Abstract
Anisotropic expansion of plant cells requires organized arrays of cortical microtubules. Mutations in microtubule-associated proteins and a particular mutation in α-tubulins were reported to cause abnormal microtubule arrays and result in helical growth in Arabidopsis thaliana. However, the way in which these mutations affect the organization of microtubules remains unknown. We here identified 32 Arabidopsis twisting mutants that have either missense or amino acid deletion mutations in α- or β-tubulins. Mutations were mapped to the GTPase-activating region in α-tubulin, intra- and interdimer interfaces of tubulin heterodimers, and lateral contact regions among adjacent protofilaments. These dominant-negative tubulin mutants were incorporated into the microtubule polymer and formed shallow helical arrays of distinct handedness along the long axis of the root epidermal cells. A striking correlation exists between the direction in which cortical helical arrays are skewed and the growth direction of elongating roots. The GTPase-activating-region mutant had left-handed helical arrays composed of highly stabilized microtubules, which could be decorated along the entire microtubule lattices with the otherwise tip-localized End Binding 1 protein. A mutation at the intradimer interface, on the other hand, generated highly dynamic microtubules and right-handed helical arrays. Cortical microtubules in wild type and these two tubulin mutants were composed mainly of 13 protofilaments. This comprehensive analysis of tubulin mutations provides insights into the mechanism by which tubulin structures influence microtubule dynamics and organization.
Keywords: dynamic instability, GTPase-activating region, helix
Differentiated plant cells show a striking diversity of shapes that are important to their functions. As a plant cell expands under the force of turgor pressure, cellulose microfibrils in the wall function as the primary load-bearing structure, and their spatial arrangement generally determines the direction of cell expansion and final cell shape (1). A long-standing paradigm proposes that plasma membrane-associated microtubules direct or constrain the movement of the cellulose-synthesizing complex and thus dictate the deposition patterns of cellulose microfibrils, although the relationship between the cortical microtubules and cell morphogenesis may be more complicated (1, 2).
Rapidly expanding cells in the roots and the etiolated hypocotyls grow longitudinally to form highly elongated cylindrical cells and possess arrays of cortical microtubules that are arranged largely perpendicular to the cell's long axis (3). The importance of transverse arrays for straight growth has been indicated by pharmacological studies and experiments with helical growth mutants of Arabidopsis thaliana. The rapidly elongating cells of these mutants skew consistently either to the right or to the left and exhibit cortical microtubule arrays that form shallow helices with fixed handedness (4). Plant-specific microtubule-associated proteins are mutated in two right-handed helical growth mutants (5–8), whereas an amino acid exchange mutation in α-tubulins resulted in left-handed helical growth (9). These results indicate that dysfunctional cortical microtubules can arrange in helical arrays, rather than normal transverse arrays, and affect the direction of growth of expanding cells.
Cortical microtubules are nucleated at the cell cortex, frequently on existing microtubules (10), and their minus ends usually detach from the original nucleation sites (11). Polymerization-biased microtubule instability at the plus ends and slow depolymerization at the minus ends make cortically attached microtubules migrate across the cell (11). Resulting interactions among microtubules affect the reorientation of microtubules and formation of bundles (12). The dynamics of and interactions among microtubules are thus predicted to contribute to the organization of particular arrays.
In this study, we identified 32 Arabidopsis tubulin mutants that twisted in a fixed orientation and analyzed microtubule dynamics and the protofilament organization in two selected mutants. These tubulin mutants provide insights into how conserved tubulin residues contribute to microtubule dynamics, and how dynamic microtubules attribute to the growth pattern of the root.
Results
Various Tubulin Mutations Cause Directional Helical Growth.
Wild-type Arabidopsis roots grow toward the left side of hard-agar plates containing 3 μM propyzamide, a microtubule-disrupting drug. We screened mutagenized M2 seedlings in the presence of propyzamide for mutants that displayed root growth distinct from the wild type (13). Candidate mutants were then tested for skewed root growth phenotypes in the absence of the drug. After genetic loci for root twisting were mapped with respect to the tubulin loci, closely linked tubulin genes were sequenced. Finally, we obtained 40 tubulin mutant plants that showed either right- or left-handed helical growth in the root and other rapidly elongating organs (Fig. 1) [also see supporting information (SI) Table 2]. Because the same mutations were found independently in several isolates, the total number of tubulin mutations was 32. The majority of tubulin mutants were semidominant and often recovered as heterozygotes from the original screening populations, whereas the remaining weak tubulin mutants behaved like recessive mutants that showed twisting phenotypes only in homozygotes in the absence of propyzamide but were semidominant in their growth responses to the drug. Several semidominant mutants exhibited severe phenotypes, such as isotropic cell expansion, imperfect cytokinesis, disconnected vasclature, and deformed root hairs and trichomes (see SI Fig. 7 and SI Text) and did not grow to the reproductive phase or set few seeds; these mutants were analyzed as heterozygotes.
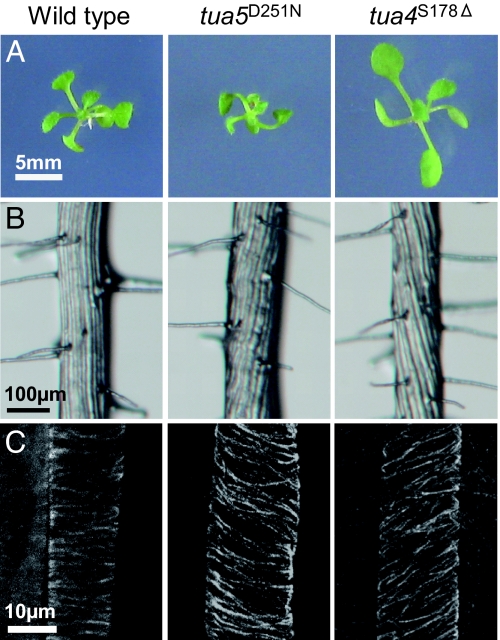
Fig. 1.
Tubulin mutants show skewed growth and helical microtubule arrays. (A) Ten-day-old seedlings. Twisting was most obvious in the cotyledon petioles. (B) Root epidermis in the differentiated zone. Epidermal cell files formed steep helices in tubulin mutants. (C) Cortical microtubules in the epidermal cells of the elongation zone.
We next examined whether mutant tubulin proteins are incorporated into the microtubule and produce dominant-negative effects (SI Fig. 8 and SI Table 2). Most mutations were individually introduced into the Arabidopsis α-tubulin 6 (TUA6) tagged with a Myc epitope at the C terminus or into the Arabidopsis β-tubulin 4 (TUB4) tagged with a multimerized Myc epitope at the N terminus. When wild-type myc-tagged tubulin proteins were expressed in Arabidopsis under the control of the cauliflower mosaic virus 35S promoter, they were incorporated into the microtubule but did not affect seedling growth or morphology. In contrast, ectopically expressed mutant tubulins reproduced the twisting phenotypes upon copolymerization with wild-type tubulins into microtubule polymers.
Molecular Modeling of Mutations on a Tubulin Heterodimer.
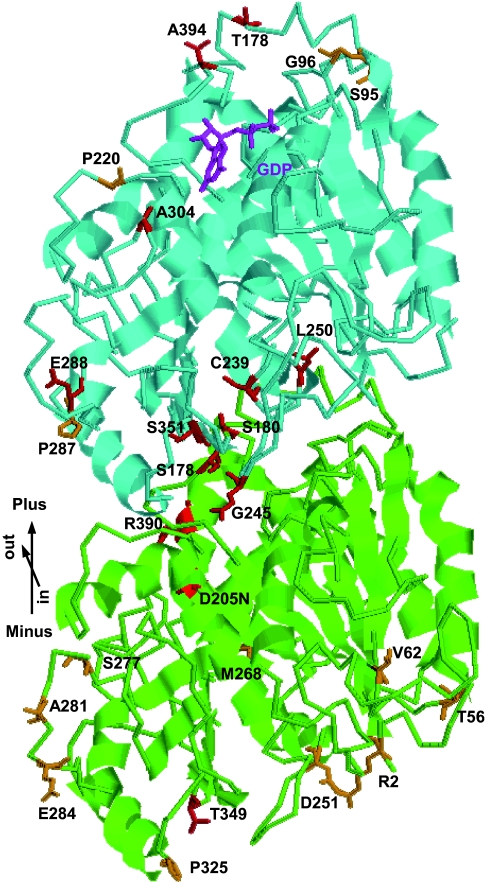
A comparison of amino acid residues among tubulin molecules from different species indicated that mutated residues are invariant or conserved (SI Fig. 9). Close inspection of the 3D structure of the αβ-tubulin heterodimer and the microtubule protofilament (14) shows that mutations are classified into five categories (Fig. 2). Tubulin mutations are described here as a combination of the tubulin isoform and the exchanged amino acids. Three left-handed helical growth mutants (tua4S178Δ, tub4L250F, and tub4S351F) had mutations at the intradimer interface between α- and β-tubulins. The TUA4S178Δ mutation is a precise one amino acid deletion in a loop preceding Helix 5 (15) and situates very close to the mutated residue in lefty1 (TUA6S180F) and lefty2 (TUA4S180F) (9). Mutations at the longitudinal interface between the αβ-tubulin subunits resulted in either right-handed helical growth (TUA6P325S, TUB1S95F, TUB4S95F, TUB4G96D, and TUB4P220S) or left-handed helical growth (TUA2T349I, TUA4T349I, TUB4T178I, and TUB1A394T). The lateral association between the protofilaments is mediated largely by the M loop and its interacting regions of the adjacent protofilament (14). Mutations at the lateral contact regions led mostly to right-handed helical growth (TUA4S277F, TUA6S277F, TUA6A281T, TUA2E284K, TUB2P287L, TUB3P287L, TUB4P287L, TUA2T56I, TUA4T56I, TUA6T56I, and TUA4V62I) but also to left-handed helical growth in one case (TUB4E288K). One right-handed mutant had a TUA5D251N mutation in the critical residue that contacts the guanine nucleotide of β-tubulin during microtubule polymerization, thus activating the inherent GTPase activity in β-tubulin (14). The remaining mutations did not map to particular functional regions defined by the structural analysis. However, because the N terminus of Arabidopsis α-tubulin has been proposed to lie close to the GTPase-activating region (16), the TUA4R2K mutations might interfere with this region.
Fig. 2.
Tubulin mutations generally map to the interacting regions. Tubulin heterodimers are viewed from the microtubule lumen side and with their β-subunits facing the plus end. Residues mutated in the right-handed helical growth mutants are shown in orange, whereas those in left-handed mutants are in red. α-tubulin, green; β-tubulin, cyan; and GDP bound in β-tubulin, magenta. The model is based on a structural model of pig tubulins (14, 15). Note that S95 in β-tubulin corresponds to S96 in TUB1 due to a single amino acid insertion in the N terminus of TUB1.
Root Growth and Microtubule Organization.
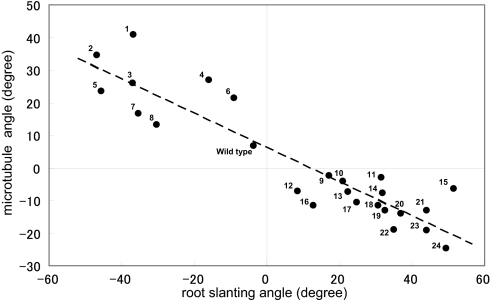
A collection of twisting tubulin mutants offered us a good opportunity to examine the relationship between the array organization of cortical microtubules and the direction of root growth. Cortical microtubules were examined in the epidermal cells at the root elongation zone. Cortical arrays of tubulin mutants were arranged in shallow helices of distinct handedness (Fig. 1C). Mean pitch angles of microtubule helices (SI Fig. 10) were plotted against the skew angles of seedling roots that were grown on the hard agar surface (SI Table 2). Fig. 3 shows a strong correlation (correlation coefficient −0.918) between the two parameters; all of the right-handed helical growth mutants had left-handed microtubule arrays, whereas all of the left-handed mutants had right-handed arrays.
Fig. 3.
Relationship between the root slanting angles and pitch angles of cortical microtubules. Roots that grew toward the right side of the hard-agar plate were assigned the plus sign, whereas roots that grew in the opposite direction had the minus sign in the root-slanting angles. The right-handed helices of cortical microtubules were assigned the plus sign, whereas the left-handed helices had the minus sign. The wild type and all tubulin mutants had the Col background. 1, tub4S351F; 2, tub4T178I; 3, tua2T349I; 4, tua3D205N; 5, tua4S178Δ; 6, tua3R390W; 7, tub4L250F; 8, tub4E288K; 9, tua4R2K; 10, tub4P220S; 11, tub3P287L; 12, tub2P287L; 13, tua6T56I; 14, tua2E284K; 15, tua6M268I; 16, tua6S277F; 17, tua6V62I; 18, tua5D251N; 19, tub1S96F; 20, tua4T56I; 21, tub4G96D; 22, tua4S277F; 23, tua6P325S; and 24, tua6A281T.
Microtubule Dynamics.
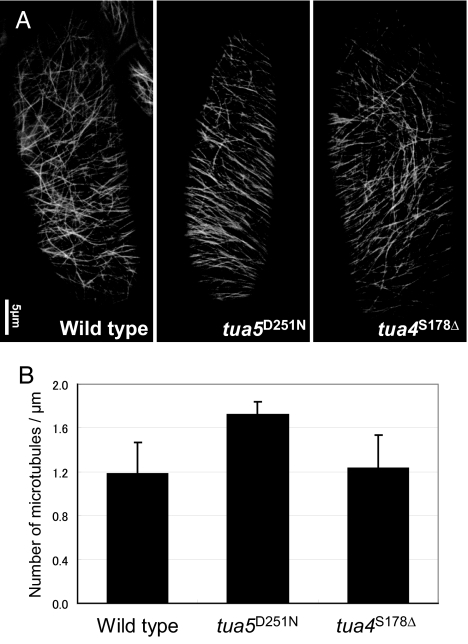
The GFP-TUB6 marker line (17) was used to analyze the dynamic behavior of cortical microtubules in the wild type and two select α-tubulin mutants. It should be noted that elongating epidermal cells of the light-grown upper hypocotyls, which were used for observation, had a more variable microtubule orientation than elongating root epidermal cells. A GTPase-activating region mutant tua5D251N and an intradimer mutant tua4S178Δ respectively represent right- and left-handed helical growth categories, because these mutations should not affect binding sites of microtubule-associated proteins and motor proteins. The microtubules or bundles of microtubules in tua5D251N were more numerous and tended to align more perpendicular to the growth axis than those in the wild type and tua4S178Δ (Fig. 4). The microtubule dynamics in tua5D251N was highly suppressed, as seen by the slower growth and shrinkage (Table 1). Remarkably, the lagging end of tua5D251N microtubules did not depolymerize appreciably and was highly stable. In tua4S178Δ microtubules, the effect was most clearly observed at the leading end, which polymerized more rapidly, resulting in a highly dynamic end. Catastrophe and rescue frequencies of these mutant microtubules were not markedly different from the wild-type frequencies at either end.
Fig. 4.
Microtubules in tua5D251N hypocotyl cells are more abundant and align more transversely. (A) Cortical microtubules in epidermal cells of 4-day-old hypocotyls, visualized by GFP-TUB6. (B) Number of microtubules in unit length. Cortical microtubules that crossed the mid-line of a cell's long axis were counted.
Table 1.
Parameters of microtubule dynamic instability
| Leading ends |
Lagging ends |
||||||
|---|---|---|---|---|---|---|---|
| Wild type | tua5D251N | tua4S178Δ | Wild type | tua5D251N | tua4S178Δ | ||
| Growth rate, μm/min | 4.68 | 2.80* | 7.46* | 1.48 | 1.56 | 2.31* | |
| Shrinkage rate, μm/min | 11.53 | 6.90* | 11.91 | 5.50 | 1.49* | 6.36 | |
| Catastrophe frequency, events/sec | 0.086 | 0.083 | 0.092 | 0.174 | 0.189 | 0.203 | |
| Rescue frequency, events/sec | 0.152 | 0.177 | 0.143 | 0.145 | 0.146 | 0.107 | |
| Time spent, growth | 76.8% | 63.4% | 52.2% | 9.1% | 14.7% | 8.4% | |
| Pause | 9.1% | 15.4% | 9.8% | 46.2% | 55.4% | 56.9% | |
| Shrinkage | 14.0% | 21.2% | 38.0% | 44.7% | 30.0% | 34.7% | |
| Dynamicity, μm/min | 4.89 | 2.65* | 8.41* | 1.89 | 0.45* | 1.56 | |
Wild type, n = 20 and t = 2,444 sec; tua5D251N, n = 10 and t = 1,260 sec; tua4S178Δ, n = 10 and t = 1,228 sec.
*Statistically significant difference from wild type (P < 0.05).
GFP-EB1 Localization.
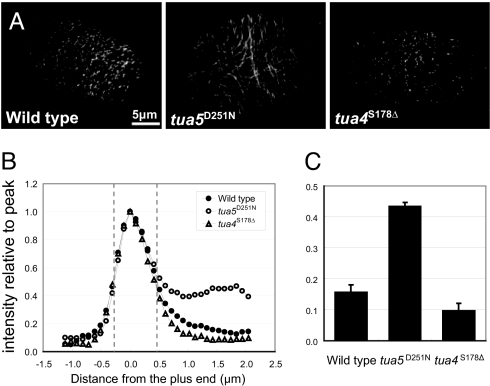
GFP fused to the Arabidopsis plus-end tracking protein EB1b (GFP-EB1; ref. 18) was next used to probe the mutant microtubules (Fig. 5 and SI Movies 1–3). The growth rates of the leading plus end were determined to be 4.54 ± 1.64, 3.77 ± 1.95, and 5.35 ± 1.65 μm/min (each from 12 microtubules observed for longer than 1,000 sec) for the wild type, tua5D251N, and tua4S178Δ, largely consistent with the analysis using GFP-TUB6. In the wild type, GFP-EB1 decorated the growing leading end of microtubules as a comet with a trailing tail. The size of the GFP-EB1 comet was slightly smaller in tua4S178Δ, whereas GFP-EB1 also labeled the entire side wall of tua5D251N microtubules. The fluorescence intensity of the wall relative to the peak intensity at the plus end was 0.16, 0.43, and 0.10 for the wild type, tua5D251N, and tua4S178Δ, respectively. Fluorescence redistribution after photobleaching showed that the photo-bleached GFP-EB1 signal on the wall of tua5D251N microtubules recovered well before the photo-bleached GFP-TUB6 signal (SI Fig. 11), indicating that GFP-EB1 in the cytoplasmic pool directly associated with the microtubule wall.
Fig. 5.
GFP-EB1 decorates tua5D251N microtubules along their entire length. (A) GFP-EB1 usually tracks the plus end of polymerizing microtubules, but in tua5D251N, the marker additionally decorated the side wall of microtubules. Epidermal cells of 4-day-old hypocotyls were analyzed. (B) Tip gradient of GFP-EB1. Mean values are plotted for fluorescence intensity along the length of the labeled microtubules, with the peak intensity being set as 1. The labeled ends grow toward the left. (C) Fluorescence intensity relative to the peak was measured at position 1–2 μm from the GFP-EB1 comet peak.
Protofilament Numbers in Cortical Microtubules.
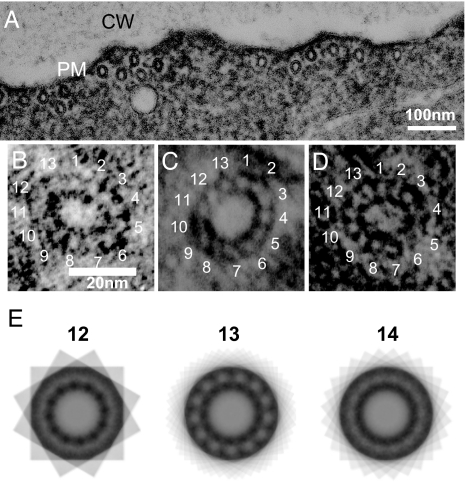
Tubulins in vitro can be polymerized to form microtubules that contain between 9 and 16 protofilaments (19), among which only 13-protofilament microtubules have unskewed protofilaments. When cortical microtubules of the root epidermal cells were examined by electron microscopy after glutaraldehyde-tannic acid fixation (20), we obtained 13 microtubule images (five from wild type, four from tua5D251N, and four from tua4S178Δ) for which the protofilament number could be determined (Fig. 6 A–D). These microtubules were all composed of 13 protofilaments. Markham's rotational analysis (21) also supported this number of 13 protofilaments (Fig. 6E). Therefore, the majority of cortical microtubules that formed either transverse or helical arrays are unlikely to have supertwisted protofilaments.
Fig. 6.
Transverse section of cortical microtubules. (A) Electron micrograph of a longitudinal section of a root epidermal cell in the elongation zone. Cortical microtubules are seen associated with the plasma membrane (PM). CW, cell wall. (B–D) Transverse sections of cortical microtubules from the wild type (B), tua5D251N (C), and tua4S178Δ (D). (E) Multiple exposures of a tua5D251N microtubule, where n = 12, 13, or 14 equal arcs of a complete circle were used. The strongest reinforcement of the ordered pattern occurred when n = 13.
Discussion
Screening for Arabidopsis mutants that displayed twisted growth of elongating organs and abnormal growth responses to propyzamide resulted in a large collection of dominant-negative tubulin mutants. The data presented here provide some of the strongest evidence to date showing conserved residues of both α- and β-tubulins directly contribute to the stability and helical pattern of cortical microtubule arrays.
Incorporation of mutant tubulins into the microtubule polymer suggests that the structure and/or dynamics of cortical microtubules are altered in these mutants. Many of the affected amino acid residues are located at the longitudinal interface of the α- and β-tubulins within and between the αβ-heterodimer and at the lateral interface between two adjacent protofilaments, indicating that the mutations generally affect the protein–protein interactions but not the stability or the folding of tubulin monomers. Several mutated residues, such as P325 and T439 in α-tubulin and S95 and G96 in β-tubulin, have indeed been implicated by atomic models of the tubulin dimer and the microtubule to make direct contact with their interacting partners in the microtubule (14).
Among six Arabidopsis α-tubulin genes, five are expressed in the vegetative stage. Mutations were found in all of the five α-tubulin genes, with a clear preference toward the TUA2/4/6 subfamily, which was expressed more strongly than the TUA3/5 subfamily in the Arabidopsis seedlings (22). Nine Arabidopsis β-tubulin genes, however, are not classified into particular subfamilies. Nine of 17 β-tubulin mutations were found in TUB4; the remaining mutations were located in TUB2, TUB3, and TUB1. The severity of the P287L mutation was greatest in the following order: TUB4>TUB3>TUB2, whereas the severity of the S96F mutation was stronger in TUB4 than in TUB1. Based on the expression profiles in the integrated microarray database Genevestigator (ref. 23; www.genevestigator.ethz.ch/at), TUB4 is expressed most strongly in Arabidopsis roots, followed by TUB2 and TUB3. Therefore, twisting mutants are frequently found in the highly expressed tubulin genes, and their expression levels in the seedling roots largely account for the severity of the mutant phenotypes. We found no evidence for the functional specialization of particular vegetative tubulin genes, at least for the organization of cortical microtubules.
Previously, two left-handed skewing lefty mutants (tua4S180F and tua6S180F) were shown to have shallow right-handed helical arrays (9). In this study using several dozen previously undescribed tubulin mutants, we presented a remarkable correlation between skewed direction of cortical helical arrays and growth direction of elongating roots. Thus, mutant roots skewed in a direction that is roughly perpendicular to the microtubule arrays. This observation is consistent with the current model in which cortical microtubule arrays guide the movement of the cellulose synthase complex in the plasma membrane and deposition of cellulose, thereby dictating the direction of cell expansion (24). This study also demonstrates that the skewed microtubule arrays are not unique to the lefty tubulins but are caused frequently by a wide range of tubulin mutations that affect interactions among tubulin subunits. Such dominant-negative tubulin mutations may underlie the evolution of the natural helical asymmetry seen in tendrils, vines, and other twisting plant organs.
The TUA5D251N mutation affects a critical residue in α-tubulin that triggers GTP hydrolysis in β-tubulin during microtubule polymerization, thus offering a predictable consequence. Hydrolysis of the GTP in β-tubulin within the microtubule would lead to a conformational change, which would strain the lattice. The energy of GTP hydrolysis thus stored in the microtubule drives microtubule dynamic instability (25). A mutation at the corresponding aspartate residue in a yeast α-tubulin inhibited microtubule depolymerization and decreased dynamics (26). We observed comparable changes in the dynamics of tua5D251N microtubules; highly suppressed depolymerization and decreased overall dynamics. Transgenic Arabidopsis plants expressing TUA6D251A/E254A also exhibited similar dynamics (16). The pronounced association of GFP-EB1 with the lateral wall of mutant microtubules may indicate that EB1 recognizes a small conformational difference between GDP-tubulin and GTP-tubulin present in the lattice. The fission yeast EB1 homolog Mal3p was recently shown to bind the microtubule lattice seam as well (27). The tua5D251N microtubules may be useful for studying ill-defined microtubule recognition modes of plant EB1 proteins.
The more dynamic microtubules in tua4S178Δ contrast with the more stable microtubules in tua5D251N. Because these mutants have contrasting microtubule arrays, with either right- or left-handed helices, we believe that microtubule dynamics is one of the major factors that contribute to the final organization of arrays. Transgenic Arabidopsis plants expressing the modified α-tubulin with an N-terminal appendage exhibited enhanced microtubule polymerization and suppressed dynamicity, had a left-handed helical array organization and right-handed skewing of the root (16), thus basically consistent with the present study. Factors other than simple dynamicity, however, may additionally contribute to the microtubule array organization. Low concentrations of microtubule depolymerization drugs cause less dynamic but more destabilized microtubules and right-handed helical arrays (17). Whether the microtubule population is biased toward polymerization or depolymerization would be another key to the array formation. Furthermore, other events may also be important, such as the outcome of intermicrotubule interactions (12) and the branching angle between the original microtubule and the newly formed microtubule (10). Rotary movements of cortical arrays may be another factor that influences the final orientation of arrays (28). Future technological advances should allow us to examine microtubule dynamics and other critical events in root epidermal cells.
To generate a helical array of fixed handedness, the left–right asymmetry may be present in the microtubule lattice itself (4). As an initial step to exploring the possible origin of the array asymmetry, we examined the protofilament numbers in cortical microtubules. Most microtubules assembled in vivo seem to be composed of 13 protofilaments, in which the protofilaments are aligned parallel to the microtubule axis, although many exceptions have been noted in different species and cell types (ref. 29 and refs. therein). In microtubules with protofilament numbers other than 13, the protofilaments are skewed along the microtubule axis, thus forming supertwists. The protofilaments in the 12-protofilament microtubule, for example, form right-handed helices; the 14-protofilament microtubule conversely contains protofilaments skewed in left-handed helices (19). Our observation of cortical microtubules in Arabidopsis root epidermal cells indicated that the wild type and the two twisting mutants had mainly 13-protofilament microtubules, as observed for cortical microtubules of Juniperus and Euphorbia plants (21). The helical asymmetry of microtubule arrays in tubulin mutants thus may not be traced back to the protofilament supertwist. It should be noted, however, that individual polymerizing microtubules in vitro can occasionally switch into energetically unfavorable configurations (30). Ideally, microtubules should be assembled in vitro from wild-type and mutant tubulins and their structure analyzed in detail by cryoelectron microscopy.
Materials and Methods
Plant Materials.
A. thaliana M2 seeds mutagenized using methanesulfonate or T-DNA insertions were obtained from Lehle Seeds (Round Rock, TX) and the Arabidopsis Biological Resource Center (Columbus, OH), respectively. Helical growth mutants were screened in the presence of 3 μM propyzamide, as described (13). All tubulin mutants were backcrossed at least twice before analysis. The majority of the tubulin mutants reported in this study will become available from the Arabidopsis Biological Resource Center.
Immunostaining.
A published protocol (16) was used with some modifications. Anti-α-tubulin antibody YOL1/34 (Millipore, Billerica, MA) and Alexa Fluor 568-conjugated anti-rat IgG (Molecular Probes, Eugene, OR) were used as primary and secondary antibodies to label microtubules at dilutions of 1:40 and 1:500, respectively. Fluorescent images were collected with a C1-ECLIPSE E600 confocal laser-scanning microscope (Nikon, Tokyo, Japan).
Transmission Electron Microscopy.
Whole seedlings were fixed in 2.5% glutaraldehyde and 8% tannic acid buffered with 0.1 M phosphate at pH 6.8 for 3.5 h at room temperature (20). After a brief wash with the buffer solution, the seedlings were postfixed in 1% osmium tetraoxide overnight at 4°C. Dehydration was done in an ethanol series at low temperature and then in propylene oxide at room temperature. Dehydrated seedlings were embedded in Spurr's resin (TAAB Laboratories, Berkshire, U.K.). Sections were cut on an ULTRACUT UCT microtome (Leica Microsystems, Wetzlar, Germany) with a diamond knife, mounted on copper grids, stained with both uranyl acetate and lead citrate, covered with carbon using a VE–1010 vacuum evaporator (Shinku Device Co., Ibaraki, Japan), and examined in an H-7100 transmission electron microscope (Hitachi High-Technologies Corp., Tokyo, Japan) at 75 kV. Wild-type sections were cut in the longitudinal direction, whereas mutant sections were cut in the oblique direction to slice skewed microtubules transversely.
Image Analysis of Microtubule Dynamics and GFP-EB1 Labeling.
Four-day-old seedlings expressing GFP-TUB6 or GFP-EB1 were used for dynamic analysis in the epidermal cells of upper hypocotyls. Microscopy was done as described (16, 17), using a DMRE microscope (Leica, Allendale, NJ) equipped with a CSU10 scanning head (Yokogawa, Tokyo, Japan) and ORCA-ER CCD camera (Hamamatsu Photonics, Shizuoka, Japan). Images were taken in the 16-bit tiff format every 4 sec during the course of 6 min for the analysis of GFP-TUB6 and during the course of 4 min for the analysis of GFP-EB1. Acquired mages were processed and analyzed using ImageJ version 1.34 (http://rsb.info.nih.gov/ij) with domestically developed plugins.
Supplementary Material
Acknowledgments
We thank T. Nakagawa (Shimane University, Nishikawatsu-cho, Matsue-shi, Japan) for pGWB2 and pGWB18, M. Tasaka (Nara Institute of Science and Technology) for a tua4R2K mutant in the Ler background, and N. Kutsuna and S. Hasezawa (University of Tokyo, Tokyo, Japan) for supplying adequate software for data analysis. The Kazusa DNA Research Institute is acknowledged for supplying a TUB4 cDNA. This work was partly supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 1702718 and 18370020 and by a grant for Ground-Based Research for Space Utilization from the Japan Space Forum.
Abbreviations
- TUA
α-tubulin
- TUB
β-tubulin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701224104/DC1.
References
- 1.Baskin TI. Annu Rev Cell Dev Biol. 2005;21:203–222. doi: 10.1146/annurev.cellbio.20.082503.103053. [DOI] [PubMed] [Google Scholar]
- 2.Wasteneys GO. Curr Opin Plant Biol. 2004;7:1–10. doi: 10.1016/j.pbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Sugimoto K, Williamson RE, Wasteneys GO. Plant Physiol. 2000;124:1493–1506. doi: 10.1104/pp.124.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishida T, Thitamadee S, Hashimoto T. J Plant Res. 2007;120:61–70. doi: 10.1007/s10265-006-0039-y. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T. Plant Cell. 2004;16:1178–1190. doi: 10.1105/tpc.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedbrook JC, Ehrhardt DW, Fisher SE, Scheible WR, Somerville CR. Plant Cell. 2004;16:1506–1520. doi: 10.1105/tpc.020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buschmann H, Fabri CO, Hauptmann M, Hutzler P, Laux T, Lloyd CW, Schäffner AR. Curr Biol. 2004;14:1515–1521. doi: 10.1016/j.cub.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 8.Shoji T, Narita NN, Hayashi K, Asada J, Hamada T, Sonobe S, Nakajima K, Hashimoto T. Plant Physiol. 2004;136:3933–3944. doi: 10.1104/pp.104.051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thitamadee S, Tuchihara K, Hashimoto T. Nature. 2002;417:193–196. doi: 10.1038/417193a. [DOI] [PubMed] [Google Scholar]
- 10.Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- 11.Shaw SL, Kamyar R, Ehrhardt DW. Science. 2003;300:1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 12.Dixit R, Cyr R. Plant Cell. 2004;16:3274–3284. doi: 10.1105/tpc.104.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naoi K, Hashimoto T. Plant Cell. 2004;16:1841–1853. doi: 10.1105/tpc.021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nogales E, Whittaker M, Milligan RA, Downing KH. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 15.Löwe J, Li H, Downing KH, Nogales E. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 16.Abe T, Hashimoto T. Plant J. 2005;43:191–204. doi: 10.1111/j.1365-313X.2005.02442.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura M, Naoi K, Shoji T, Hashimoto T. Plant Cell Physiol. 2004;45:1330–1334. doi: 10.1093/pcp/pch300. [DOI] [PubMed] [Google Scholar]
- 18.Mathur J, Mathur N, Kernebeck B, Srinivas BP, Hülskamp M. Curr Biol. 2003;13:1991–1997. doi: 10.1016/j.cub.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Chrétien D, Kenney JM, Fuller SD, Wade RH. Structure (London) 1996;4:1031–1040. doi: 10.1016/s0969-2126(96)00110-4. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara K, Linck RW. Methods Cell Biol. 1982;24:217–233. doi: 10.1016/s0091-679x(08)60657-3. [DOI] [PubMed] [Google Scholar]
- 21.Ledbetter MC, Porter KR. Science. 1964;144:871–874. doi: 10.1126/science.144.3620.872. [DOI] [PubMed] [Google Scholar]
- 22.Abe T, Thitamadee S, Hashimoto T. Plant Cell Physiol. 2004;45:211–220. doi: 10.1093/pcp/pch026. [DOI] [PubMed] [Google Scholar]
- 23.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paredez AR, Somerville CR, Ehrhardt DW. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 25.Nogales E, Wang HW. Curr Opin Struct Biol. 2006;16:221–229. doi: 10.1016/j.sbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Anders KR, Botstein D. Mol Biol Cell. 2001;12:3973–3986. doi: 10.1091/mbc.12.12.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandblad L, Busch KE, Tittmann P, Gross H, Brunner D, Hoenger A. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 28.Chan J, Calder G, Fox S, Lloyd C. Nat Cell Biol. 2007;9:171–175. doi: 10.1038/ncb1533. [DOI] [PubMed] [Google Scholar]
- 29.Chrétien D, Wade RH. Biol Cell. 1991;107:161–174. doi: 10.1016/0248-4900(91)90062-r. [DOI] [PubMed] [Google Scholar]
- 30.Chrétien D, Fuller SD. J Mol Biol. 2000;298:663–676. doi: 10.1006/jmbi.2000.3696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.