cell viability after 8-1 and 3TC treatment in HepG2 and HepG2(2.2.15) cells. Open squares, 8-1, HepG2 (2.2.15); filled squares, 8-1, HepG2; open circles, 3TC, HepG2 (2.2.15); filled circles, 8-1, HepG2. All data are presented as the means of three or more independent experiments; error bars indicate standard deviation. VC, vehicle control.
Results
Anti-HBV Activity of 8-1.
The anti-HBV activities of 8-1 were examined by using various HepG2 hepatoma-derived cell lines that stably replicate HBV: HepG2(2.2.15) [ayw serotype, wild-type (WT) genome], HepW10 (adr, WT), and HepD2 (adr, 3TC-resistant with double mutations at rtM204V and rtV180M) (5). In addition, we used an HBV transgenic mouse-derived hepatocyte cell line, HBV-Met (ayw, WT) (6). Southern, Northern, and Western blot analyses were used to measure HBV DNA, RNA, and core protein levels after treatment with 8-1. As shown in Fig. 1B, the various forms of the HBV intracellular DNA replication intermediates were potently inhibited by 8-1 in a concentration-dependent manner as exemplified in HepG2(2.2.15) cells. 3TC was used as a positive control and also inhibited HBV DNA synthesis effectively (7). 8-1 also inhibited HBV DNA expression in other HBV-producing cell lines, with IC50 values similar to those observed in HepG2(2.2.15) cells (Table 1).
Table 1.
Anti-HBV activity and cytotoxicity of 8-1 and 3TC
| Cells | IC50, μM |
CC50, μM |
||||
|---|---|---|---|---|---|---|
| DNA |
pgRNA* |
2.4/2.1 RNA* |
8-1 | 3TC | ||
| 8-1 | 3TC | 8-1 | 8-1 | |||
| HepG2(2.2.15) (ayw, WT) | 0.08 ± 0.02 | 0.05 ± 0.01 | 0.3 ± 0.1 | 0.9 ± 0.1 | 9 ± 1.4 | >100 |
| HepW10 (adr, WT) | 0.2 ± 0.1† | 0.1 ± 0.1† | 0.4 ± 0.2 | >3 | 13 ± 4 | >100 |
| HepD2 (adr, double mutant) | 0.03 ± 0.01† | >3† | 0.09 ± 0.02 | 1.5 ± 0.8 | 12 ± 2 | >100 |
| HBV-Met (ayw, WT) | 0.3 ± 0.2 | 0.2 ± 0.1 | 0.5 ± 0.1 | 1 ± 0.2 | ND | ND |
| HepG2 | 29 ± 3 | ≈100 | ||||
Data are mean values of two or more independent experiments. ND, not determined.
*3TC did not inhibit HBV RNA and protein synthesis in the cell lines studied.
†Determined by inhibition of extracellular HBV DNA by using real-time PCR (3).
HBV RNA expression was also potently inhibited by 8-1 in HepG2(2.2.15) cells (Fig. 1C). The IC50 for the 3.5-kb pregenomic RNA (pgRNA) inhibition was 0.3 ± 0.1 μM, whereas the 2.4/2.1-kb S-RNAs had a slightly higher IC50 of 0.9 ± 0.1 μM. In contrast, viral RNA levels were unchanged after 3TC treatment as expected (Fig. 1C). In HepW10 and HepD2 cells expressing the adr serotype, HBV RNA expression is much greater than HBV DNA. The IC50 for pgRNA inhibition was 0.4 ± 0.2 and 0.09 ± 0.02 μM in HepW10 and HepD2 cells, respectively (Table 1). In fact, RNA from the 3TC-resistant mutant virus was even more sensitive to 8-1 than WT virus (Table 1). Further studies will be necessary to determine whether this is a unique feature of this particular cell line or is a general property of all 3TC-resistant viruses.
HBV core protein expression was also inhibited in a dose-dependent manner by 8-1 with an IC50 of 0.04 ± 0.01 μM in HepG2(2.2.15) cells (Fig. 1D). Again, as expected, 3TC did not inhibit viral core protein synthesis. Core protein synthesis was also inhibited in HepW10 and HepD2 cells (data not shown).
An HBV-transgenic mouse-derived cell line, HBV-Met, was also used to study the antiviral activity of 8-1 (Table 1). This cell line was derived from immortalized murine hepatocytes and produces HBV after differentiation in 2% DMSO (6). The IC50 of 8-1 for HBV DNA in HBV-Met cells was 0.3 ± 0.2 μM, which was slightly higher than in HepG2(2.2.15) cells (Table 1). As with the human-derived cell lines, HBV pgRNA (IC50 = 0.5 ± 0.1 μM) and protein (IC50 = 0.1 ± 0.1 μM) were also inhibited in these cells.
Cytotoxicity of 8-1.
The cytotoxicity of 8-1 was studied by using MTT conversion assays in the HBV-transfected HepG2 cells (Table 1). The concentration of 50% cytotoxicity (CC50) was ≈10 μM in HepG2(2.2.15) cells. Compared with its anti-HBV DNA IC50 (0.08 μM), the cell culture selective index was >100. In HepG2 cells, the CC50 of 8-1 was 29 μM (Fig. 1E), ≈3-fold more than virus-harboring HepG2(2.2.15) cells. The CC50 values of 8-1 in HepW10 and HepD2 were also examined and found to be 13 ± 4 and 12 ± 2 μM, respectively, much higher (30- to 130-fold) than the IC50 for HBV RNA inhibition in these cells. 3TC did not exhibit any significant cytotoxicity even at concentrations as high as 100 μM (Table 1).
Time Course Study of 8-1 Action.
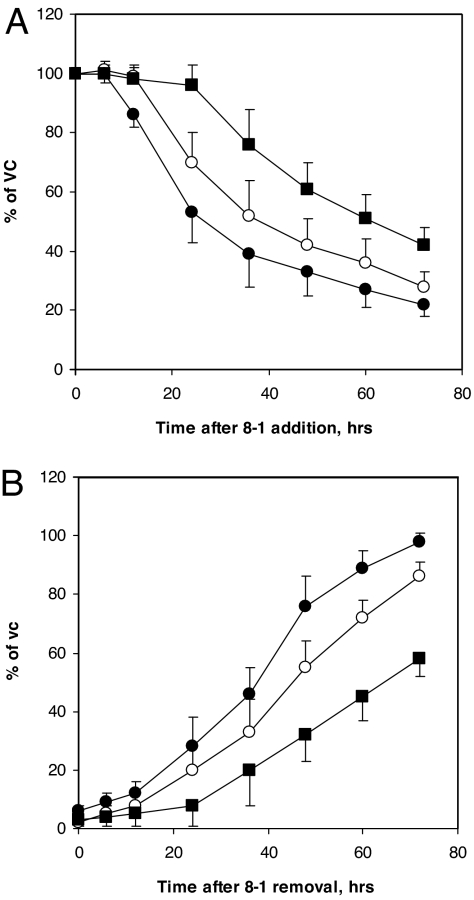
To explore the mechanism of the unique anti-HBV activity of 8-1, a time course study of the inhibition of HBV DNA, pgRNA, and core protein was performed by using HepG2(2.2.15) cells. 8-1 (2 μM) was added to cultures for the indicated time periods (Fig. 2), after which cells were harvested for analysis of viral DNA, RNA, and core protein. We found a significant decrease of viral RNA beginning 18–24 h after incubation with 8-1, followed by a decrease in viral protein levels (Fig. 2A). The HBV DNA declined only after a delay of ≈48 h after drug addition (Fig. 2A). The time course of the rebound of these viral markers after termination of a 6-day 8-1 treatment was also studied. The sequence was similar to the time course of inhibition, that is, viral RNA was the first to rebound 24–48 h after the cessation of treatment, followed by the rebound of viral protein, and lastly the viral DNA (Fig. 2B).
Fig. 2.
Time course of HBV DNA, RNA, and protein expression after 8-1 treatment. (A) HBV DNA, RNA, and protein levels were measured at the indicated time points after addition of 2 μM 8-1 to HepG2(2.2.15) cells. HBV RNA decreased first, followed by protein, and then DNA. (B) Rebound of HBV 3.5 kb RNA, core protein, and DNA upon removal of 8-1 in HepG2(2.2.15) cells treated with 2 μM 8-1 for 6 days. HBV RNA was the first to rebound, followed by protein, and last viral DNA. Filled circles, 3.5 kb pgRNA; open circles, core protein; filled squares, DNA. Data are presented as the mean of three independent experiments; error bars represent standard deviation. VC, vehicle control.
Stability Study of Viral RNA and Protein After 8-1 Treatment.
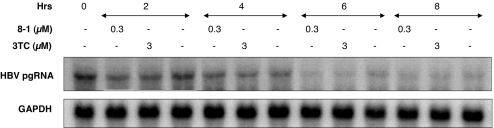
We next determined whether the viral RNA decrease observed in Northern blot analysis was the result of enhanced degradation of viral RNA. A cell line that expresses the HBV 3.5-kb RNA under control of a tetracycline-responsive promoter was used for these studies (8). When cells are cultured without doxycycline, viral pgRNA synthesis is not initiated. When doxycycline (2 μg/ml) is added to the culture medium, HBV pgRNA transcription is induced and accumulates over time (maximal by 4 days). When doxycycline is removed from the medium, transcription of pgRNA terminates, and the existing pgRNA is subject to time-dependent degradation. When an antiviral concentration of 8-1 (0.3 μM) was added to the cells after doxycycline removal, the half-life of the viral pgRNA was not enhanced, similar to that observed with 3 μM 3TC (Fig. 3).
Fig. 3.
8-1 does not impact virus RNA stability. pgRNA production was induced in Met-On-TRE-HBV cells by adding 2 μg/ml doxycycline for 4 days. After doxycycline was removed, 8-1 (0.3 μM) or 3TC (3 μM) was added to the cultures. The cells were collected at the indicated time points, and HBV pgRNA expression was analyzed by Northern blot and compared with GAPDH used as a loading control.
We also assessed whether 8-1 would enhance viral protein degradation. Cycloheximide (Sigma, St. Louis, MO) was added to HepG2(2.2.15) cell cultures at a concentration of 100 μg/ml for 1 h to terminate the cellular protein synthesis. Two concentrations of 8-1 (0.3 and 1 μM) were then added to the cultures for 3, 6, and 9 h, and cell lysates were analyzed by Western blot to examine core protein levels. As with the viral RNA, no enhanced degradation of the viral core protein was observed after 8-1 treatment (data not shown).
8-1 Inhibits HBV Promoter Activity.
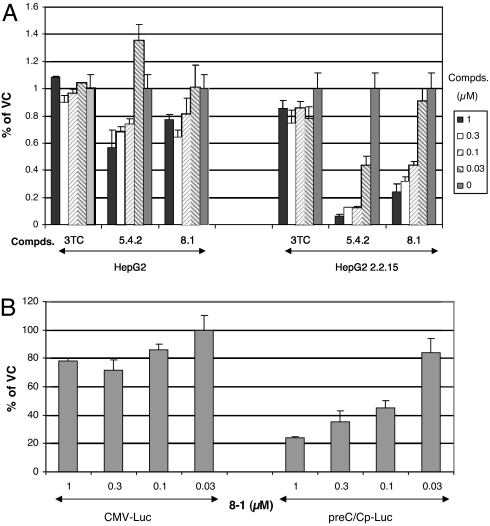
The preceding analysis suggested that the target of action of 8-1 is at the viral RNA transcription step and not at the HBV DNA polymerase, as with other anti-HBV nucleos(t)ide analogues. Therefore, we constructed plasmids containing the promoters (preC/Cp, Xp, pSp, or Sp) for the four different HBV transcripts followed by the luciferase reporter gene to examine the effect of 8-1 on HBV promoter activity. After transient transfection into HepG2(2.2.15) and HepG2 cells, 8-1, 5-4-2, or 3TC was added to the transfected cell cultures, and viral promoter activity was examined. 8-1 and 5-4-2, but not 3TC, inhibited all of the HBV promoters in a dose-dependent manner, as exemplified by the preC/C promoter shown in Fig. 4A. The inhibition was more pronounced in virus-harboring HepG2(2.2.15) than in virus negative HepG2 cells (Fig. 4A). The IC50 was 0.08–0.1 μM in HepG2(2.2.15) cells (similar to the IC50 for DNA inhibition), but was >1 μM in HepG2 cells. This suggests that viral components may play some role in the action of 8-1 in inhibiting HBV promoter activity. The sensitivity of the other HBV promoters to 8-1 treatment was approximately the same as preC/C promoter (data not shown). In contrast, CMV promoter-driven luciferase gene expression was not largely inhibited at a concentration as high as 1 μM of 8-1 in HepG2(2.2.15) cells (Fig. 4B). The inhibition of the viral promoter–luciferase expression was also time-dependent, becoming pronounced only after treatment for a 16–24 h incubation (data not shown).
Fig. 4.
Inhibition of HBV transcription by 8-1. (A) 8-1 and 5-4-2 inhibited preC/Cp-Luc activity in HepG2(2.2.15), but not in HepG2, cells after a 6-day treatment with the drug. (B) 8-1 selectively inhibited the HBV promoter (preC/Cp), but not the CMV promoter, in HepG2(2.2.15) cells. Data are presented as the mean of triplicate experiments (each with duplicate samples) normalized to vehicle-treated cells. Error bars represent standard deviation. VC, vehicle control.
8-1 Modifies the Interaction of Hepatocyte Nuclear Factors (HNFs) with HBV Promoters.
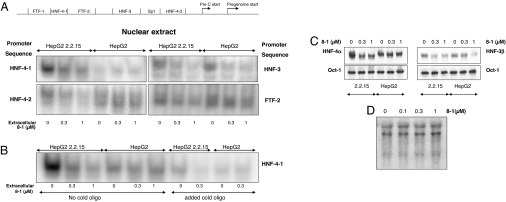
It has been reported that hepatocyte nuclear transcriptional factors, together with viral proteins, bind to the HBV promoters and modulate viral promoter activity (see ref. 9 for a review; see also refs. 10 and 11). DNA oligonucleotides corresponding to the HBV precore/core promoter/Enh II sequence HNF-4, HNF-3, Fetoprotein transcription factor (FTF), SP1 were synthesized and radiolabeled for electrophoretic mobility-shift assays (EMSAs) (Fig. 5A). Nuclear extracts from untreated HepG2(2.2.15) or HepG2 cells were incubated with the radiolabeled probes and 8-1 to determine whether 8-1 altered the binding of nuclear proteins. Even at a dose as high as 100 μM, binding of nuclear proteins was not altered (data not shown). Next, the nuclear extracts from HepG2(2.2.15) and HepG2 cells pretreated with 8-1 for 6 days (as with the HBV assays) were incubated with the probes. The binding to the HNF-4-1, HNF-4-2, HNF-3, and FTF-2 probes was significantly decreased at a dose as low as 0.3 μM (Fig. 5A). The binding to SP1 DNA probe was not affected by 8-1, and the binding of the nuclear extract to FTF-1 did not yield a distinctive shift (data not shown). The decreased binding to HNF-4-1 and HNF-4-2 sequences was only observed in HepG2(2.2.15) cells treated with 8-1, but not in HepG2 cells (Fig. 5A Left). However, the decreased binding to HNF-3 and FTF-2 was observed in both 8-1-treated HepG2(2.2.15) and HepG2 cells, but to varying degrees (Fig. 5A Right). Excessive unlabeled oligomer competed with the radioactive oligomer and extinguished the binding signals, demonstrating specificity (Fig. 5B).
Fig. 5.
Interaction of transcription factors with HBV promoter elements. (A) EMSA analysis of the interaction of defined oligomer DNA sequences of HBV preC/C-Enh II with nuclear extracts from 8-1-treated HepG2(2.2.15) and HepG2 cells. (Left) The decreased binding to HNF-4-1 and HNF-4-2 was only observed in HepG2(2.2.15) cells, but not in HepG2. (Right) The decreased binding to HNF-3 and FTF-2 was observed in both 8-1-treated HepG2(2.2.15) and HepG2 cells, but to a different extent. (B) Unlabeled oligomer added to the EMSA competed with the radiolabeled oligomer and reduced the binding signals. (C) Western blot analysis of the HNF-4 (by using HNF-4α Ab) and -3 (by using HNF-3β Ab) in the 8-1-treated HepG2(2.2.15) and HepG2 cells. Oct-1 was used as an internal control. (Left) HNF-4α was down-regulated in HepG2(2.2.15) cells, but not in HepG2 cells. (Right) HNF-3β was down-regulated in both HepG2(2.2.15) and HepG2 cells, but to a lesser extent in HepG2. (D) Northern blot analysis of HNF-4α mRNA in HepG2(2.2.15) cells treated by 8-1 for 6 days. The HNF-4α mRNA expression was not altered by 8-1.
8-1 Down-Regulates HNF Expression in HBV-Harboring Cells.
To determine whether the decreased binding observed in EMSA assays was due to a decrease in the amount of the HNFs, Western blot analysis was performed by using HNF-4α- and HNF-3β-specific antibodies (Abs) (Santa Cruz Biotechnology, Santa Cruz, CA). As shown in Fig. 5C, the amount of HNF-4α was decreased when cells were exposed to doses of as low as 0.3 μM 8-1 in HepG2(2.2.15) cells, but not in HepG2 cells. However, HNF-3β was down-regulated in both HepG2(2.2.15) and HepG2 cells, but to a lesser extent in HepG2 cells (Fig. 5C Right). There was no impact on the transcriptional factor Oct-1, which served as an internal control and has no binding site at the HBV promoters. Thus, the inhibition of the viral promoter activity by 8-1 could be due to the down-regulation of HNF-4α and -3β (as well as FTF) proteins in the HBV-expressing cells. The combined interaction of this down-regulation likely results in decreased HBV transcription. The decrease in HNF-4α and -3β expression in HepG2(2.2.15) cells could also explain the inhibition of other viral protein promoters (pSp, Sp, and Xp) observed, because all these viral protein promoters have HNF-4α and/or HNF-3β binding sequences (9).
We also studied whether the down-regulation of HNF was at the transcriptional or translational level. HNF-4α mRNA in 2.2.15 cells treated for 6 days with 8-1 was examined by Northern blot analysis. All three isoforms of the HNF-4α mRNA were not down-regulated by 8-1 treatment (Fig. 5D). Therefore, the down-regulation of HNF-4α protein in viral harboring cells is at the posttranscriptional level.
Discussion
8-1 exhibited unique anti-HBV activity by decreasing the expression of all viral macromolecules, i.e., HBV DNA, RNA, and protein, in WT virus (ayw and adr) and a 3TC-resistant variant (adr) with little cytotoxicity. Preliminary studies have indicated that it also has potent activity against duck HBV using established cell culture system Duck HBV DStet cells (12). HBV promoter activity was inhibited by 8-1, and this inhibition was more pronounced in cells harboring virus. The inhibition was also time-dependent and had similar effects on all HBV promoters (preC/Cp, Xp, pSp, and Sp). Treatment with 8-1 did not influence viral RNA stability or viral protein degradation. The binding activity of HNFs (i.e., HNF-4, HNF-3, FTF) in HepG2(2.2.15) cells was significantly decreased in cells treated with 8-1. The decrease of binding was due to the down-regulation of the HNFs, including HNF-4α and -3β, in viral-harboring cells by a posttranscriptional mechanism.
The down-regulation of HNF-3 would not be predicted to affect cellular physiology because this factor is only indispensable for the earliest stage of liver development and differentiation (13). However, this factor may play a very critical role in HBV infection, replication, and mutagenesis (14), as well as interacting with other transcriptional factors to regulate virus replication (15). HNF-4 has been reported to be required for expression of many adult and fetal genes (16), and its knockout influenced survival of the mice (17). Thus HNF-4 down-regulation might be detrimental to cell viability. However, 8-1 down-regulated HNF-4 only in virus-harboring cells, but not in HBV-negative cells. This fact may provide an appreciable selectivity to specifically eliminate virus-infected cells. This selectivity may also explain why 8-1 was more cytotoxic in HepG2(2.2.15) cells than in the parental HepG2 cells. The down-regulation of HNF-4α was not due to a decrease of its mRNA and is likely a posttranscriptional event possibly related to decreased protein synthesis and/or increased protein degradation.
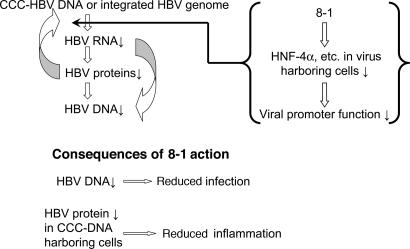
Our current hypothesis of the mechanism of 8-1's anti-HBV action is that 8-1 first inhibits HBV RNA production by inhibiting the transcription of viral RNAs through decreasing the amount of host hepatocyte nuclear transcription factors required for initiation of viral transcription (Fig. 6). This down-regulation only or largely occurs in virus-harboring cells, but not or only slightly in the absence of the virus. This observation suggests that viral factors play important roles in this selective decrease of these hepatocyte nuclear transcription factors. Because of this decrease, the initiation of the HBV RNA transcription apparatus becomes ineffective, and HBV RNA production is inhibited. This in turn decreases HBV protein production and viral capsid assembly. HBV pgRNA reverse transcription and DNA polymerization are thus aborted because these processes occur inside the viral capsid. The effect of reduced viral protein will also feed back to further decrease viral RNA transcription because that process requires viral core or X protein to enhance the initiation of pregenomic 3.5-kb RNA (18) (Fig. 6).
Fig. 6.
Hypothesized mechanism for the inhibition of HBV replication and the consequence of this action. By specific down-regulation of the HNFs in virus-infected cells, 8-1 diminishes HBV replication by blocking HBV RNA expression, leading to reduced protein and DNA levels. This mechanism of action may attenuate the inflammation caused by HBV infection in the liver of patients by reducing the amount of viral antigens present in hepatocytes.
The antiviral mechanism of 8-1 is completely different from typical HBV reverse transcriptase/polymerase inhibitors. Different antiviral mechanism endows 8-1 with a totally different viral resistance profile. Some HBV variants are resistant to polymerase inhibitors but should still retain sensitivity to 8-1-like drugs. Therefore, this class of drug might be a novel approach to combat viral resistance and selectively treat HBV infection and its associated diseases. The consequence of the antiviral action of 8-1 in patients could be multiple. First, diminishing virus replication would prevent new infection of hepatocytes. Second, inflammation caused by HBV infection would be attenuated because viral protein expression is decreased in HBV infected cells (Fig. 6). Finally, 8-1 may also selectively eliminate virus-harboring cells [as seen from more cytotoxic effect in HepG2(2.2.15) cells than in HepG2 cells]. This could raise the possible concern that 8-1 may cause liver failure in patients with limited hepatic reserve or a high percent of infected hepatocytes if the dosage regimen is not optimized. Therefore, in vivo studies will be necessary to address these concerns. Nevertheless, the further development of this class of drugs is warranted.
Materials and Methods
Compounds.
3TC was obtained from GlaxoSmithKline (Research Triangle Park, NC). 8-1 and 5-4-2 were synthesized by H. P. Hsieh (National Health Research Institute, Taiwan, Republic of China).
Cell Cultures.
HepG2, HepG2(2.2.15), HepW10, and HepD2 cells were cultured at 37°C in a humidified 5% CO2/air atmosphere in MEME supplemented with 10% (vol/vol) FCS, and 50 μg/ml kanamycin. Cells were subcultured once a week by detaching the cells with pancreatin (0.5 mg/ml) followed by change of medium on the following day. HBV-Met and Met-On-TRE-HBV (6, 8) cells were cultured on collagen-coated flasks at 37°C in a humidified 5% CO2/air atmosphere in RPMI medium 1640 supplemented with 10% (vol/vol) FCS, 5 mM l-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, 10 μg/ml EGF (BD Biosciences, Franklin Lakes, NJ), 10 μg/ml insulin (Sigma), and 10 μg/ml IGF-2 (Sigma). Cells were subcultured once a week, and fresh medium was added every other day.
Determination of HBV Replication.
HepG2(2.2.15), HepW10, and HepD2 cells were seeded in six-well culture plates at a density of ≈5 × 105 cells per well. HBV-Met or Met-On-TRE-HBV was seeded on collagen-coated 10-cm dishes. Every 2 days, medium was changed. At day 6 after seeding, compounds were added to the cell cultures, and fresh medium was fed every other day for another 6 days. Cells were collected for Southern, Northern, or Western blot analysis for viral DNA, RNA, and protein, respectively. For Southern blot analysis, total DNA was isolated by using DNeasy kits (Qiagen, Valencia, CA) following the manufacturer's instructions. Twenty micrograms of DNA were separated by electrophoresis with 1% agarose and transferred to Hybond-N+ membrane (Amersham Biosciences, Bucks, U.K.) by using the capillary transfer method with 20× SSC overnight. After UV-crossing linked, the blot was prehybridized with 1 mg/ml salmon sperm DNA diluted in QuickHyb reagent (Invitrogen, Carlsbad, CA) for 1 h at 65°C. Random 32P-labeled HBV full-length probe was then added to the prehyb reagent and hybridized overnight at 65°C. The blot was washed with 2× SSC/0.1% SDS twice for 5 min at room temperature (RT) and 0.2× SSC/0.1× SDS three times for 20 min at 65°C. The membrane was exposed to BioMax MR Film (Kodak, Rochester, NY) at −70°C overnight or detected by using a PhosphorImager S1 (Molecular Dynamics, Sunnyvale, CA). For detection of extracellular DNA, a real-time PCR assay was used as described (3).
For Northern blot analysis, total RNA was isolated by using RNeasy kits (Qiagen) following the manufacturer's instructions. First, 20 μg of RNA were separated by electrophoresis on 1% agarose with 5% formaldehyde (Sigma) and then transferred to Hybond-N+ membrane (Amersham Biosciences) as above. The prehybridization and hybridization were performed identically to the Southern blot analysis.
For Western blot analysis, cells were lysed by using 200 μl of 2× lysis buffer (100 mM Tris·HCl, pH 6.8/200 mM DTT/4% SDS/0.2% bromophenol blue/20% glycerol). Next, 10 μl of sample was electrophoresed by 10% SDS/PAGE and then transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA) with transfer buffer (20% methanol/39 mM glycine/48 mM Tris/0.037% SDS) at 110 V for 2 h at 4°C. The membrane was blocked with 5% nonfat dry milk in PBS for 1 h and probed by using a polyclonal Ab specific for HBV core antigen (DakoCytomation, Carpinteria, CA) at 4°C overnight. After incubation with the secondary Ab for 2 h at RT and washing 3 × 20 min using PBS with 0.2% Tween 20, the membrane was subjected to chemiluminescent detection by using routine procedures.
Cytotoxicity Assay.
Cells were seeded in 24-well culture plates at a density of 2 × 104 cells per well. Culture medium was removed 1 day later and replaced with medium supplemented with (or without) the compounds under study. After 3 days of culture, the viability of the cells was determined by Methylene Blue assay. Briefly, the medium was removed from the cells, and the plates were air dried. Then 300 μl of Methylene Blue (Sigma) (2.5%, vol/wt) was added for 2 h at RT. Plates were washed with water and again air dried, and then 0.5 ml of 1% sarkosyl was added to each well and rotated at RT for 3 h. Next, 150-μl samples were transferred to 96-well plates, and the absorbance of the samples at A595 was measured by using an automatic plate reader (Argus 300; Packard, Downers Grove, IL).
HBV Promoter Luciferase Reporter Assay.
The promoter regions of HBV core [nucleotides (nt) 1501–1822], X(nt 831-1280), preS (nt 2461–2820), or S (nt 2871–30) were cloned upstream of the luciferase gene of pTA-Luc (BD Biosciences), respectively. HepG2 and HepG2(2.2.15) were transiently transfected with the reporter vector in a 10-cm2 dish by using Fugene 6 reagent according to the manufacturer's instructions (Roche Applied Science, Indianapolis, IN). Cells were split into 48-well plates 24 h after transfection and continuously cultured for 6 days. Compounds were then added to the medium for 6 days (similar to an antiviral assay). Transcriptional activity was determined by measuring luciferase activity in a multiwell plate luminometer (Tecan US, Durham, NC) using the Luciferase Reporter Assay System (Promega, Madison, WI).
Nuclear Extract Preparation.
HepG2 and HepG2(2.2.15) cell nuclear extract preparations were prepared based on the procedure described by Dignam et al. (19). Cells in 75-cm2 flasks were washed with ice-cold PBS and detached with pancreatin. The cell pellets were washed with 25 ml of cold PBS once and suspended in 1 ml of Dignam A buffer [10 mM Hepes, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.1% Nonidet P-40/0.5 mM PMSF/0.5 mM DTT/one tablet proteinase inhibitors cocktail (Roche Diagnostics, Mannheim, Germany)] on ice for 10 min. The cells were centrifuged for 1 min at 12,000 rpm at 4°C, and the supernatant was collected and stored at −80°C. The nuclear pellets were resuspended in Dignam C buffer (20 mM Hepes, pH 7.9/420 mM NaCl/0.2 mM EDTA/25% glycerol/1.5 mM MgCl2/0.5 mM DTT, with proteinase inhibitors added) and conducted for short ultrasonic treatment. The nuclei were left on ice for 1 h and then centrifuged as above. The supernatant was collected and aliquoted as 10 μl per vial and stored at −80°C. The protein content was measured with a Bio-Rad protein assay.
EMSA.
Nuclear extracts prepared as above were incubated with α-[32P]ATP labeled oligonucleotides in gel-shift binding buffer (10 mM Tris·HCl, pH 7.9/50 mM NaCl/1 mM EDTA/0.05% nonfat dry milk/5% glycerol/0.01% saturated bromophenol blue/50 μg/ml poly-dIdC) for 40 min at RT. Samples were separated by using 5% native polyacrylamide gels followed by autoradiography. Competition assays were performed by incubating the nuclear extracts with unlabeled oligonucleotides on ice for 30 min before the addition of the radiolabeled probe. The sequence of the oligomers used was as follows: HNF-4-1, 5′-GAGGACTCTTGGACTCTCA-3′ (nt 1660–1678); HNF-4-2, 5′-TTAGGTTAAAGGTCTTTGT-3′ (nt 1755–1773); HNF-3, 5′-TCAAAGACTGTGTGTTTAAGGAC-3′ (nt 1710–1732); FTF-1, 5′-GCCCAAGGTCTTACATAAGA-3′ (nt 1642–1661); FTF-2, 5′-AATGTCAACGACCGACCTTGAGG-3′ (nt 1681–1703); SP1, 5′-CTGGGAGGAGCTGGGGGAGGAGATT-3′ (nt 1732–1756).
HNF-4α mRNA Examination.
Total RNA from 8-1-treated HepG2(2.2.15) cells was isolated and subjected to Northern blot analysis similar to HBV RNA determination. A HNF-4α probe was prepared by random labeling a HNF-4α cDNA fragment (1,412 bp) obtained by RT-PCR by using the following primers: forward, 5′-TCTCCAAAACCCTCGTCGACAT-3′; reverse, 5′-TAACTTCCTGCTTGGTGATGGT-3′.
Acknowledgments
We thank Francis V. Chisari (The Scripps Research Institute, La Jolla, CA) for providing HBV Met and Met-On-TRE-HBV cells; Christoph Seeger (Fox Chase Cancer Center, Philadelphia, PA) for Duck HBV DStet cells; Dr. Heing-Pang Hsieh (National Health Research Institute, Taiwan, Republic of China) for synthesizing 8-1 and other helioxanthin analogues; Chuan-Jen Wang, Lam Wing, and Elisabeth A. Gullen for excellent technical assistance; and Ginger Dutschman and Scott Bussom for critical reading and discussing the manuscript. This work was supported by National Institutes of Health Grant R01 AI73299 (to Y.-C.C.) and Yale Liver Center Pilot Project Grant VIP 1426376 (to C.Y.). Y.-C.C. is a Fellow of the National Foundation for Cancer Research.
Abbreviations
- CC50
concentration of 50% cytotoxicity
- FTF
Fetoprotein transcription factor
- HBV
hepatitis B virus
- HNF
hepatocyte nuclear factor
- pgRNA
pregenomic RNA
- RT
room temperature.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, et al. Science. 2003;299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 2.Yeo H, Li Y, Fu L, Zhu JL, Gullen EA, Dutschman GE, Lee Y, Chung R, Huang ES, Austin DJ, Cheng YC. J Med Chem. 2005;48:534–546. doi: 10.1021/jm034265a. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Fu L, Yeo H, Zhu JL, Chou CK, Kou YH, Yeh SF, Gullen E, Austin D, Cheng YC. Antivir Chem Chemother. 2005;16:193–201. doi: 10.1177/095632020501600305. [DOI] [PubMed] [Google Scholar]
- 4.Cheng YC, Ying CX, Leung CH, Li Y. J Clin Virol. 2005;34(Suppl 1):S147–S150. doi: 10.1016/s1386-6532(05)80026-5. [DOI] [PubMed] [Google Scholar]
- 5.Fu L, Cheng YC. Antimicrob Agents Chemother. 2000;44:3402–3407. doi: 10.1128/aac.44.12.3402-3407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasquetto V, Wieland SF, Uprichard SL, Tripodi M, Chisari FV. J Virol. 2002;76:5646–5653. doi: 10.1128/JVI.76.11.5646-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doong SL, Tsai CH, Schinazi RF, Liotta DC, Cheng YC. Proc Natl Acad Sci USA. 1991;88:8495–8499. doi: 10.1073/pnas.88.19.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. Proc Natl Acad Sci USA. 2005;102:9913–9917. doi: 10.1073/pnas.0504273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moolla N, Kew M, Arbuthnot P. J Viral Hepat. 2002;9:323–331. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 10.Raney AK, Johnson JL, Palmer CN, McLachlan A. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert S, Galarneau L, Lamontagne A, Roy S, Belanger L. J Virol. 2000;74:5032–5039. doi: 10.1128/jvi.74.11.5032-5039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo J-T, Pryce M, Wang X, Barrasa MI, Hu J, Seeger C. J Virol. 2003;76:1885–1893. doi: 10.1128/JVI.77.3.1885-1893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sund NJ, Ang SL, Sackett SD, Shen W, Daigle N, Magnuson MA, Kaestner KH. Mol Cell Biol. 2000;20:5175–5183. doi: 10.1128/mcb.20.14.5175-5183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Xie Y, Wu X, Kong Y, Wang Y. Virology. 1995;214:371–378. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 15.Tang H, McLachlan A. J Virol. 2002;76:8572–8581. doi: 10.1128/JVI.76.17.8572-8581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watt AJ, Garrison WD, Duncan SA. Hepatology. 2003;37:1249–1253. doi: 10.1053/jhep.2003.50273. [DOI] [PubMed] [Google Scholar]
- 17.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon JA, Rho HM. Biochem Cell Biol. 2002;80:445–455. doi: 10.1139/o02-133. [DOI] [PubMed] [Google Scholar]
- 19.Dignam JD, Martin PL, Shastry BS, Roeder RG. Methods Enzymol. 1983;101:582–698. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]