Abstract
Manganese toxicity is a major problem for plant growth in acidic soils, but cellular mechanisms that facilitate growth in such conditions have not been clearly delineated. Established mechanisms that counter metal toxicity in plants involve chelation and cytoplasmic export of the metal across the plasma or vacuolar membranes out of the cell or sequestered into a large organelle, respectively. We report here that expression of the Arabidopsis and poplar MTP11 cation diffusion facilitators in a manganese-hypersensitive yeast mutant restores manganese tolerance to wild-type levels. Microsomes from yeast expressing AtMTP11 exhibit enhanced manganese uptake. In accord with a presumed function of MTP11 in manganese tolerance, Arabidopsis mtp11 mutants are hypersensitive to elevated levels of manganese, whereas plants overexpressing MTP11 are hypertolerant. In contrast, sensitivity to manganese deficiency is slightly decreased in mutants and increased in overexpressing lines. Promoter-GUS studies showed that AtMTP11 is most highly expressed in root tips, shoot margins, and hydathodes, but not in epidermal cells and trichomes, which are generally associated with manganese accumulation. Surprisingly, imaging of MTP11–EYFP fusions demonstrated that MTP11 localizes neither to the plasma membrane nor to the vacuole, but to a punctate endomembrane compartment that largely coincides with the distribution of the trans-Golgi marker sialyl transferase. Golgi-based manganese accumulation might therefore result in manganese tolerance through vesicular trafficking and exocytosis. In accord with this proposal, Arabidopsis mtp11 mutants exhibit enhanced manganese concentrations in shoots and roots. We propose that Golgi-mediated exocytosis comprises a conserved mechanism for heavy metal tolerance in plants.
Keywords: Golgi, heavy metal transport, metal tolerance protein, metal trafficking, manganese transporter
Transition metals are required by living systems where they perform a wide variety of functions as cofactors for enzymes and transcription factors. Transition metals are also present in many environments at potentially toxic concentrations, and this has led to the evolution of mechanisms that counter toxicity. In plants exposed to high concentrations of transition metals in the soil, binding of the metals to phytochelatins in the cytosol lowers metal activity (1). Additionally, metals can be removed from the cytosol through the action of metal transporters. Transporters involved in metal tolerance localize to the plasma membrane, thereby removing metals from the cell, or to the vacuolar membrane, where the metal can be sequestered into a large and metabolically relatively inert intracellular compartment (2).
Manganese is the second most prevalent transition metal, after iron, in the Earth's crust and an essential micronutrient for all organisms, including humans and plants (3). In addition to being a cofactor for a variety of enzymes (including various decarboxylases of the tricarboxylic acid cycle, RNA polymerases, and numerous glycosyl transferases), the metal is a constituent of mitochondrial manganese superoxide dismutase. As an integral part of the water-splitting enzyme in photosystem II, manganese is essential for photosynthesis. In biological systems manganese occurs in a variety of oxidation states. We use here the generalized abbreviation Mn.
Excessive exposure to Mn evokes toxicity symptoms. In humans, Mn toxicity manifests itself primarily in the central nervous system and is symptomatically similar to Parkinson's disease (4). In plants, Mn toxicity is a widespread phenomenon on acid and waterlogged soils because soil Mn becomes more available at low pH and in reducing conditions (3). On acid soils, which cover 30% of the Earth's surface (5), Mn toxicity is, together with Al and proton toxicity, a main limiting factor for agriculture and forestry.
Manganese toxicity in plants typically manifests itself as chlorosis, brown specks, necrosis, and crinkled leaves (6). These symptoms appear to result from inhibition of chlorophyll synthesis, Mn and polyphenol accumulation in cell walls, and interference with Ca2+ homeostasis (3). However, symptoms of Mn toxicity vary widely among plant species, as do critical Mn concentrations at which such symptoms are expressed (6–8). Plants that tolerate high Mn concentrations can exhibit distinct compartmentation patterns, such as accumulation in the epidermal cell layer (9) and deposition in trichomes (10). In conditions of high Mn supply, plants accumulate high concentrations of the metal in vacuoles (11) and ectopic expression of vacuolar Mn transporters can increase the Mn tolerance of plants (12, 13).
Compartmentation at tissue and cellular levels is likely to determine the level of Mn tolerance. However, our understanding of Mn homeostasis in plants is still rudimentary (14). Nevertheless, some transport systems have been described as mediating Mn transport across plant membranes. Manganese is able to cross the root plasma membrane nonspecifically through Ca2+-permeable channels (15), although the molecular identity of these channels has yet to be established. In addition, the divalent cation transporter IRT1 is able to translocate Mn actively into the cell (16). Intracellularly, the P-type ATPase ECA1 is believed to supply the endoplasmic reticulum with Mn (17). In accord with a role of the vacuole in Mn sequestration, two Mn transporters have been localized to the vacuolar membrane: AtCAX2 and ShMTP1. In Arabidopsis, AtCAX2, a member of a Ca2+ transporter family, is a H+-driven cation antiporter that is weakly selective among Mn2+, Ca2+, and Cd2+ (12, 18, 19). Although a modest increase in Mn tolerance was conferred by ectopic expression of AtCAX2 in tobacco, overexpressing lines (12) or knockout mutants (18) for this gene did not exhibit an altered Mn tolerance.
The second vacuolar Mn transporter, ShMTP1, is a member of the cation diffusion facilitator (CDF) family of heavy metal transporters and was cloned from the highly Mn-tolerant plant Stylosanthes hamata (13). This protein increased Mn tolerance and accumulation when expressed in yeast or Arabidopsis (13). The CDF gene family is phylogenetically very widely distributed with representatives in bacteria, yeast, plants, and humans (20, 21). The encoded transporters are powered by H+ or K+ antiport (22). Closely related family members usually have similar ionic selectivities but can have different subcellular localizations. The highly similar human Zn2+ transporters Zn-T1, Zn-T2, and Zn-T3, for example, are targeted to plasma membrane, intracellular vesicles, and synaptic vesicles (23). There are 12 CDF members in the Arabidopsis genome (24). The Zn2+-transporting subfamily has been studied most extensively in plants (24–29).
Here we show that an Arabidopsis CDF, AtMTP11 (At2g39450), mediates Mn transport and is able to restore Mn tolerance to a Mn-hypersensitive yeast strain. We demonstrate that AtMTP11 is a determinant of Mn tolerance in plants. Unlike other plant transporters involved in metal tolerance, AtMTP11 localizes neither to the plasma membrane nor to the vacuole, but to a Golgi-associated compartment. This finding suggests a mechanism for metal tolerance involving membrane trafficking. In accord with this hypothesis, mtp11 knockout mutants accumulate more Mn. Because two orthologous genes from poplar are also able to complement the yeast and Arabidopsis mutants and localize identically to AtMTP11, we conclude that this mechanism is conserved and is of general importance in herbaceous and woody species.
Results
Arabidopsis and Poplar MTP11 Genes Complement the Mn-Hypersensitive Phenotype of a pmr1Δ Yeast Mutant and Transport Mn.
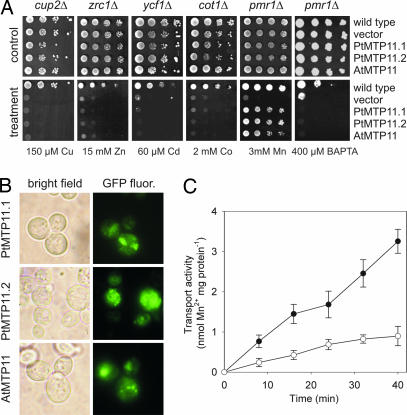
PMR1 encodes a yeast secretory pathway Ca/Mn-ATPase that is located in a Golgi-like compartment (30). pmr1Δ deletion strains are Mn-hypersensitive and accumulate Mn to high concentrations (31). Fig. 1A shows that Mn tolerance of a pmr1Δ strain was restored after transformation with the Arabidopsis CDF member AtMTP11 (At2g39450). The pmr1Δ mutant is also unable to grow on Ca2+-depleted media because the Pmr1 protein is essential for loading of Ca2+ into the ER and Golgi apparatus (32, 33). Fig. 1A shows that AtMTP11 was not able to complement this phenotype, suggesting that AtMTP11 is selective for Mn over Ca2+. To examine whether AtMTP11 affects homeostasis of yeast with respect to exposure to other metals, we tested for complementation of yeast mutants sensitive to Cu2+ (cup2Δ), Zn2+ (zrc1Δ), Cd2+ (ycf1Δ), and Co2+ (cot1Δ) (Fig. 1A). In no case did AtMTP11 alter the phenotypes of these hypersensitive mutants, suggesting that AtMTP11 may be a Mn-specific transporter. Localization in yeast of AtMTP11 with a C-terminal GFP fusion revealed a punctate pattern (Fig. 1B) that resembled that of the Pmr1 protein (30). To examine whether MTP11 has functional orthologs in woody plants, we cloned two homologous genes from poplar, PtMTP11.1 and PtMTP11.2 [supporting information (SI) Text and SI Fig. 7A]. Phenotypes (Fig. 1A) and localization (Fig. 1B) in yeast of both genes were identical to those of AtMTP11, indicating that those genes have a similar function.
Fig. 1.
The Arabidopsis thaliana and Populus trichocarpa MTP11 genes specifically complement the Mn-hypersensitive phenotype of a pmr1Δ yeast mutant. (A) Dilution series of wild-type and mutant yeast strains transformed with PtMTP11.1, PtMTP11.2, AtMTP11, or the empty vector were spotted onto plates supplemented with metals or BAPTA as indicated. (B) Localization in yeast of PtMTP11.1, PtMTP11.2, and AtMTP11 fused to GFP. (C) AtMTP11 mediates 54Mn transport. Microsomal vesicles from pmr1Δ yeast transformed with AtMTP11 (filled circles) or the empty vector (open circles) were assayed for 54Mn2+ uptake activity as described in Materials and Methods. Aliquots (≈100 μg of protein) of the vesicles were preincubated in 500 μl of reaction medium for 5 min at 25°C. MnCl2 containing 54Mn2+ was added at 0 min to a total Mn2+ concentration of 100 μM. The presented data show net uptake subtracted for background activity in the absence of ATP. The experiment was conducted on three different microsomal preparations with similar results. The means ± SE of four independent samples of one microsomal preparation are shown.
To determine Mn transport activity of AtMTP11, we prepared microsomal membrane vesicles from AtMTP11-expressing or empty vector-transformed pmr1Δ yeast. ATP-dependent 54Mn uptake was significantly higher in the AtMTP11-expressing strain (Fig. 1C). After 40 min of 54Mn exposure, vesicles from the AtMTP11-expressing strain had accumulated 3.3 nmol of Mn (mg of protein)−1, compared with 0.9 nmol of Mn (mg of protein)−1 in the control vesicles.
Atmtp11 Insertional Mutants Are Hypersensitive to High Mn.
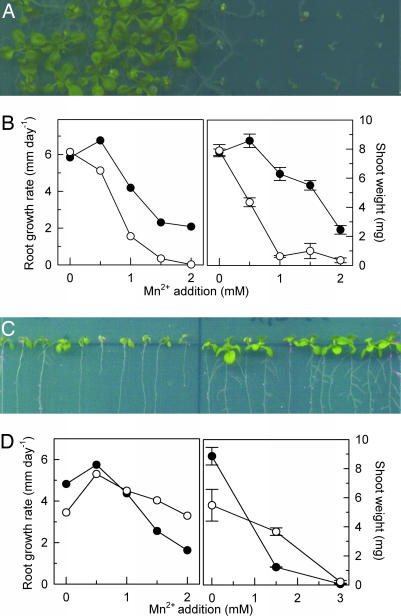
To examine the role of AtMTP11 in planta, we obtained three independent T-DNA insertion lines for the encoding gene (SI Fig. 7C). Two lines, mtp11-1 and mtp11-3, were devoid of full-length transcript, whereas AtMTP11 expression was root-specifically knocked down by 70% in mtp11-2 (data not shown). Shoot growth and root elongation of mtp11-1 (Fig. 2 A and B) and mtp11-3 (SI Fig. 8A) were hypersensitive to elevated levels of Mn. Growth was largely abolished at a concentration of 1 mM Mn, at which wild-type shoot and root growth is only mildly compromised. Complementation of the mtp11-1 mutant with the wild-type MTP11 gene restored its Mn tolerance (SI Fig. 9). Mn sensitivity of the mtp11-2 knockdown line was also increased, albeit less than in the fully knocked-out lines (data not shown). By contrast, we were unable to detect a mutant phenotype with respect to a known Mn-dependent process, i.e., protein glycosylation (SI Fig. 10), suggesting that the principal role of MTP11 relates to Mn tolerance at high concentrations rather than provision of essential Mn. The results indicate that MTP11 plays a central role in Mn tolerance and that there is little functional redundancy with respect to other tolerance mechanisms.
Fig. 2.
The Arabidopsis mtp11-1 mutant is hypersensitive to Mn, whereas an MTP11 overexpressor is Mn-hypertolerant. (A) Growth of wild-type (Left) and mtp11-1 (Right) seedlings on a half-strength MS plate supplemented with 1 mM MnSO4. (B Left) Effect of Mn on root elongation of wild type (filled circles) and mtp11-1 mutant (open circles). Each data point is a linear regression of root elongation of 30 seedlings measured at 4, 6, 8, 11, and 13 days after germination. Regression coefficients of all points are >0.95. (B Right) Effect of Mn on shoot fresh weight of 17-day-old wild-type (filled circles) and mtp11-1 mutant (open circles) seedlings grown on horizontal plates. Data are the means ± SEM of three plates (25 seedlings per line and plate). (C) Growth of Col 0 (Left) and 35S::AtMTP11 line 8 (Right) on a near-vertical half-strength MS plate supplemented with 1.5 mM MnSO4. (D Left) Effect of Mn on root elongation of wild type (filled circles) and 35S::AtMTP11 line 3 (open circles). Experimental details are as in B. (D Right) Effect of Mn on shoot fresh weight of 13-day-old wild type (filled circles) and 35S::AtMTP11 line 3 (open circles) grown on near-vertical plates. Data are the means ± SEM of three plates (10 seedlings per line and plate). For A–D, experiments were repeated twice with similar results.
MTP11 Overexpressors Are Hypertolerant to High Mn.
Because the absence of AtMTP11 caused Mn hypersensitivity, we examined whether overexpression of MTP11 would confer hypertolerance. As determined by quantitative real-time RT-PCR, transformation of Col 0 wild-type Arabidopsis with an MTP11 cDNA under control of a CaMV35S promoter increased MTP11 expression levels in mature leaves 3- to 20-fold (10 independent lines tested; data not shown). Lines 35S::MTP11-3 and 35S::MTP11-8, both showing 20-fold overexpression, were chosen for further examination. Fig. 2 C and D and SI Fig. 8B show that root elongation and shoot growth of seedlings on standard half-strength Murashige and Skoog (MS) medium were reduced in both lines, perhaps as a result of disruption of Mn homeostasis in the overexpressing lines. However, neither shoot nor root growth was significantly affected by 1.5 mM Mn, a concentration that was detrimental to the growth of wild-type plants. Similarly, Mn tolerance of Arabidopsis Col 0 wild type was increased by transformation with CaMV35S-driven PtMTP11.1 and PtMTP11.2 (data not shown). Despite the positive effect of increased MTP11 expression on Mn tolerance, AtMTP11 transcript level was not increased in Col 0 plants exposed to short-term or long-term Mn stress (SI Fig. 11 A and B).
An Arabidopsis mtp11 Mutant Can Be Complemented by Poplar MTP11 Genes.
To examine whether poplar MTP11 proteins have a similar function in planta as their Arabidopsis homologs, we stably expressed both PtMTP11.1 and PtMTP11.2 under control of the CaMV35S promoter in the mtp11-1 mutant. The results in SI Fig. 8C demonstrate that both poplar MTP11s were able to complement the Mn-hypersensitive phenotype of this mutant, indicating that MTP11 plays a similar role in Arabidopsis and poplar.
AtMTP11 Affects Dry Matter Accumulation During Mn Deficiency.
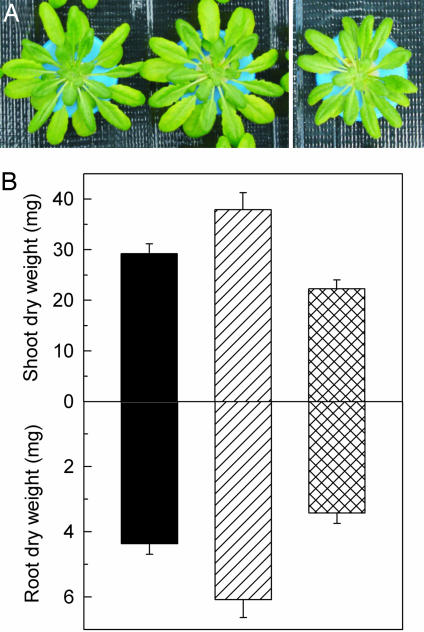
Because AtMTP11 regulates Mn homeostasis in high-Mn conditions, we examined whether AtMTP11 also determines Mn use efficiency of Mn-starved plants. Interestingly, Mn starvation had opposing effects on mutant and overexpression lines to those observed in response to Mn oversupply. Thus, Fig. 3 shows that, relative to wild-type plants, dry matter of both shoots and roots of Mn-starved plants was decreased in a 35S::MTP11 overexpressor and increased in the mtp11-1 knockout mutant, indicating that AtMTP11 negatively affects Mn use efficiency. AtMTP11 expression in Col 0 plants was not decreased during Mn stress (SI Fig. 11C).
Fig. 3.
AtMTP11 affects root and shoot dry-matter accumulation of Mn-deficient plants. (A) Examples of Mn-deficient wild-type, mtp11-1, and 35S::MTP11 plants. (B) Dry-weight accumulation of Col 0 (black bars), mtp11-1 mutant (hatched bars), and 35S::AtMTP11 overexpressor (line 8; crossed bars) plants grown for 34 days in Mn-depleted nutrient solution. Data are the means ± SEM of at least six plants per data point. Differences between mtp11-1 and 35S:MTP11 are highly significant (P < 0.001).
The AtMTP11 Promoter Is Active in Distinct Tissues.
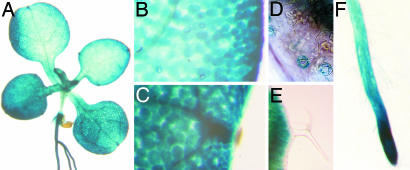
Activity of the AtMTP11 promoter was visualized by fusion to the GUS gene and histochemical detection of GUS activity. GUS expression was visible in shoots and roots (Fig. 4A), but staining was specific to certain tissues. Generally, GUS staining was increased toward the leaf margins (Fig. 4B) and very pronounced in hydathodes (Fig. 4C). In the epidermis, only stomata showed weak GUS staining (Fig. 4D). Interestingly, epidermal pavement cells and trichomes, which are often associated with the accumulation of heavy metals, including Mn, did not stain (Fig. 4E). In roots, staining was most pronounced at the root tip (Fig. 4F). The GUS staining pattern suggested that AtMTP11 does not confer Mn tolerance through Mn sequestration because expression was low in tissues commonly involved in accumulation (trichomes) but high in tissues associated with secretion (hydathodes).
Fig. 4.
Activity of the AtMTP11 promoter is tissue-specific. PrAtMTP11::GUS-transformed seedlings were grown for 2–3 weeks on half-strength MS plates and stained for GUS activity. (A) GUS is expressed in roots and shoots. (B) In leaves, expression is more pronounced at the leaf margin. (C) Expression is particularly high in hydathodes. (D) In the epidermal cell layer, only guard cells show GUS staining. (E) Trichomes do not show GUS staining. (F) In roots, staining is strongest at the root tip. Data for line 5 are shown; five other lines gave comparable results.
Arabidopsis and Poplar MTP11 Proteins Are Targeted to a Golgi-Like Compartment.
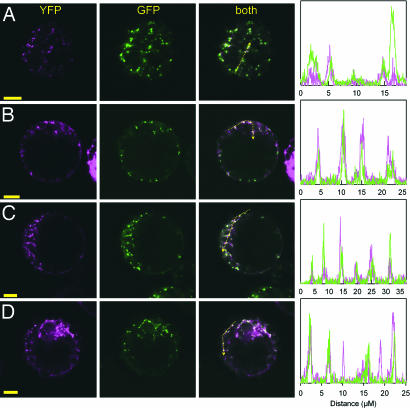
To elucidate the cellular mechanism by which AtMTP11 may confer Mn tolerance, we fused the Arabidopsis and poplar MTP11 cDNAs C-terminally to mGFP5. Transient expression of all MTP11 fusion proteins in Arabidopsis mesophyll protoplasts gave rise to a punctate pattern of GFP-derived fluorescence (SI Fig. 12 A–C). To define the compartment to which MTP11 is targeted, we fused the MTP11 cDNAs to EYFP and transiently expressed these constructs in mesophyll protoplasts from Arabidopsis lines stably transformed with GFP markers. Spectral unmixing allowed the complete separation of GFP and YFP signals without bleed-through between channels (SI Fig. 12D). Expression of AtMTP11–EYFP in a line stably expressing GFP fusions targeted to mitochondria (34) or endoplasmic reticulum (35) clearly showed that AtMTP11 is not targeted to either of those compartments (SI Fig. 12 E and F). In contrast, the AtMTP11–EYFP-derived fluorescence pattern overlapped strongly, but not completely, with that of a trans-Golgi marker, sialyl transferase-GFP (35) (Fig. 5A). In accord with a Golgi-like localization of MTP11, AtMTP11-mGFP5 fluorescence lost its punctate appearance and became clustered when cells were treated with the Golgi-disrupting agent brefeldin A (30–50 μM) (SI Fig. 12 G–H). To assure that the localization was not caused by EYFP or mGFP5 masking a potential C-terminal signal peptide, we constructed an N-terminal EYFP–AtMTP11 fusion. The fluorescence pattern of this construct was indistinguishable from that of the C-terminal fusion protein (Fig. 5B). Localization of both poplar transporters, PtMTP11.1 and PtMTP11.2, fused C-terminally to EYFP was also identical to that of their Arabidopsis ortholog (Fig. 5 C and D).
Fig. 5.
MTP11 attached to fluorescent proteins localizes to a Golgi-like compartment. Confocal images of Arabidopsis mesophyll protoplasts stably expressing Golgi-targeted GFP (sialyl transferase-GFP) and transformed with AtMTP11–EYFP (A), EYFP–AtMTP11 (B), PtMTP11.1–EYFP (C), or PtMTP11.2–EYFP (D). (Scale bars: 10 μm.) Diagrams show intensity plots of EYFP and GFP fluorescence of the profile sketched in the merged image.
Stable expression of the Arabidopsis and poplar MTP11-mGFP5 constructs in the mtp11-1 mutant complemented its Mn-sensitive phenotype to a high degree (data not shown), although GFP fluorescence was not detectable in those lines. However, GFP was detectable by immunoblotting crude membrane fractions of complemented plants (data not shown). To determine the subcellular localization of MTP11-mGFP5 in those lines, microsomal membranes were fractionated by sucrose density gradient centrifugation. Proteins of each fraction were separated by SDS/PAGE, blotted onto nitrocellulose membranes, and probed with a monoclonal GFP-specific antibody. SI Fig. 13 shows that the GFP distribution profile very closely resembled that of latent inosine diphosphatase activity. Because latent inosine diphosphatase is exclusively associated with Golgi membranes (36), this result further supports the Golgi localization of MTP11.
MTP11 Mediates Mn Exclusion.
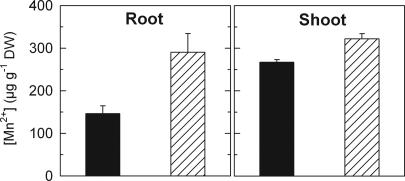
The ability of MTP11 to complement the pmr1Δ phenotype in yeast (Fig. 1A) raises the possibility that MTP11 detoxifies Mn by a mechanism similar to that of Pmr1, i.e., through loading of secretory vesicles and subsequent exocytosis. Accordingly, accumulation of Mn in log phase cultures of pmr1Δ yeast growing in medium containing 1 mM Mn was less than half in AtMTP11-expressing strains [22.7 ± 1.8 pmol of Mn (106 cells)−1] as compared with the vector control [53.5 ± 5.4 pmol of Mn (106 cells)−1] (n = 4). The subcellular localization of AtMTP11 to the Golgi network in plants raises the possibility that MTP11 mediates Mn exocytosis in planta in a similar way. If so, then it would be predicted that mtp11 mutant plants should have a higher tissue content of Mn than wild type despite the intracellular location of the transporter. We therefore measured tissue Mn levels in wild type and mutant. Fig. 6 demonstrates that mutants do indeed contain higher Mn levels than wild type, particularly in root tissue.
Fig. 6.
The mtp11-1 knockout mutant accumulates more Mn in roots and shoots. Col 0 wild-type (black bars) and mtp11-1 mutant (hatched bars) plants were grown hydroponically for 34 days as described in Materials and Methods. Data are the means ± SEM of six plants per data point. The experiment was repeated three times with comparable results. Differences between Col 0 and mtp11-1 are significant (roots, P < 0.05; shoots, P < 0.01).
Discussion
Manganese is an essential transition metal for plants, playing crucial roles as cofactor of many enzymes. However, the metal is required in very small quantities only, and exposure to elevated levels of Mn, which frequently occurs in acidic soils, causes various toxicity symptoms. Thus, like most organisms, plants have developed mechanisms to cope with excess Mn.
At the cellular level, metal toxicity is commonly manifested in the cytoplasm through interference with specific enzyme functions. Consequently, cytosolic free Mn concentrations are likely to be kept in the submicromolar range (11). Metal detoxification mechanisms are therefore typically based on lowering the free concentration of metals in the cytosol. Two well documented strategies in plants are the extrusion of metals via plasma membrane-located transporters and the deposition of (complexed) metals in the vacuole via tonoplast-located transporters. The latter include secondary, proton-coupled systems such as AtCAX2 and ShMTP1. AtCAX2 has been proposed to function in the vacuolar compartmentation in Arabidopsis of transition metals such as Mn and Cd (12, 18, 19), whereas ShMTP1 contributes to Mn tolerance in S. hamata (13).
In yeast and humans, a further metal detoxification pathway has been described that relies on exocytosis via metal-containing vesicles. In this pathway, secretory pathway Ca/Mn-ATPases are believed to confer Mn tolerance by sequestering the metal into secretory vesicles and subsequent exocytosis (31, 37). Although analogous mechanisms have not yet been described for plants, our data regarding characterization of the Arabidopsis and poplar CDFs AtMTP11 and PtMTP11s strongly suggest that a similar strategy is used by plants.
MTP11 Encodes a Mn Transporter That Complements a Yeast Secretory Pathway Ca/Mn-ATPase Deletion Mutant.
Expression of AtMTP11 or PtMTP11s in yeast mutants that are affected in the homeostasis of different metals showed that the MTP11 gene products do not affect yeast capacity to grow in the presence of high concentrations of Cu, Zn, Cd, or Co. In contrast, MTP11 expression specifically restored yeast Mn tolerance in the Mn-hypersensitive pmr1Δ strain. In yeast, MTP11 mediates Mn transport activity, and GFP-labeled MTP11 proteins are targeted to endomembranes in a manner similar to the secretory pathway Ca/Mn-ATPase Pmr1p (30). Thus, the data strongly suggest that, when heterologously expressed in yeast, MTP11 can substitute Pmr1 function for Mn detoxification in a Golgi-associated compartment.
MTP11 Expression Levels Affect Mn Sensitivity in Planta.
Using Atmtp11 loss-of-function mutants and MTP11-overexpressing lines of Arabidopsis grown in the presence of elevated Mn concentrations, we found that, compared with wild-type plants, Mn sensitivity was enhanced in the mutants whereas overexpression led to an improvement of Mn tolerance. Thus, these phenotypes are fully in agreement with MTP11 functioning as a primary determinant of Mn detoxification. Interestingly, in conditions of Mn deficiency, mtp11 mutants performed better, and AtMTP11 overexpressors performed worse, than wild type. This negative impact of AtMTP11 on Mn use efficiency in Mn-limiting conditions is compatible with a function that excludes Mn from the cytosol and hence from access to sites where the metal performs essential metabolic functions. Accordingly, we found that MTP11 expression in wild-type plants was not decreased during Mn starvation.
MTP11 Expression Patterns Point to a Role in Mn Secretion.
Deposition of inorganic and complexed forms of metals in vacuoles constitutes a major mechanism of detoxification (2). A large degree of tissue specificity has been observed in this property, with harmful metals typically accumulating in epidermal tissues (9) and specialized structures such as trichomes (10). Perhaps surprisingly, MTP11 expression is undetectable in these tissues, with expression predominantly in the root tip and leaf hydathodes. Such an expression pattern suggests that MTP11 achieves Mn detoxification through cellular exclusion rather than compartmentation. Thus, root tip cells that are predominantly devoid of large central vacuoles would therefore have to rely on Mn extrusion into the apoplast. Similarly, hydathodes are typically found at the end of vascular tissues and participate in the secretion of water that contains excess salts, including transition metals (38). In contrast, vacuolar deposition of heavy metals is predominant in leaf and root epidermal tissue where MTP11 expression is relatively low.
Our data clearly point to a role of MTP11 in cellular extrusion of Mn. In plants, cellular metal extrusion pathways have hitherto been associated with plasma membrane-located efflux mechanisms, such as the heavy metal ATPase HMA2 (39). However, our localization studies strongly argue against an analogous mode of action of MTP11. We never observed MTP11 localization to the plasma membrane with either the Arabidopsis or poplar isoforms and irrespective of the terminus at which the fluorescent reporter was positioned. In contrast, our data show that both AtMTP11 and PtMTP11s are located on the Golgi network. This localization pattern is compatible with a Mn extrusion function only if the metal is sequestered into vesicles that traffic to the plasma membrane to release the metal from the cell via exocytosis. This mechanism for metal detoxification in plants explains why Atmtp11 mutants have elevated Mn levels, especially in roots where cellular extrusion effectively refluxes the metal into the rhizosphere.
In conclusion, the present set of data including phenotypic characterization of Arabidopsis mutants and overexpressors, yeast complementation, tissue expression, and membrane localization of Arabidopsis and poplar MTP11 are suggestive of a secretory pathway-mediated mechanism of heavy metal detoxification in plants.
Materials and Methods
Details.
The following can be found in SI Text: sequence analysis; yeast strains, plasmids, transformation, and growth methods; Arabidopsis lines, plasmids, and transformation; MTP11 expression analysis; determination of protein glycosylation; and subcellular fractionation, immunoblotting, and inosine diphosphatase assay.
Arabidopsis Growth Methods.
To assess root and shoot growth of seedlings, sterilized seeds were placed onto half-strength MS medium (M0404, pH 6.5; Sigma, St. Louis, MO), solidified with 8 g·liter−1 agar (A1296; Sigma), and stratified for 3 days at 4°C. Plates were placed horizontally or near-vertically into a growth cabinet set to 16/8-h photoperiod at 100–150 μmol·m−2·s−1, 22°C (day)/17°C (night), and 65% relative humidity. In plate assays, each plate contained all lines to be compared to ensure equal growth conditions. For hydroponic culture of plants, seedlings grown for 2 weeks on near-vertical half-strength MS plates were transferred to aerated nutrient solution (30 plants in 750 ml) containing 2 mM KNO3, 0.5 mM (NH4)2SO4, 2 mM CaCl2, 0.5 mM MgSO4, 0.3 mM KH2PO4, 42.5 μM FeNaEDTA, 3.5 μM MnSO4, 0.125 μM CuSO4, 0.25 μM ZnSO4, 17.5 μM H3BO3, 0.05 μM NaMoO4, and 0.0025 μM CoCl2 (pH 6.5). After 7 days, seedlings were transferred into new containers containing the same solution (16 plants in 6 liters). In Mn-deficiency experiments, MnSO4 was omitted from the nutrient solution. Solutions were prepared from salts of the highest purity available (puriss p.a.; Fluka, Buchs, Switzerland). Nutrient solutions were changed every 3–4 days. Hydroponically grown plants were cultivated in short day length (10/14-h photoperiod) and otherwise identical conditions as described for plate cultures.
Elemental Analysis.
For determination of Mn, dried plant tissue was digested by using an open-vessel microwave system (MARS 5; CEM, Mathews, NC) following the protocol in ref. 40. Mn concentration was determined by atomic absorption spectroscopy (SpectrAA 20; Varian, Palo Alto, CA).
Imaging.
GFP and YFP fluorescence of transiently transformed protoplasts were observed by confocal laser scanning microscopy with a Zeiss LSM510 Meta head based on an Axiovert 200M microscope (Zeiss, Jena, Germany). GFP and YFP fluorescence were determined by operating the microscope in lambda mode and spectrally unmixing the images with prerecorded spectra of GFP, YFP, and autofluorescence. Power of the 488-nm laser and amplification were adjusted to avoid saturation of the signal. There was no bleed-through between obtained GFP and YFP signals (SI Fig. 12D).
Mn Transport Assay.
Yeast microsomes were isolated from log-phase cultures of pmr1Δ pFL61 and pmr1Δ pFL61-AtMTP11 yeast according to ref. 41. Vesicles were stored at −80°C in resuspension buffer containing 5 mM BTP/Mes (pH 7.5), 300 mM sorbitol, 5 mM MgCl2, 1 mM DTT, 10 μM PMSF, 1 mg·liter−1 leupeptin, and 2 mg·liter−1 pepstatin. The uptake experiment was performed by the filtration method as described (41) with minor modifications. The experiment was initiated by preincubating 100 μg of microsomal protein in 500 μl of uptake buffer (5 mM BTP/Mes, pH 7.5/300 mM sorbitol/25 mM KCl/1 mM ATP/5 mM MgCl2/1 mM DTT) for 5 min at 25°C. MnCl2 labeled with 54Mn2+ (286.16 GBq·mg−1; PerkinElmer Life Science, Waltham, MA) was added to a final concentration of 100 μM Mn2+. At specific time intervals, aliquots were removed and filtered through buffer-premoistened cellulose acetate microspin filters (0.45 μm). The filters were washed two times with 500 μl of ice-cold washing buffer consisting of 5 mM BTP/Mes (pH 7.5), 300 mM sorbitol, 25 mM KCl, and 1 mM MnSO4. The γ-emission trapped on the filter was measured by using a Ge(Li) detector (Princeton Gamma-Tech, Princeton, NJ). Background values resulting from unspecific adsorption were determined in uptake buffer without ATP, and these values were subtracted from the corresponding values in uptake buffer with ATP to yield the net Mn uptake.
Supplementary Material
Acknowledgments
We thank Tina Peiter-Volk, Sarah Kilmartin, and Frédéric Guinet for excellent technical support. The mtp11-3 T-DNA mutant was generated in the context of the GABI-Kat program and provided by Bernd Weisshaar (Max Planck Institute for Plant Breeding Research, Cologne, Germany). We thank David Logan (University of St. Andrews, St. Andrews, U.K.) and Chris Hawes (Oxford Brookes University, Oxford) for the gift of GFP marker lines. This work was supported by grants from the Biotechnology and Biological Sciences Research Council (to D.S. and F.J.M.M.), the European Union (to D.S.), and the Agence Nationale de la Recherche (to M.C. and D.B.); a postdoctoral fellowship from the Ministère Délégué à l'Enseignement Supérieur et à la Recherche (to B.M.); and the IFR 110 (Génomiques, Ecophysiologie et Ecologie Fonctionnelle).
Abbreviations
- CDF
cation diffusion facilitator
- MS
Murashige and Skoog.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EF453693 (Populus trichocarpa MTP11.1) and EF453694 (Populus trichocarpa MTP11.2)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0609507104/DC1.
References
- 1.Cobbett CS. Plant Physiol. 2000;123:825–832. doi: 10.1104/pp.123.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemens S, Palmgren MG, Krämer U. Trends Plants Sci. 2002;7:309–315. doi: 10.1016/s1360-1385(02)02295-1. [DOI] [PubMed] [Google Scholar]
- 3.Marschner H. Mineral Nutrition of Higher Plants. London: Academic; 1995. [Google Scholar]
- 4.Crossgrove J, Zheng W. NMR Biomed. 2004;17:544–553. doi: 10.1002/nbm.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Uexküll HR, Mutert E. Plant Soil. 1995;171:1–15. [Google Scholar]
- 6.Bergmann W. Nutritional Disorders of Plants. Jena, Germany: Gustav Fischer; 1992. [Google Scholar]
- 7.Peiter E, Yan F, Schubert S. J Plant Nutr. 2000;23:617–635. [Google Scholar]
- 8.El-Jaoual T, Cox DA. J Plant Nutr. 1998;21:353–386. [Google Scholar]
- 9.González A, Lynch JP. Aust J Plant Physiol. 1999;26:811–822. [Google Scholar]
- 10.Blamey FPC, Joyce DC, Edwards DG, Asher CJ. Plant Soil. 1986;91:171–180. [Google Scholar]
- 11.Quiquampoix H, Loughman BC, Rathcliffe RG. J Exp Bot. 1993;44:1819–1827. [Google Scholar]
- 12.Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ. Plant Physiol. 2000;124:125–133. doi: 10.1104/pp.124.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR. Plant Cell. 2003;15:1131–1142. doi: 10.1105/tpc.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pittman JK. New Phytol. 2005;167:733–742. doi: 10.1111/j.1469-8137.2005.01453.x. [DOI] [PubMed] [Google Scholar]
- 15.White PJ, Bowen HC, Demidchik V, Nichols C, Davies JM. Biochim Biophys Acta. 2002;1564:299–309. doi: 10.1016/s0005-2736(02)00509-6. [DOI] [PubMed] [Google Scholar]
- 16.Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. Plant Mol Biol. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, Liang F, Hong B, Young JC, Sussman MR, Harper JF, Sze H. Plant Physiol. 2002;130:128–137. doi: 10.1104/pp.004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittman JK, Shigaki T, Marshall JL, Morris JL, Cheng N-H, Hirschi KD. Plant Mol Biol. 2004;56:959–971. doi: 10.1007/s11103-004-6446-3. [DOI] [PubMed] [Google Scholar]
- 19.Schaaf G, Catoni E, Fitz M, Schwacke R, Schneider A, von Wirén N, Frommer WB. Plant Biol. 2002;4:612–618. [Google Scholar]
- 20.Paulsen IT, Saier MH. J Membr Biol. 1997;156:99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 21.Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. BMC Genomics. 2007 doi: 10.1186/1471-2164-8-107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guffanti AA, Wei Y, Rood SV, Krulwich TA. Mol Microbiol. 2002;45:145–153. doi: 10.1046/j.1365-2958.2002.02998.x. [DOI] [PubMed] [Google Scholar]
- 23.Liuzzi JP, Cousins RJ. Annu Rev Nutr. 2004;24:151–172. doi: 10.1146/annurev.nutr.24.012003.132402. [DOI] [PubMed] [Google Scholar]
- 24.Blaudez D, Kohler A, Martin F, Sanders D, Chalot M. Plant Cell. 2003;15:2911–2928. doi: 10.1105/tpc.017541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desbrosses-Fonrouge A-G, Voigt K, Schröder A, Arrivault S, Thomine S, Krämer U. FEBS Lett. 2005;579:4169–4174. doi: 10.1016/j.febslet.2005.06.046. [DOI] [PubMed] [Google Scholar]
- 26.Persans MW, Nieman K, Salt DE. Proc Natl Acad Sci USA. 2001;98:9995–10000. doi: 10.1073/pnas.171039798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Gustin JL, Lahner B, Persans MW, Baek D, Yun D-J, Salt DE. Plant J. 2004;39:237–251. doi: 10.1111/j.1365-313X.2004.02126.x. [DOI] [PubMed] [Google Scholar]
- 28.Arrivault S, Senger T, Krämer U. Plant J. 2006;46:861–879. doi: 10.1111/j.1365-313X.2006.02746.x. [DOI] [PubMed] [Google Scholar]
- 29.van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JAC, Hooykaas PJJ. Plant Physiol. 1999;119:1047–1055. doi: 10.1104/pp.119.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antebi A, Fink GW. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lapinskas PJ, Cunningham KW, Liu XF, Fink GW, Culotta VC. Mol Cell Biol. 1995;15:1382–1388. doi: 10.1128/mcb.15.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strayle J, Pozzan T, Rudolph HK. EMBO J. 1999;18:4733–4743. doi: 10.1093/emboj/18.17.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dürr G, Strayle J, Plemper R, Elbs S, Klee SK, Catty P, Wolf DH, Rudolph HK. Mol Biol Cell. 1998;9:1149–1162. doi: 10.1091/mbc.9.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Logan DC, Leaver CJ. J Exp Bot. 2000;51:865–871. [PubMed] [Google Scholar]
- 35.Hawes C, Satiat-Jeunemaitre B. Biochim Biophys Acta. 2005;1744:93–107. doi: 10.1016/j.bbamcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Nagahashi J, Nagahashi SL. Protoplasma. 1982;112:174–180. [Google Scholar]
- 37.Xiang M, Mohamalawari D, Rao R. J Biol Chem. 2005;280:11608–11614. doi: 10.1074/jbc.M413116200. [DOI] [PubMed] [Google Scholar]
- 38.Neumann D, zur Nieden U, Schwieger W, Leopold I, Lichtenberger O. J Plant Physiol. 1997;151:101–108. [Google Scholar]
- 39.Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang L, Bell RW, Dell B, Woodward J. Commun Soil Sci Plant Anal. 2004;35:427–440. [Google Scholar]
- 41.Ueoka-Nakanishi H, Tsuchiya T, Sasaki M, Nakanishi Y, Cunningham KW, Maeshima M. Eur J Biochem. 2000;267:3090–3098. doi: 10.1046/j.1432-1033.2000.01343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.