Short abstract
A discussion of experimental approaches and theoretical difficulties in the identification of ribozymes with novel catalytic functions.
Abstract
New regulatory RNAs with complex structures have recently been discovered, among them the first catalytic riboswitch, a gene-regulatory RNA sequence with catalytic activity. Here we discuss some of the experimental approaches and theoretical difficulties attached to the identification of new ribozymes in genomes.
Catalysis by RNA was discovered a quarter of a century ago. The discoveries that certain introns were capable of self-splicing [1] and that the RNA moiety of bacterial ribonuclease P (RNase P) on its own could process precursor tRNAs [2] were the first indications that catalytic remnants of a postulated RNA world had persisted until the present day. By the late 1980s, the catalytic scope of RNA had been extended by the discovery of the so-called small nucleolytic ribozymes (or RNA-based enzymes). This family consists of four members: the hammerhead [3], the hairpin [4,5], the hepatitis delta virus (HDV) [6,7] and the Neurospora crassa Varkud satellite (VS) [8,9] ribozymes. All the small nucleolytic ribozymes are involved in the processing of RNA replication intermediates and catalyze a simple RNA cleavage or ligation reaction.
Most present-day ribozymes have as their substrates the conventional 3',5'-phosphodiester bonds in RNA [10]. In arguably the simplest such reaction, the RNA moiety of RNase P catalyzes the hydrolysis of precursor tRNAs (Figure 1a). More frequently, however, ribozymes catalyze a transesterification reaction, as do the small nucleolytic ribozymes, (Figure 1b) and the self-splicing introns (Figure 1c,d). The small nucleolytic ribozymes catalyze the one-step cleavage of a 3',5'-phosphodiester bond, with the formation of a 2',3'-cyclic phosphate and a 5'-hydroxyl in the cleavage products (Figure 1b). Despite having the same reaction mechanism, the small nucleolytic ribozymes differ dramatically from each other in their architecture and exhibit significant variation in the pH profiles of their catalytic activity and in the metal ions required for catalysis [11]. It seems likely that this reaction mechanism is best suited to a simple and single RNA cleavage, as in the processing of multimeric replication intermediates into monomers. Other RNA-cleaving entities that use this mechanism are the in vitro selected leadzyme [12], the protein RNase A [13], and the recently discovered catalytic riboswitch glmS [14], an RNA element that controls gene expression via its ribozyme activity.
Figure 1.
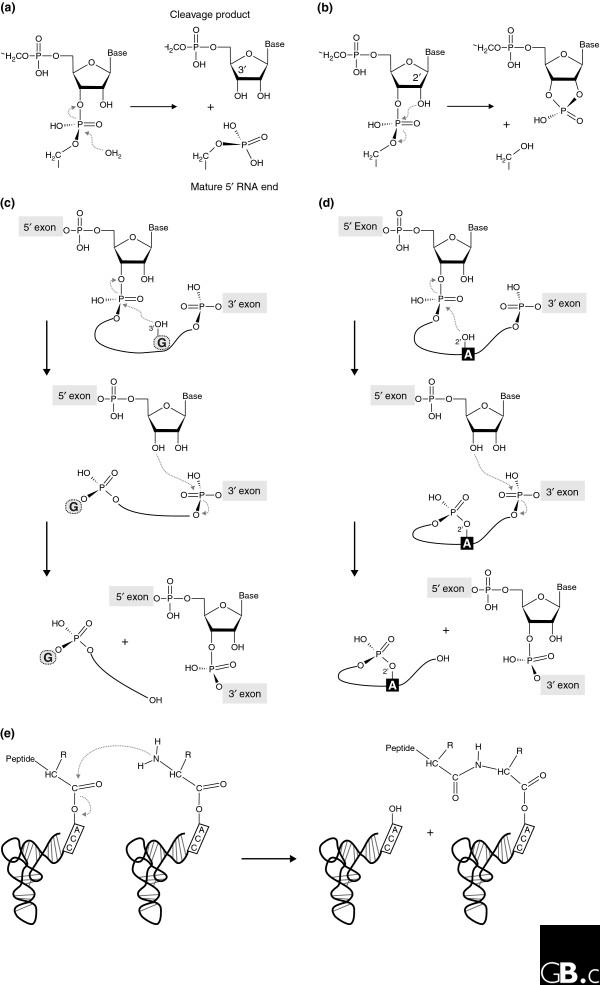
Biochemical reactions naturally catalyzed by RNA. (a) Precursor tRNA hydrolysis by bacterial RNase P yields a phosphate-containing 5' end of the mature tRNA and a 3'-hydroxyl group at the 5' cleavage product. (b-d) Transesterification reactions catalyzed by (b) the small nucleolytic ribozymes, (c) group I introns, and (d) group II introns, in which different chemical groups serve as the attacking nucleophile. In the small nucleolytic ribozymes (b), a defined 2'-hydroxyl attacks the neighboring 3',5'-phosphodiester bond, resulting in a 2',3'-cyclic phosphate and a 5'-hydroxyl in the respective cleavage products. In the first step of group I intron splicing (c), the 3'-hydroxyl of the exogenous guanosine (G) cofactor attacks the 5'-exon-intron junction and sets the 5' exon free, which leads to the covalent attachment of the cofactor to the 5' end of the intron. In a second transesterification reaction, the 5' exon forms a conventional 3',5' bond with the 3' exon, releasing the linear intron with the additional guanosine [1]. In group II introns (d), the conserved branch-point adenosine (A) serves as the nucleophile, leading to the formation of a lariat intron. (e) Peptide-bond formation catalyzed by the ribosome.
In contrast to this simple reaction, self-splicing of the group I and group II introns involves two consecutive reaction steps (Figure 1c,d). The first frees the 3'-OH of the 5' exon, which allows, in the second step, an attack of the phosphodiester at the junction between the last residue of the intron and the first residue of the 3' exon. Self-splicing group I introns make use of the 3'-hydroxyl of an exogenous guanosine as the initial attacking nucleophile; the guanosine is phosphorylated in the reaction and released (Figure 1c). In the self-splicing group I introns, the formation of an intermediate with a 2',3'-cyclic phosphodiester bond has not been observed, probably because that might entail a loss of structural integrity in the spliced exons by the formation of 2',5'-phosphodiester connectivity in the second reaction step [15]. A similar two-step strategy is adopted by the self-splicing group II introns [16,17], but in this case the attacking nucleophile is the 2'-hydroxyl of the conserved intronic branchpoint adenosine (Figure 1d). While this forms an RNA lariat in the intron, the structural integrity of the connected exons is ensured. It should be noted that the splicing of tRNA introns in the Eukarya and the Archaea does not result from self-splicing as in the Bacteria, but starts with the action of an endonuclease, a protein enzyme, which leaves 2',3'-cyclic phosphate termini [18-20].
The persistence of the RNA world has been splendidly confirmed by the demonstration that the ribosome is a ribozyme - that is, the ribosomal RNA components are the catalytically active elements in polypeptide synthesis [21] - placing ribozyme activity at the heart of modern cells and showing that ribozymes could catalyze reactions other than the cleavage and ligation of RNA (Figure 1e). The first indications of catalytic RNA in the ribosome came from biochemical data [22] that showed persistence of ribosome catalytic activity after digestion and denaturation of the ribosomal proteins. The final proof that rRNA is the catalyst in protein biosynthesis came from crystallographic work that showed that the peptidyltransferase reaction center of the ribosome is devoid of any protein component, and is made up exclusively of rRNA residues [21].
In the past few years, a number of new catalytic RNA molecules have been discovered, including a catalytic riboswitch, and known elements have been detected at new genomic locations. Table 1 lists the currently known naturally occurring catalytic RNAs. Do we now know the full spectrum of the diversity and versatility of catalytic RNAs, or are there yet more to be discovered? In this article we will focus on the approaches used to identify novel catalytic RNA species and on the accompanying experimental and bioinformatic difficulties. To solve some of these problems, new bioinformatic tools that better integrate our current understanding of RNA architecture, molecular biology and evolution will have to be developed.
Table 1.
The natural occurrence of ribozymes and riboswitches
| Type | Species* | Rfam accession number† | |
| Ribozyme | Group I intron | Thermus thermophila [1] | RF00028 |
| More than 20,000 sequences from all three kingdoms‡ | |||
| Didymium iridis (branching enzyme, group I intron derivative) [46] | |||
| Group II intron | Saccharomyces cerevisiae mitochondria [17,18] | RF00029 | |
| More than 8,000 sequences from all three kingdoms‡ | |||
| Hammerhead | Tobacco ringspot virus satellite RNA (sTRSV) [3] | RF00008 | |
| Several additional satellite RNAs of plant viruses§ | RF00163 | ||
| Viroids of the Avsunviroidae family [99,100] | |||
| Carnation small viroid-like RNA (CarSV RNA) [101] | |||
| Satellite DNAs of various amphibian species [102,103], Schistosoma mansoni [30] and Dolichopoda cave crickets [31] | |||
| Arabidopsis thaliana genome [33] | |||
| Hairpin | Tobacco ringspot virus satellite RNA (sTRSV) [4] | RF00173 | |
| Two additional satellite RNAs of plant viruses: sCYMV and sARMV [104] | |||
| HDV | Human hepatitis delta virus RNA [6] | RF00094 | |
| Homo sapiens genome (intronic) [29] | |||
| RNase P | Escherichia coli [2] | RF00010 | |
| More than 1,000 sequences from various Bacterial phyla¶ | RF00011 | ||
| Archeal phyla: Crenarchaeota, Euryarchaeota [105] | RF00373 | ||
| VS | Neurospora crassa Varkud satellite [8] | NL | |
| Catalytic riboswitch | Glms riboswitch | Bacillus subtilis [14] | RF00234 |
| Bacterial phyla: Actinobacteria, Firmicutes | |||
| Riboswitch | Adenine¥ | B. subtilis [106] | RF00167 |
| Bacterial phyla: Proteobacteria, Firmicutes | |||
| Coenzyme B12 | E. coli and Salmonella typhimurium btuB mRNAs [107] | RF00174 | |
| Bacterial phyla: Actinobacteria, Proteobacteria, Deinococcus-thermus, Bacteroidetes, Spirochaetes, Chloroflexi, Firmicutes, Fusobacteria, Cyanobacteria, Thermogales | |||
| Flavin mononucleotide (FMN) | 20 Gram-positive and Gram-negative bacteria [23,24,108] | RF00050 | |
| Bacterial phyla: Actinobacteria, Deinococcus-thermus, Thermus/deinococcus group, Proteobacteria, Firmicutes, Thermotogae, Fusobacteria, Thermogales | |||
| Guanine¥ | B. subtilis [25] | RF00167 | |
| Bacterial phyla: Proteobacteria, Firmicutes | |||
| Glycine | B. subtilis [109] | RF00154 | |
| Bacterial phyla: Actinobacteria, Proteobacteria, Fusobacteria, Firmicutes | |||
| Lysine | B. subtilis [25,110-112] | RF00168 | |
| Bacterial phyla: Proteobacteria, Thermogales, Firmicutes | |||
| Intracellular magnesium | S. enterica [113] | NL | |
| S-adenosylmethionine (SAM) | B. subtilis [114-116] | RF00162 | |
| Bacterial phyla: Cyanobacteria, Actinobacteria, Proteobacteria, Firmicutes | |||
| Thiamine pyrophosphate (TPP) | Rhizobium etli [117] | RF00059 | |
| Bacterial phyla: Actinobacteria, Deinococcus-thermus, Bacteroidetes, Proteobacteria, Thermus/deinococcus group, Spirochaetes, Chloroflexi, Firmicutes, Fusobacteria, Cyanobacteria, Thermogales | |||
| Eukaryal phyla: Metazoa, Cercozoa, Fungi, Viridiplantae | |||
| Archeal phyla: Euryarchaeota |
*For each type, the first entry represents the species in which the ribozyme or riboswitch was originally discovered. †Rfam accession numbers for detailed sequence listings [118]; NL, not listed in Rfam. ‡Detailed sequence listings also at [119]. §Detailed sequence listings also at [120]. ¶Detailed sequence listings also at [121]. ¥Adenine and guanine riboswitches are listed together as purine riboswitch in Rfam.
The discovery of riboswitches and new catalytic RNAs
Riboswitches are bimodular RNAs that are made up of a ligand-binding region (an aptamer) and a domain that controls gene expression. They are usually located in the 5' untranslated regions of bacterial mRNAs, where they control the expression of the gene by binding a low molecular weight metabolite that triggers a conformational change in the RNA [23-26]. In recent years, many of these genetic control elements have been discovered, and it has become clear that they are structurally and functionally highly diverse [27,28]. Riboswitches control gene expression at both the transcriptional and translational levels, and can act as 'on' or 'off' switches. The majority of riboswitches are negative control elements, and among these, the first catalytic riboswitch discovered - glmS [14] - employs the ultimate method of switching off gene expression: when it binds its cognate ligand it cleaves itself, thus destroying the function of the mRNA of which it is a part.
The biological function of other recently discovered catalytic RNAs is less clear. Using an ingenious in vitro selection scheme, Szostak and co-workers [29] recently discovered an HDV-ribozyme-like element in an intron of a human mRNA and have demonstrated its biochemical activity. In this scheme, a library of uniformly sized, small circular DNAs was used as templates for rolling-circle transcription; self-cleaving RNAs can thus be identified by the appearance of unit-length RNA fragments. Cedergren and co-workers identified and biochemically characterized hammerhead ribozymes in the genomes of schistosomes [30] and cave crickets [31], and, using database searching, our group recently identified novel examples of hammerhead ribozymes [32] and found two hammerhead sequences encoded at distinct loci in the genome of Arabidopsis thaliana that we have characterized as catalytically active in vitro and in vivo [33].
Ribozyme topology versus sequence conservation
To carry out RNA-based chemical catalysis, some parts of the ribozyme molecule must adopt very precise relative positions and orientations. In addition to specific recognition, there must be dynamic mechanisms for substrate binding and product release. With the notable exception of the ribosome, present-day ribozymes act on the phosphodiester backbone linking two consecutive nucleotides. Although the catalytic processes of such reactions are basically similar, they can be achieved in diverse ways and, in addition, as chemical convergence is pervasive, ribozymes display a rich repertoire of architectures that position the reactants appropriately. Furthermore, the number of conserved nucleotides and their dispersion throughout the molecule vary considerably from one ribozyme to the other: for example, the hammerhead ribozyme and the group I introns have about the same number of conserved residues - around seven - although the latter can be up to four times as large [34,35]. The positions and relative dispositions of the conserved structural elements with respect to the beginning and end of the ribozyme motif also vary (Figure 2). Most families of ribozymes can be subdivided into classes distinguished by their highly non-homologous peripheral elements [36-38]. However, the three-dimensional architectures of the ribozyme cores belonging to the same family are expected to be similar because they are maintained by tertiary constraints which, despite the conservation of short sequence segments, can form in diverse ways.
Figure 2.
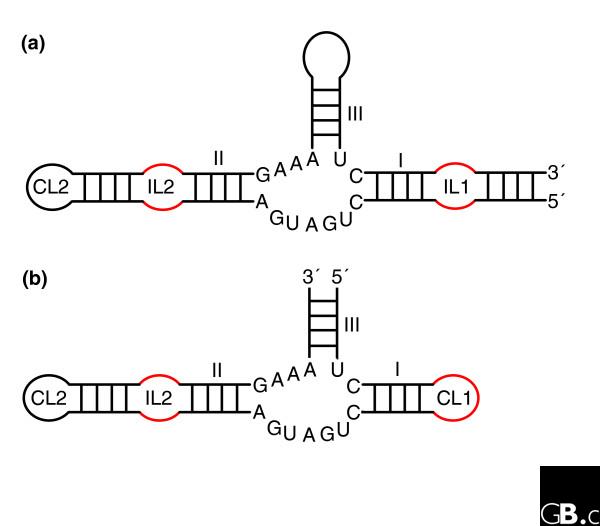
The hammerhead ribozymes are based on a three-way junction and there are two main types. (a) Type I has the ends of the single-stranded RNA on stem I; (b) type III has the ends of the single-stranded RNA on stem III. For unknown reasons, potential type II ribozymes (ends of the single-stranded RNA on stem II) have never been observed. The three-dimensional architecture is maintained by coaxial stacking of stems II and III, which, through constraints in the conserved three-way junction residues [92], orients stem I so that loop-loop interactions between stems I and II form (Figure 3) [40,42]. The internal loop of stem II (IL2) is often replaced by a capping loop (CL2); similarly, CL1 in type III can be replaced by an internal loop (IL1) followed by another hairpin. Although only one structure has been fully characterized, sequence alignments show that the loop-loop interactions (mainly constituting non-Watson-Crick pairs) are very diverse.
The hammerhead ribozyme well illustrates the difficulties of identifying new ribozymes either experimentally or by in silico approaches. Indeed, an incomplete catalytic RNA fold, which did not include tertiary contacts between elements away from the catalytic site of cleavage and sequence conservation, was accepted for a long time, until the full hammerhead ribozyme was (re)discovered [39-41]. A recent crystal structure [42] shows how the presence of tertiary contacts between loops far removed from the catalytically conserved region induces conformational changes in the core that promote the active state of the ribozyme. Importantly, all those contacts involve networks of non-Watson-Crick base pairing with patterns of evolution unlike those of Watson-Crick base pairs [40,43]. Fully biologically active hammerhead ribozymes possess structural complexity and strict sequence requirements (Figure 3b), but because of the non-Watson-Crick pairings, this is not immediately apparent from the sequence alone. In contrast, because of its convoluted pseudoknotted topology based on Watson-Crick pairs, the HDV ribozyme reveals most of its complexity immediately (Figure 3a). Incomplete hammerhead ribozymes without peripheral elements and with low sequence and structural complexity display reduced catalytic activities. Indeed, in vitro evolution, starting from random libraries, produced structurally diverse ribozymes with low activity, which contained some hammerhead variants [44]. Another experiment selecting in vitro for self-cleaving motifs with hammerhead-type biochemical activity [45] led to the conclusion that the hammerhead motif makes the most common ribozyme fold and suggested that this motif has had multiple independent origins. The long-range interactions were not considered in those two in vitro selection schemes, as their importance had not been recognized at the time. The sequences collected during the second selection scheme would enable optimization of hammerhead ribozyme activity.
Figure 3.
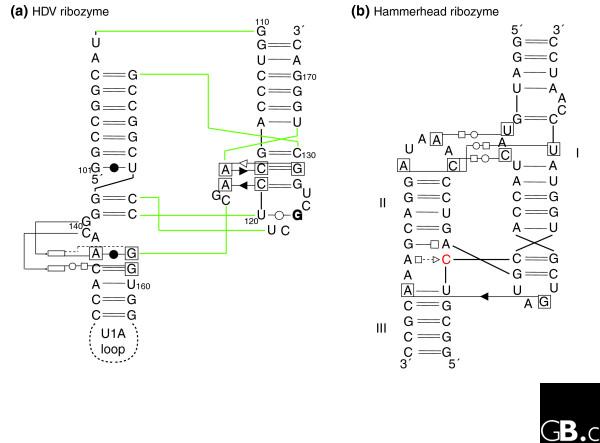
Schematic diagrams of the interaction networks maintaining the three-dimensional architecture of two different ribozymes. (a) The HDV ribozyme [7,93]; (b) the active hammerhead ribozyme [42]. The HDV ribozyme has a convoluted pseudoknotted topology: the color lines indicate the path of the sugar-phosphate backbone. The nomenclature is as follows [75]. Each nucleotide has three edges with hydrogen bonding possibilities: the Watson-Crick edge (denoted by a circle), the Hoogsteen edge (denoted by a square) and the sugar edge (denoted by a triangle). A pairwise base-base interaction can be formed either with the attached sugar moieties on the same side of the line of approach (cis-configuration, the symbols are closed) or with the sugars on either sides of the line of approach (the trans-configuration, the symbols are open). To avoid ambiguities, when annotating tertiary contacts, the nucleotides that are involved have been boxed. When the base of a nucleotide is in the syn-conformation with respect to the sugar it is marked in bold. The rectangles indicate the position actually occupied in space by a nucleotide. In (b), the cleavage occurs 3' of the red C.
Ribozyme topology versus sequence variability
A ribozyme with a new branching activity, GIR1, has recently been experimentally identified in slime molds [46]. On the basis of its secondary structure, this ribozyme belongs to the group I intron family. It carries out the first cleavage step of a group II intron, however, leading to the formation of a small lariat with a 2',5'-linkage at the 5' end of the endonuclease mRNA of which it forms a part, thereby protecting the message from exonuclease degradation. Thus, in this case, a similar secondary structure scaffold is the basis for two ribozymes catalyzing different chemical reactions: activation of an internal O2'-hydroxyl group in the case of the new ribozyme compared with activation of an O3'-hydroxyl group of an external cofactor for the rest of the group I intron family (Figure 1c). This is yet another example of the fact that similar RNA sequences can assume two different folds and catalyze two different chemical reactions, as shown by Schultes and Bartel [47]. Minor variations could convert a starting sequence into either of these highly active ribozymes, demonstrating that the evolving paths of RNA sequence can easily cross in sequence space. Similarly, RNA folds recognizing different ligands may be very close in sequence space [48]: for example, a small series of 'neutral' mutations (that is, mutations that have no effect on secondary structure) transformed a flavin-binding aptamer into a GMP-binding aptamer [49]. Extensive networks of neutral variation in sequence space interconnect RNA regions with similar function and structure [50,51], as confirmed by the recent elucidation of more three-dimensional RNA structures (see [43,52] for reviews).
It is now recognized that the most common RNA-RNA binding contact is the so-called A-minor motif [53]. This occurs between two contiguous adenines in one partner RNA and the shallow/minor groove side of two stacked Watson-Crick pairs in the other. An analysis of tertiary contacts shows that the contiguous adenines can originate from a variety of local environments (for example, bulging, apical or internal loops) and that the only molecular recognition requirement in the receptor RNA is the presence of two Watson-Crick base pairs [54,55]. In other words, coupled to the vast shape space accessible through mutations neutral for secondary structure, there are weak but crucial sequence constraints imposed by the tertiary contacts. In RNA architectures, the additional structural constraints originate from the topology of the secondary structure (junctions of helices, number of base pairs within helices, and so on). In short, RNA sequences (and thus their structure and function) are characterized by neutrality at all levels from molecular recognition between motifs to secondary structure and three-dimensional architecture.
This complex interplay between sequence conservation and neutral evolution on the one hand, and diversity in folds despite conservation in interaction protocols on the other, is central to the theoretical and experimental difficulties in identifying key regulatory RNA sequences from genomic sequence. For example, group I introns are characterized by an invariant core onto which is grafted a variety of peripheral elements [36,56]. Long-range contacts between those non-homologous peripheral elements are necessary for biological activity. All known group I introns contain a tertiary contact between two specific paired-segment regions (regions 5 and 9; Figure 4). However, the examples that have been crystallized (for a review, see [57]) show that in each case, the contacts are achieved through different local topologies (Figure 4), each with different sequence constraints. Interestingly, in a first attempt at modeling the lariat-forming group I-like intron, GIR1, from slime molds, it was not possible to construct the usual intramolecular contacts between regions 5 and 9 [58].
Figure 4.
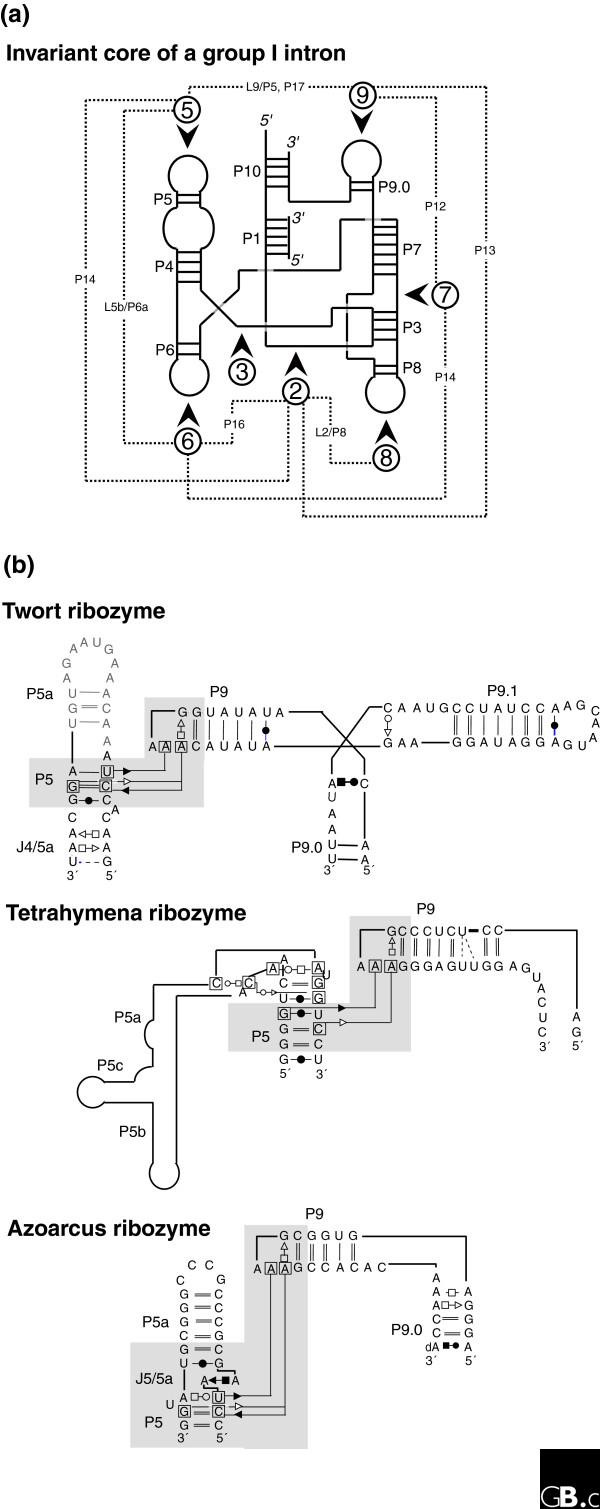
Different local topologies can give rise to similar tertiary contacts in group I introns. (a) The invariant core of a group I intron [36,94] is illustrated in schematic form with the paired segments indicated by P and the loop regions by L. The dashed lines indicate the contacts between the peripheral elements, which are indicated by the numbers in circles. (b) Three different group I introns illustrate distinct ways of achieving a similar tertiary contact (involving non-Watson-Crick A-minor base-base interactions between a GAAA tetraloop and two stacked pairs) connecting distant regions. In each case region 9 folds towards region 5 (as indicated by the shaded region) but, in the Twort ribozyme [95] this is via a three-way junction, in the Tetrahymena ribozyme [96], it is via a large bend (this is not the natural junction, however), and in the Azoarcus ribozyme [97], it is via a kink-turn. Each motif has a different sequence and set of structural constraints [77,92].
Are there more ribozymes that catalyze 2',5'-phosphodiester bond formation or cleavage to be discovered? Scattered evidence of the occurrence of 2',5'-bonds exists throughout the literature. A 2',5'-phosphodiester bond was observed in vitro [59] and in vivo [60] during circularization of the genome of the peach latent mosaic viroid, and the HDV ribozyme, unlike the hammerhead ribozyme, has been shown to cleave 2',5'-linkages efficiently [61].
Searching genomes for ribozymes and riboswitches
Novel catalytic RNA entities can, in principle, be looked for either by database searches using defined consensus motifs from a given ribozyme or by experimentally testing candidate RNAs for biochemical activity. Both approaches have advantages and disadvantages. Database searches require RNA sequence alignments (as produced, for example, by Rfam [62]) coupled with covariance analysis [63-67]. The quality of the sequence alignment is central to this process, however, and not many databases are as carefully hand-curated as the RNase P database [68]. In database screening, the definition of what we consider to be the consensus motif of a given catalytic RNA is crucial. Even if a catalytic RNA motif is well defined, searches are complicated by the requirement to combine a complex assembly of structural (hairpin) and sequence information, which prevents simple solutions such as purely sequence-based homology searches. Generally, the tools available adequately identify isolated hairpins [69]. Given a pattern description for a catalytic RNA motif, several programs, such as PatScan [70] or RNAMOT [71], can be used to screen the public databases. Hits from such searches require further analysis, and initially, a calculation of the secondary structure is necessary - although usually not sufficient. A secondary structure, calculated using a program such as RNAfold [72], is predictive if the required helical elements of the RNA motif under consideration will form in the hit sequence. Secondary-structure prediction programs have difficulty in accurately predicting large structures, however, and can also produce vast numbers of alternative structures when scanning whole genomes [73,74].
For individual sequences found in a database search, a test of their particular biochemical activity (Figure 1) might be sufficient. However, functionally similar RNA molecules frequently exhibit numerous and highly divergent sequence insertions or deletions that interrupt the pattern of secondary-structure motifs and render the computer description of a given motif inadequate for finding sequences with similar activity. Furthermore, the use of pattern-description programs is incomplete if the complexity of the RNA structure - which goes way beyond the Watson-Crick base pairing [75] - is not taken into consideration. These issues, and whether the additional, essential tertiary interactions of a given RNA motif will form, can be addressed by a combination of comparative analysis of similar ribozymes with isostericity matrices, which give the geometrically equivalent base pairs for each particular type of base-base interaction [76]. All pairwise base-base interactions present in nucleic acids have been classified into 12 families, where each family is a 4 × 4 matrix of the bases A, G, C, and U [75]. This classification allows the deduction of all possible geometrically equivalent base pairs in a given family. The isostericity matrices have been verified for several RNA motifs using structural alignments anchored in crystal structures [77]. Thus, for assumed structurally homologous positions in an RNA motif, one can compare the resulting pairwise interactions with the known isostericity matrices to assess the validity of an RNA motif assignment in an alignment [78]. As this type of analysis is an iterative process, it is worth noting that it might also lead to refinement and extension of the pattern of the consensus motif that the search was started with. If applied to large assemblies of sequence information, as has been done for the kink-turn and C-loop RNA motifs [77], this approach allows a broader description (the comprehensiveness of which is currently unknown) and refinement of a given motif.
The analysis of co-variation of nucleotides in sequence alignments underlies most manual or automated secondary-structure determination. However, high sequence conservation (which is usually considered a marker for conservation of function) leads to serious ambiguities and difficulties in deriving secondary structures. The catalytic riboswitch glmS is a good example: the crystal structure [79] presents a different secondary structure from that deduced from sequences. The new helices involve pairings between segments, conserved at more than 95% in sequence, and thus giving no co-variation signal. The requirement for a well-defined RNA motif in database searches is also an intrinsic limitation of this approach.
While novel genomic locations of a known catalytic RNA can be identified from sequence similarities, novel activities cannot be so readily discovered. For this purpose, a recently introduced in vitro selection scheme [29] can be applied. Interestingly, from the human genome, a close variant of a known catalytic RNA motif was selected and characterized as a HDV-ribozyme-like sequence rather than a new catalytic RNA. It is intriguing that these sequences were discovered by their biochemical activity and not by in silico approaches (as an active HDV ribozyme can be made by both the 'genomic' sequence and its complementary sequence despite its intricate pseudoknotted secondary structure). Furthermore, additional, as-yet structurally uncharacterized, sequences were reported [29], and so this new selection scheme might actually have identified new self-cleaving RNA entities. The selection scheme described in [29] uses DNA minicircles that cover the genomic sequence of a given organism as the templates for rolling-circle RNA transcription (Figure 5). It can thus readily monitor RNA self-cleavage, but other activities will be missed. Thus, to assess the prevalence of a given catalytic RNA motif, the combined approach using sequence and three-dimensional structure information described above is most suitable, while novel in vitro selection schemes might be designed to discover activities other than RNA cleavage in a given organism.
Figure 5.
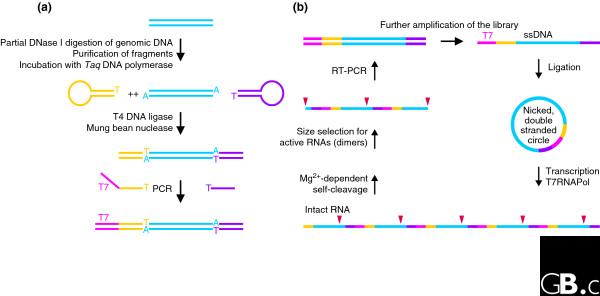
Identification of catalytic RNA from a genomic library. (a) Preparation of the genomic library. Genomic DNA is first partially digested and fragments of approximately 150 bp (blue) are gel-purified and incubated with Taq polymerase to give them 3' A overhangs. In the next step, ligation of covalently closed oligonucleotides (yellow and purple) to the library prevents the unwanted combination of DNA fragments. After removal of DNA hairpins, a T7 promoter (magenta) is then added by PCR, yielding an amplified linear library. (b) The in vitro selection scheme. The library is further amplified by PCR using a 5'-phosphorylated reverse primer and a biotinylated forward primer that allows the isolation of the phosphorylated strand using streptavidin beads. Single strands are individually circularized by ligation with a splint oligonucleotide and the second strand is added by incubation with Taq polymerase and deoxynucleoside triphosphates. The resulting nicked double-stranded library is suitable for rolling-circle transcription by T7 polymerase [98], yielding multimeric RNA species potentially encoding sites of self-cleavage (red triangles). The RNA is then incubated for self-cleavage, and active molecules (dimers) are size-selected. The scheme is completed by preparation of the next-generation DNA library using reverse transcription-PCR (RT-PCR). Modified from [29].
As pointed out earlier, most of the reactions known to be naturally catalyzed by RNA (Figure 1) involve the breakage or formation of 3',5' (and occasionally 2',5') phosphodiester bonds. RNA has the potential to catalyze other chemical reactions, however. As well as peptide formation in the ribosome, Diels-Alder cycloaddition [80] and Michael addition [81] can be catalyzed by RNA, as shown by in vitro Darwinian evolution. Thus, reactions catalyzed by RNA in nature might be more diverse than currently known. The discovery of such activities is likely to be serendipitous and made by keen observers of RNA molecular behavior.
New small or large noncoding RNAs are regularly being discovered in both bacteria and mammals. Recent evidence shows that most of the mammalian genome is transcribed in complex patterns, producing tens of thousands of novel transcripts [82,83]. Novel RNAs are regularly predicted on the basis of their sequence conservation or secondary-structure elements [84-87]. But these predictions do not utilize information on the non-Watson-Crick base pairing or tertiary structure so crucial to the activity of many ribozymes, and, as discussed above, these features are often not well conserved in the sequence. Nor do the predictive algorithms used give any indication of what the RNA function might be. Vertebrate genomes contain a large number of conserved noncoding elements (CNEs) or ultraconserved elements [88,89], whose biological functions and mechanisms of action remain to be established. The evidence for transcription of most of these conserved elements is, however, still scanty [89-91]. In any case, the recent additions to the list of natural catalytic RNAs indicate that there are likely to be many more to come; new algorithms will be required that use all available information to identify and classify them.
Acknowledgments
Acknowledgements
We thank François Michel (CGM, CNRS, Gif-sur-Yvette) and Michael Pheasant (IMB, University of Queensland, St Lucia) for constructive comments on the manuscript and Neocles Leontis (Bowling Green, OH) for numerous discussions. CH acknowledges support by the Deutsche Forschungsgemeinschaft (grant HA3459-3) and by the EU-STREP Fosrak and EW support by grants ANR-05-BLAN-0331-04 and CEE BAC RNA:LSHG-CT-2005-018618.
References
- Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. doi: 10.1038/323349a0. [DOI] [Google Scholar]
- Rupert PB, Ferre-D'Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Wu H-N, Lin YJ, Lin FP, Makino S, Chang M-F, Lai MMC. Human hepatitis d virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci USA. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-d'Amaré AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- Lilley DM. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- Lilley DM. Structure, folding and mechanisms of ribozymes. Curr Opin Struct Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Pan T, Uhlenbeck OC. A small metalloribozyme with a two-step mechanism. Nature. 1992;358:560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]
- Roberts GC, Dennis EA, Meadows DH, Cohen JS, Jardetzky O. The mechanism of action of ribonuclease. Proc Natl Acad Sci USA. 1969;62:1151–1158. doi: 10.1073/pnas.62.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Semlow DR, Silverman SK. Parallel selections in vitro reveal a preference for 2'-5' RNA ligation upon deoxyribozyme-mediated opening of a 2',3'-cyclic phosphate. J Mol Evol. 2005;61:207–215. doi: 10.1007/s00239-004-0326-y. [DOI] [PubMed] [Google Scholar]
- Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- van der Veen R, Arnberg AC, van der Horst G, Bonen L, Tabak HF, Grivell LA. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986;44:225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]
- Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/S0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/S0092-8674(03)00391-X. [DOI] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genome-wide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–3888. doi: 10.1128/mcb.18.7.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 2000;28:4037–4043. doi: 10.1093/nar/28.20.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräf S, Przybilski R, Steger G, Hammann C. A database search for hammerhead ribozyme motifs. Biochem Soc Trans. 2005;33:477–478. doi: 10.1042/BST0330477. [DOI] [PubMed] [Google Scholar]
- Przybilski R, Gräf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann C. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005;17:1877–1885. doi: 10.1105/tpc.105.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Lisacek F, Diaz Y, Michel F. Automatic identification of group I intron cores in genomic DNA sequences. J Mol Biol. 1994;235:1206–1217. doi: 10.1006/jmbi.1994.1074. [DOI] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns - a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Costa M, Deme E, Jacquier A, Michel F. Multiple tertiary interactions involving domain II of group II self-splicing introns. J Mol Biol. 1997;267:520–536. doi: 10.1006/jmbi.1996.0882. [DOI] [PubMed] [Google Scholar]
- De la Pena M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Westhof E. A tale in molecular recognition: the hammerhead ribozyme. J Mol Recognit. 2007;20:1–3. doi: 10.1002/jmr.806. [DOI] [PubMed] [Google Scholar]
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acids Res. 2006;34:6587–6604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Breaker RR. Structural diversity of self-cleaving ribozymes. Proc Natl Acad Sci USA. 2000;97:5784–5789. doi: 10.1073/pnas.97.11.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Szostak JW. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Westhof E, Johansen S. An mRNA is capped by a 2',5' lariat catalyzed by a group I-like ribozyme. Science. 2005;309:1584–1587. doi: 10.1126/science.1113645. [DOI] [PubMed] [Google Scholar]
- Schultes EA, Bartel DP. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- Huang Z, Szostak JW. Evolution of aptamers with a new specificity and new secondary structures from an ATP aptamer. RNA. 2003;9:1456–1463. doi: 10.1261/rna.5990203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held DM, Greathouse ST, Agrawal A, Burke DH. Evolutionary landscapes for the acquisition of new ligand recognition by RNA aptamers. J Mol Evol. 2003;57:299–308. doi: 10.1007/s00239-003-2481-y. [DOI] [PubMed] [Google Scholar]
- Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proc Biol Sci. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- Fontana W, Schuster P. Continuity in evolution: on the nature of transitions. Science. 1998;280:1451–1455. doi: 10.1126/science.280.5368.1451. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. The A-minor motifs in the decoding recognition process. Biochimie. 2006;88:993–999. doi: 10.1016/j.biochi.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Michel F, Jacquier A, Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982;64:867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Einvik C, Nielsen H, Westhof E, Michel F, Johansen S. Group I-like ribozymes with a novel core organization perform obligate sequential hydrolytic cleavages at two processing sites. RNA. 1998;4:530–541. doi: 10.1017/S1355838298971758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote F, Perreault JP. Peach latent mosaic viroid is locked by a 2',5'-phosphodiester bond produced by in vitro self-ligation. J Mol Biol. 1997;273:533–543. doi: 10.1006/jmbi.1997.1355. [DOI] [PubMed] [Google Scholar]
- Cote F, Levesque D, Perreault JP. Natural 2',5'-phosphodiester bonds found at the ligation sites of peach latent mosaic viroid. J Virol. 2001;75:19–25. doi: 10.1128/JVI.75.1.19-25.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih IH, Been MD. Ribozyme cleavage of a 2,5-phosphodiester linkage: mechanism and a restricted divalent metal-ion requirement. RNA. 1999;5:1140–1148. doi: 10.1017/S1355838299990763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL. Consensus folding of aligned sequences as a new measure for the detection of functional RNAs by comparative genomics. J Mol Biol. 2004;342:19–30. doi: 10.1016/j.jmb.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Yao Z, Weinberg Z, Ruzzo WL. CMfinder - a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- Weinberg Z, Ruzzo WL. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics. 2006;22:35–39. doi: 10.1093/bioinformatics/bti743. [DOI] [PubMed] [Google Scholar]
- Brown JW. The Ribonuclease P Database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/S0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- Laferriere A, Gautheret D, Cedergren R. An RNA pattern matching program with enhanced performance and portability. Comput Appl Biosci. 1994;10:211–212. doi: 10.1093/bioinformatics/10.2.211. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Giegerich R. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics. 2004;5:140. doi: 10.1186/1471-2105-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/S1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossinet F, Westhof E. Sequence to structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon-carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- Sengle G, Eisenfuhr A, Arora PS, Nowick JS, Famulok M. Novel RNA catalysts for the Michael reaction. Chem Biol. 2001;8:459–473. doi: 10.1016/S1074-5521(01)00026-6. [DOI] [PubMed] [Google Scholar]
- Teixeira A, Tahiri-Alaoui A, West S, Thomas B, Ramadass A, Martianov I, Dye M, James W, Proudfoot NJ, Akoulitchev A. Autocatalytic RNA cleavage in the human beta-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- Carninci P. Tagging mammalian transcription complexity. Trends Genet. 2006;22:501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Puerta-Fernandez E, Barrick JE, Roth A, Breaker RR. Identification of a large noncoding RNA in extremophilic eubacteria. Proc Natl Acad Sci USA. 2006;103:19490–19495. doi: 10.1073/pnas.0607493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The functional genomics of noncoding RNA. Science. 2005;309:1527–1528. doi: 10.1126/science.1117806. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Doudna JA. Crystallization and structure determination of a Hepatitis delta virus ribozyme: Use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- Cech TR, Damberger SH, Gutell RR. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1994;1:273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- Golden BL, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubendiek SL, Kool ET. Generation of catalytic RNAs by rolling transcription of synthetic DNA nanocircles. Nat Biotechnol. 1997;15:273–277. doi: 10.1038/nbt0397-273. [DOI] [PubMed] [Google Scholar]
- Flores R, Delgado S, Gas ME, Carbonell A, Molina D, Gago S, De la Pena M. Viroids: the minimal non-coding RNAs with autonomous replication. FEBS Lett. 2004;567:42–48. doi: 10.1016/j.febslet.2004.03.118. [DOI] [PubMed] [Google Scholar]
- Tabler M, Tsagris M. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 2004;9:339–348. doi: 10.1016/j.tplants.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Daros JA, Flores R. Identification of a retroviroid-like element from plants. Proc Natl Acad Sci USA. 1995;92:6856–6860. doi: 10.1073/pnas.92.15.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LM, Gall JG. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987;48:535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Epstein LM. Cloning and characterization of extended hammerheads from a diverse set of caudate amphibians. Gene. 1996;172:183–190. doi: 10.1016/0378-1119(96)00126-6. [DOI] [PubMed] [Google Scholar]
- DeYoung MB, Siwkowski AM, Lian Y, Hampel A. Catalytic properties of hairpin ribozymes derived from Chicory Yellow Mottle virus and Arabis Mosaic virus satellite RNAs. Biochemistry. 1995;34:15785–15791. doi: 10.1021/bi00048a024. [DOI] [PubMed] [Google Scholar]
- Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/S1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/S0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 2003;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+). Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rfam http://www.sanger.ac.uk/Software/Rfam/index.shtml
- Gutell lab comparative RNA web site http://www.rna.icmb.utexas.edu
- Subviral RNA database http://subviral.med.uottawa.ca
- RNase P database http://www.mbio.ncsu.edu/RNaseP/home.html


