Short abstract
A search of the literature reveals that researchers study relatively few genes out of the total human genome.
Abstract
Gene annotation, as measured by links to the biomedical literature and funded grants, is governed by a power law, indicating that researchers favor the extensive study of relatively few genes. This emphasizes the need for data-driven science to accomplish genome-wide gene annotation.
Following the completion of the primary sequence of the mouse and human genomes, one of the key challenges for the biomedical community is the functional annotation of all genes [1]. With more than 650,000 citations indexed in Medline in 2005 alone, it is tempting to assume that our understanding of gene function is steadily and uniformly progressing.
As one method of quantifying our progress toward this ambitious goal of genome-wide gene annotation, we analyzed links into the biomedical literature as curated and indexed in the Entrez Gene database of the National Center for Biotechnology Information (NCBI) [2]. At the time of our study, there were 40,822 human genes in the database. We observe that the probability P(k) that a gene has k references decays by a power law, P(k) ~ k-a, (a = 1.31) (Figure 1a). Simply put, over all human genes in the Entrez Gene database, the most common number of linked citations is zero (16,346 entries; not used in calculation), the next most common is one linked citation (6,325 genes), followed by two linked citations (3,959 genes), and so on. The occurrence of very well cited genes is relatively rare, with only 64 human genes with more than 200 citations in Entrez Gene. This distribution of citations is also reflected in an analysis of mouse genes (a = 1.40; data not shown). Among the most highly referenced entries are well studied genes with known roles in important biological processes. For example, the top two cited genes in both human and mouse are the tumor suppressor p53 and the gene for the pleiotropic cytokine tumor necrosis factor (TNF). This power-law relationship is also observed when searching for gene symbols and aliases directly in abstracts and titles in the PubMed database (Figure 1b).
Figure 1.
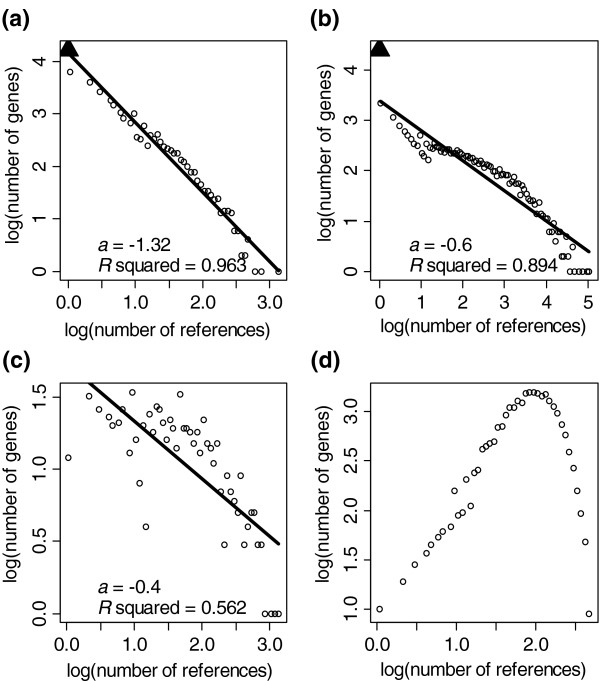
Power-law-like distributions. (a) The relationship between the probability P(k) of observing a human gene with k references in Entrez Gene decays according to a power law P(k) ~ k-a. This trend has also been observed for mouse genes (data not shown) as indexed in the Entrez Gene database. (b) This distribution is also observed when directly searching symbols and aliases in Medline abstracts. The number of genes with zero references is shown in (a) and (b) as black triangles, but were not used in the power-law calculation. (c) Analysis of the CRISP database of NIH-funded grants in 2005 also reveals a power-law relationship. (d) A gamma distribution is most consistent with the research community's goal of genome-wide gene annotation. In this example, gamma-distribution parameters were shape = 2 and scale = 50. Axes are shown in log10 scale.
Evidence of power-law relationships has been observed in many aspects of biology and natural systems - populations in cities, metabolic networks, protein-protein interactions, and the topology of the Internet (see, for example [3-5]). The observation of this pattern in the biomedical literature probably reflects an underlying natural principle. Researchers studying scale-free networks showed that a power-law relationship in the connectivity of nodes was a consequence of new nodes being preferentially attached to well connected nodes [5]. In information science [6], this has been termed the 'principle of least effort', and we suggest that the power law manifests itself here on the basis of researchers' natural tendency to study that which is easy to study, previously studied genes.
If the pattern of citations in the biomedical literature is an accurate reflection of historical patterns of research, then an analysis of recent grants funded by the National Institutes of Health (NIH) will probably reveal future trends. We therefore examined the CRISP database [7] for all grants funded by the NIH in 2005. Because grants are not indexed by gene name, we identified CRISP keywords that correspond to gene names through manual curation and comparison with Entrez Gene. Although fewer gene keywords were identified, which resulted in a noisier picture, we again found that the number of grant citations per gene also decays according to a power law (a = 0.39) (Figure 1c). Similar analyses based on keyword searches of grant abstracts, based on investor initiated (RO1) grant information from 2003 and 2004, all resulted in qualitatively similar results.
Understanding the function of all the genes in the mammalian genome is a goal shared by researchers and funding agencies alike. Success in achieving this goal will require concerted efforts to fight the power law and the principle of least effort. Specifically, these efforts will require the transformation of the observed exponential distributions to something that better approximates a normal distribution (or more precisely, a gamma distribution as shown in Figure 1d). This ideal distribution would indicate that the majority of genes have some minimal non-zero degree of gene annotation, with tails that extend in both directions. Recent progress in data-driven research and ongoing advances in genome-scale gene annotation are important steps toward achieving this transformation. These emerging techniques include gene and protein expression analysis, protein-protein interactions, and high-throughput screening using overexpression and RNA interference methodologies. Historically unbiased methods such as genetics will also contribute as candidate genomic loci are refined to the resolution of individual genes.
In summary, we have shown power-law-like distributions in gene annotation (measured by links to the biomedical literature) and research funding (measured by gene references in funded grants). This shows that the research community is still far from understanding the function of all mammalian genes, and instead focuses most of its effort on relatively few. While recent advances in data-driven and genome-scale research are promising, recognition of this phenomenon and a dramatic shift in the pattern of both scientific publishing and funding will be required for our goal of genome-wide gene annotation to be realized.
References
- Collins FS, Green ED, Guttmacher AE, Guyer MS, US National Genome Research Institute A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;(33 Database):D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipf GK. Human Behavior and the Principle of Least Effort. Cambridge, MA: Addison-Wesley; 1949. [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Mann T. A Guide to Library Research Methods. New York, NY: Oxford University Press; 1987. [Google Scholar]
- CRISP http://crisp.cit.nih.gov


