Abstract
The diversity of the T cell receptor (TCR) repertoire is limited, because of the processes of positive and negative T cell selection. To obtain T cells with specificities beyond the immune system's capacity, we have developed a strategy for retroviral TCR display. In this approach, a library of T cell variants is generated in vitro and introduced into a TCR-negative murine T cell line by retroviral transfer. We document the value of TCR display by the creation of a library of an influenza A-specific TCR and the subsequent in vitro selection of TCRs that either recognize the parental influenza epitope or that have acquired a specificity for a different influenza A strain. The resulting in vitro selected TCRs induce efficient T cell activation after ligand recognition and are of equal or higher potency than the in vivo generated parent receptor. TCR display should prove a useful strategy for the generation of high-affinity tumor-specific TCRs for gene transfer purposes.
T cells, the prime mediators of adaptive cellular immunity, specify their action through the T cell receptor (TCR)-mediated recognition of a peptide epitope bound to a MHC molecule. The immune system contains a large collection of T cells that covers a broad range of peptide–MHC specificities and, thereby, can identify subtle changes in MHC–epitope presentation. However, self-tolerance leads to the removal of the high-affinity T cell repertoire specific for self antigens (1, 2), and this removal will include T cells with desirable specificities, such as many self antigens expressed on tumor tissues. Because of the potential therapeutic value of TCRs with such tumor/lineage specificities, we set out to develop an in vitro strategy for TCR selection that can be used to bypass in vivo tolerance.
For the in vitro isolation and generation of mAbs, antibody-phage display has proven to be a useful technology to replace hybridoma technology and animal immunization (3). Analogous to this technology, TCRs have been expressed as single-chain fragments (scTCRs) on the surface of both phage (4) and yeast (5). Recently, yeast TCR display was shown to be a successful strategy for the in vitro selection of variant scTCR fusion proteins with a dramatic increase in affinity for an allogeneic peptide–MHC target (6). Such high-affinity scTCRs may be of significant use as probes for the detection of specific peptide–MHC complexes. However, it is unclear whether yeast- or phage-based TCR display systems will prove equally useful to change the fine-specificity of TCRs. Specifically, the ability of T cells to discriminate between closely related ligands seems to be related directly to the property of TCR/CD3 complexes to cluster after encountering their cognate ligands (7, 8), and it may prove difficult to copy this process in these systems. We present here a T cell-based in vitro TCR selection strategy that can be used to isolate αβ heterodimeric TCRs with increased affinities or altered specificities. This strategy for TCR display closely mimics the in vivo situation by retroviral insertion of a TCR library into a TCR-negative T cell host. Such T cell line-displayed TCR libraries not only allow the selection of desirable TCRs by biochemical means but also offer the possibility to test the functional behavior of selected TCRs directly.
Through the generation and screening of an in vitro T cell library based on a murine influenza A-specific TCR, we have isolated variant TCRs that are either specific for the parental viral strain or that have acquired a specificity for a variant influenza epitope. These in vitro selected TCRs recognize peptide–MHC complexes on target cells with high efficiency and high specificity. The ability to control TCR fine specificity in a direct manner by retroviral display provides a general strategy for the generation of T cells with specificities that could not be obtained previously . In addition, retroviral TCR display offers a powerful strategy to dissect structure-function relationships of the TCR in a physiological setting.
Materials and Methods
Preparation of H-2Db Tetramers.
Peptides were produced by using standard fluorenylmethoxycarbonyl chemistry. Soluble allophycocyanin (APC)-labeled H-2Db tetramers were produced as described (9, 10) and stored frozen in Tris-buffered saline/16% (vol/vol) glycerol/0.5% BSA.
Cell Lines.
The 34.1Lζ cell line is derived from the cell line 34.1L, a day 14 fetal thymus-derived prethymocyte cell line (11). Because initial experiments indicated that the expression of the CD3ζ TCR component was limiting in 34.1L, a CD3ζ-encoding vector was introduced by retroviral gene transfer: CD3ζ cDNA was amplified by PCR with primers CD3ζtop (CCCAAGCTTATGAAGTGGAAAGTGTCTGTTC) and CD3ζbottom (ATAAGAATGCGGCCGCTTACTGGTAAAGGCCATCGTG) (Isogen Bioscience BV, The Netherlands) subcloned into the retroviral-vector pMX, and this construct was used to retrovirally transduce 34.1L cells. The resulting 34.1Lζ cells were cloned, and expression of the transduced CD3ζ chain was assessed by reverse transcription –PCR. CD8+ 34.1Lζ cells were generated by transduction of 34.1Lζ cells with a retroviral vector encoding the CD8α chain. CD8 expression was assessed by mAb staining.
The EL4 tumor cell line is a murine thymoma cell line of the H-2b haplotype (12). The EL4PR and EL4NT cell lines were obtained by transduction of EL4 cells with a retrovirus encoding the enhanced green fluorescent protein (eGFP) with the A/PR/8/34 and A/NT/60/68 cytotoxic T lymphocyte (CTL) epitope as a C-terminal fusion, respectively, and transduced cells were isolated by fluorescence-activated cell sorting of eGFP-expressing cells. All cell lines were grown in Iscove's modified Dulbecco's medium (Life Technologies, Paisley, Scotland) supplemented with 5% (vol/vol) FCS (BioWhittaker)/0.5 μM β-mercaptoethanol (Merck)/100 units/ml penicillin/100 μg/ml streptomycin (Roche Molecular Biochemicals).
Production of Retroviral Supernatants and Retroviral Transduction.
Plasmid DNA was transfected into Phoenix-A cells, derivatives of the human embryonic kidney cell line 293T, by pfx-2 lipid transfection (Invitrogen). After transfection, the cells were cultured for 48 h before the transduction procedure. A recombinant human fibronectin fragment CH-296 transduction procedure (RetroNectin; Takara Shuzo, Otsu, Japan) was used based on a method developed by Hanenberg et al. (13). Nontissue culture-treated Falcon Petri dishes (3-cm diameter; Becton Dickinson) were coated with 2 ml of 30 μg/ml recombinant human fibronectin fragment CH-296 at room temperature for 2 h. The CH-296 solution was removed and replaced with 2 ml of 2% (vol/vol) BSA (Sigma) in PBS for 30 min at room temperature. The target cells were plated on RetroNectin-coated dishes (0.5 × 106 cells per Petri dish) in 1 ml of retroviral supernatant. Cells were cultured at 37°C for 24 h, washed, and transferred to 25-cm2 culture flasks (Becton Dickinson).
Construction of the F5 TCR Complementarity Determining Region 3 (CDR3) Library.
TCR cDNAs were generated from F5 TCR transgenic T cells by reverse transcriptase reaction (Roche Molecular Biochemicals). The F5 TCRα cDNA was amplified by PCR with primers F5α-top (GGGGGATCCTAAACCATGAACTATTCTCCAGCTTTAGTG) and F5α-bottom (GGAAGGGGGCGGCCGCTCAACTGGACCACAGCCTCAG) (Perkin–Elmer) and ligated into the pMX-IRES-enhanced GFP vector. The F5 TCRβ cDNA was amplified by PCR with primers F5β-top (GGGGGATCCTAAACCATGGCCCCCAGGCTCCTTTTC) and F5β-bottom (GGAAGGGGGCGGCCGCTCAGGAATTTTTTTTCTTGACCATGG) and ligated into the pMX vector.
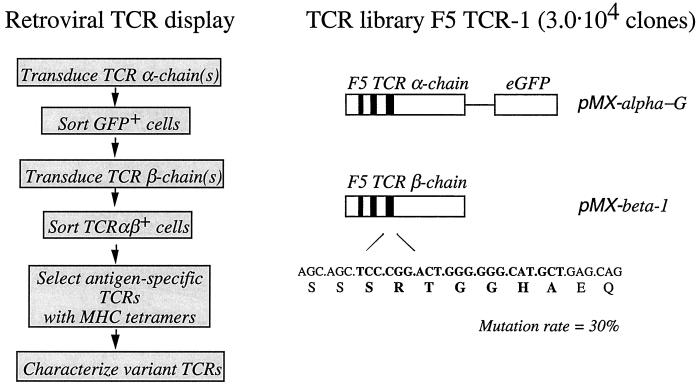
To diversify the CDR3 region of the F5 TCRβ chain, the F5β-CDR3-HM primer (CTGGTCCGAAGAACTGCTCAGCATGCCCCCCAGTCCGGGAGCTGCTTGCACAAAGATACAC) was synthesized. The CDR3 coding sequence contains 70% of the original nucleotide (underlined) and 10% of each of the other three nucleotides. A 5′ fragment of the F5 TCRβ was amplified by PCR with F5β-top and F5β-CDR3-HM primers, and a 3′ fragment was amplified with F5β-CDR3–3′-top (GAGCAGTTCTTCGGACCAG) and F5β-bottom primers. Both resulting F5 TCRβ fragments were assembled by PCR (14) in the presence of F5β-top and F5β-bottom primers, and this TCRβ CDR3 DNA library was ligated into the pMX vector. Ligation products were introduced into Escherichia coli MC1061 cells by electroporation to generate a TCRβ CDR3 library with a complexity of 3 × 106 clones. This DNA library was used to create retroviral supernatant, which was used to transduce 34.1Lζ cells. TCR-expressing cells were sorted by flow cytometry. Based on the sequence requirements for TCR expression (see Results), ≈25% of total library TCRs (0.8 × 106 clones) will fulfill the requirements for surface expression. Because this number is significantly greater than the number of retrovirally TCR-expressing cells that were sorted to produce the T cell library (3.0 × 104), we have taken the latter as the size-limiting step in library production.
Flow Cytometric Analysis and TCR CDR3 Library Screening.
Aspecific staining to 34.1L cells was blocked with 0.5 μg/ml anti-FcγRII/III mAb (clone 2.4G2). Cells were stained with phycoerythrin-conjugated anti-TCRβ chain (H57–597) mAb (PharMingen) or MHC tetramers at 4°C (unless indicated otherwise). Propidium iodide (1 μg/ml; Sigma) was included before analysis. Data acquisition and analysis were performed on a FacsCalibur (Becton Dickinson) with CELLQUEST software. Cell sorting was performed on a FACStar Plus (Becton Dickinson) with LYSIS II software.
34.1Lζ Stimulation Assay.
The self-inactivating [nuclear factor of activated T cells (NFAT)]6-YFP retroviral construct was produced as described (15). TCR-expressing 34.1Lζ cells were transduced with the self-inactivating retroviral construct. Transduced cells, as revealed by YFP expression after overnight phorbol 12-myristate 13-acetate (10 ng/ml; Sigma) and ionomycin (1.67 μg/ml; Sigma) stimulation, were isolated by flow cytometry. Transduced 34.1Lζ cells were incubated overnight at 37°C with target cells at an effector:target ratio of 1:10 (unless indicated otherwise) in the presence of peptides at the indicated concentrations. The percentage of YFP-expressing 34.1Lζ cells was determined by flow cytometric analysis.
Determination of MHC-TCR Dissociation Rates.
34.1Lζ TCR-expressing cells were stained with their cognate APC-labeled peptide/H-2Db tetramers for 20 min at 4°C, washed once with PBS/0.5% (vol/vol) BSA/0.02% NaN3, and subsequently exposed to an excess of homologous unlabeled H-2Db monomers (10 μM) at 25°C. Decay of H-2Db tetramer staining was measured by flow cytometry and is plotted as the percentage of maximum staining as follows: (FIexp − FI0)/(FImax − FI0) × 100%. Simultaneous addition of H-2Db tetramers and 10 μM unlabeled homologous H-2Db monomers during cell labeling prevents the binding of tetrameric MHCs (not shown).
Results and Discussion
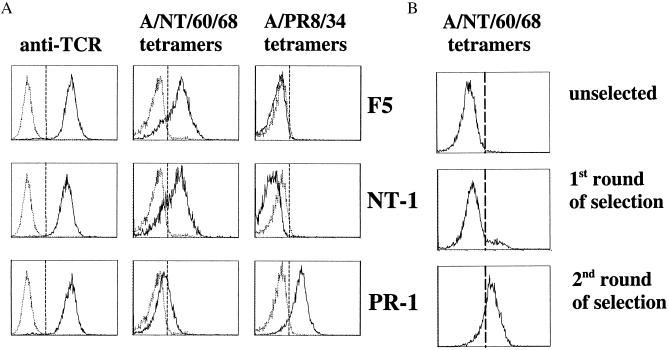
As a host for a T cell line-displayed TCR library, an immature T cell line that does not express endogenous TCR α and β chains was created. This cell line, named 34.1Lζ, expresses all CD3 components required for TCR assembly, but is devoid of CD4 or CD8 coreceptor expression. As a first target for retroviral TCR display, we used a high-affinity murine TCR, of which the antigen specificity is well established (16). This F5 TCR (Vα4;Vβ11) specifically recognizes the immunodominant H-2Db-restricted CTL epitope nucleoprotein (NP)366–374 (ASNENMDAM) of the influenza A/NT/60/68 NP (17). After introduction of the F5 TCR in the 34.1Lζ cell line by retroviral transduction, the transduced cell line expresses high levels of the introduced F5 TCR as measured by anti-TCRβ and MHC tetramer flow cytometry (Fig. 2A).
Figure 2.
MHC tetramer analysis of in vitro selected TCRs. (A) Flow-cytometric analysis of 34.1Lζ cells expressing the F5 (Top), NT-1 (Middle), or PR-1 TCRs (Bottom). The left panels represent staining with phycoerythrin-conjugated anti-TCRβ chain (H57–597) mAb. The middle panels represent staining with APC-labeled tetrameric H-2Db complexes containing the A/NT/60/68 NP epitope (ASNENMDAM), and the right panels represent staining with APC-labeled H-2Db tetramers containing the A/PR8/34 NP epitope (ASNENMETM). Tetramer staining was performed at 37°C (37). (B) Selection of influenza A-reactive TCRs from in vitro TCR libraries. The panels represent staining of the TCRβ CDR3 library with APC-labeled tetrameric H-2Db complexes containing the A/NT/60/68 NP epitope before screening (Top) and after 1 (Middle) and 2 (Bottom) sorts with A/NT/60/68 H-2Db tetramers. Tetramer selections were performed at 4°C.
To test the feasibility of in vitro selection of TCRs with defined specificities, we aimed to isolate novel TCRs with either the same specificity as the parental TCR or receptors that have acquired a specificity for a variant influenza epitope. To modify the peptide specificity of TCRs without generating variant TCRs that are broadly cross-reactive, mutation of only those areas of the TCR that primarily interact with the antigenic peptide is preferred. Structural analysis of four different human and mouse αβTCRs in complex with their cognate peptide–MHC class I all point to the CDR3 loops of the TCR α and β chain as the major determinants of peptide specificity (18–21). In all cases examined, the TCR binds diagonally across the peptide–MHC class I complex such that the CDR3 of the TCRα chain is primarily in contact with the N-terminal part of the MHC-bound peptide, whereas the TCRβ CDR3 mainly interacts with the C-terminal part. Because in the current set of experiments we were interested primarily in obtaining TCRs that can discriminate between epitopes that differ in the C-terminal half of the peptide (see below), a TCR library was manufactured such that its structural diversity is directed toward the TCRβ CDR3 loop exclusively. Through PCR assembly, an F5 TCRβ DNA library that contains a 30% mutational frequency in its 7-amino acid CDR3 was generated (Fig. 1). The 34.1Lζ cell line was transduced with F5 TCRα DNA and the TCRβ library DNA to generate a library of T cells with variant CDR3β loops, and 3.0 × 104 surface TCR-expressing cells were isolated by flow cytometry. Sequence analysis of single-cell clones from TCR-expressing cells was used to provide an estimate of the structural requirements for TCR cell surface expression. These data indicate that the serine on position 1 in the CDR3β is conserved and that for the glycine pair on positions 4 and 5 only conservative amino acid substitutions (alanine/serine) are allowed for all mutant TCRs that are expressed at the cell surface (data not shown).
Figure 1.
(Left) Schematic representation of the generation and screening of retroviral TCR display libraries. (Right) Generation of the TCR library F5 TCR-1. CDRs of the TCR α and β chains are depicted as solid boxes. The CDR3 DNA sequence of the β chain targeted in the current experiments is depicted in bold.
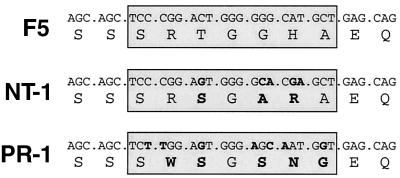
To examine whether variant TCRs could be obtained that retain the ligand specificity of the parental F5 TCR, the T cell library was screened for binding of tetrameric H-2Db complexes containing the A/NT/60/68 NP CTL epitope (ASNENMDAM; Fig. 2B). After a first selection round, a population of A/NT/60/68 H-2Db tetramer-reactive cells was isolated by flow cytometry. Sequence analysis of the CDR3β loops within this population revealed that although this population is diverse, at most positions within the CDR3β only conservative amino acid mutations are allowed for recognition of the A/NT/60/68 NP366–374 tetramers (data not shown). To enrich for TCRs with the highest affinity for the A/NT/60/68 epitope, a subsequent, more stringent selection round was performed in which tetramer-high, TCR-low cells were isolated. In this population two different clones persisted: the parental F5 clone and a variant clone named NT-1. The CDR3β DNA sequence of the NT-1 TCR contains five mutations that result in three conservative amino acid substitutions (Table 1). This variant TCR binds A/NT/60/68 NP366–374 tetramers with efficiency similar to that of the F5 TCR (Fig. 2A).
Table 1.
Selection of variant T cell receptors by retroviral TCR display
A/NT/60/68 and A/PR/8/34 nucleoprotein-specific T cell receptors were selected from the TCR library F5 TCR-1. Sequences of the CDR3 of the F5 and variant TCRβ chains are boxed. Mutations and resulting amino acid substitutions are indicated in bold.
The TCRβ CDR3 library was subsequently screened for the presence of TCRs that bind H-2Db tetramers containing a variant influenza A-NP epitope. This variant NP366–374 epitope (ASNENMETM), derived from the influenza A/PR8/34 strain, differs from the A/NT/60/68 CTL epitope by two conservative amino acid substitutions in the C-terminal half of the peptide and is not recognized by the F5 TCR (ref. 16; Fig. 2A). The TCRβ CDR3 library was subjected to four rounds of selection with H-2Db tetramers that contain the variant epitope to select for the TCR clone(s) that exhibits the highest affinity for this epitope. From this TCR library selection, a single TCR clone emerged (named PR-1) that avidly binds to the A/PR8/34 NP366–374 tetramers (Fig. 2A). Interestingly, although in this library screen we did not select against reactivity with the A/NT/60/68 T cell epitope, the PR-1 TCR has lost the ability to recognize the parental epitope, as judged by MHC tetramer staining (Fig. 2A). Sequence analysis of the PR-1 TCR reveals seven nucleotide mutations in its CDR3β DNA sequence compared with the parental F5 TCR. These mutations result in four conservative amino acid changes and one nonconservative Arg to Trp substitution (Table 1).
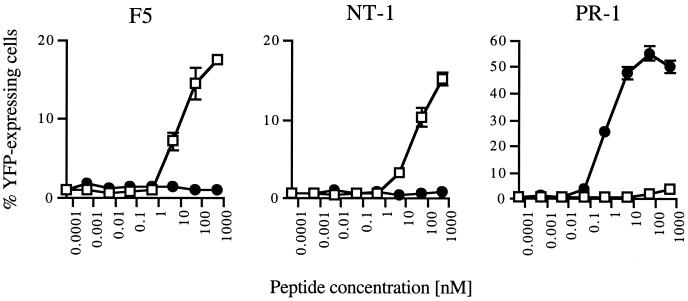
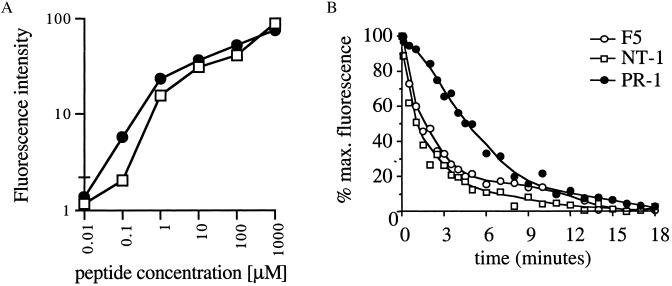
To examine whether in vitro selected variant TCRs can evoke T cell activation after peptide recognition, ligand-induced IL-2 gene transcription was measured. To this purpose, we used a self-inactivating retroviral vector containing multiple NFAT-binding sites upstream of a minimal IL2 promoter and the reporter gene YFP (15, 22). 34.1Lζ cells expressing the F5, NT-1, or PR-1 TCR were virally transduced with the NFAT-YFP reporter construct, and the transduced cells were exposed to target cells in the presence of different concentrations of either the A/NT/60/68 or A/PR8/34 T cell epitope. Both variant clones NT-1 and PR-1 efficiently induce T cell activation after specific antigen recognition with an absolute specificity for the epitope used during the in vitro selections (Fig. 3). Remarkably, the PR-1 TCR shows a dramatically increased sensitivity for its ligand as compared with the recognition of the A/NT/60/68 epitope by the F5 TCR. This high TCR sensitivity is not a consequence of elevated levels of TCR cell surface expression (Fig. 2A) and can only be explained to a minor extent by the small difference in H-2Db-binding affinity between the A/NT/60/68 peptide and A/PR8/34 peptide (Fig. 4A). To examine whether the PR-1 TCR displays an increased sensitivity caused by improved MHC-TCR binding kinetics, MHC-TCR dissociation rates were determined (23–25). Measurement of the decay in peptide/H-2Db-tetramer staining after the addition of an excess of competing ligand reveals that the PR-1 TCR displays an ≈4-fold increase of TCR/MHC half-life as compared with the parental F5 TCR (Fig. 4B). In line with the functional data, the off-rate of the NT-1/MHC complex is similar to that of the high-affinity F5 TCR.
Figure 3.
Signaling function of in vitro selected TCRs. TCRαβ-expressing 34.1Lζ cells transduced with the NFAT-YFP construct were exposed to EL4 target cells in the presence of different concentrations of either the A/NT/60/68 (▫) or A/PR8/34 (●) T cell epitope. Sensitivity and specificity of the different TCRs were determined by flow cytometric analysis of the percentage of YFP-expressing 34.1Lζ cells. In accordance with previous results, the distribution of YFP expression after stimulation is bimodal (22, 38), and T cell activation after stimulation with phorbol 12-myristate 13-acetate (10 ng/ml) and ionomycin (1.67 mg/ml) results in 60–65% YFP-expressing cells (not shown). Data shown are means of triplicates ± SD.
Figure 4.
TCR-MHC off-rates of in vitro selected TCRs. (A) Comparison of H-2Db-binding affinity of the A/NT/60/68 (▫) and A/PR8/34 (●) peptide. TAP-deficient RMA-S cells (39) were incubated overnight at 37°C with peptides at the indicated concentrations. Cells were stained with biotin-conjugated anti-mouse H-2Db antibodies (PharMingen) and phycoerythrin-conjugated streptavidin and analyzed by flow cytometry. Fluorescence intensity is calculated as: FIexp − FI0. Data shown are means of triplicates ± SD. (B) Determination of MHC-TCR dissociation rates. 34.1Lζ TCR-expressing cells were stained with their cognate APC-labeled peptide/H-2Db tetramers at 4°C and subsequently exposed to an excess of homologous unlabeled H-2Db monomers at 25°C. Decay of H-2Db-tetramer staining was measured by flow cytometry and is plotted as the percentage of maximum staining.
These experiments show that in vitro selection of variant TCRs by retroviral TCR display can yield receptors with high potency, as revealed by both biochemical means and functional assays. This outcome occurs despite the fact that the diversity of the library used in these experiments (3 × 104 independent clones) was relatively modest. We estimate that through optimization of transduction and sorting strategies, retroviral TCR display libraries 106-107 in size are technically achievable in this system. Such in vitro TCR libraries will enclose a diversity that approaches that of the total human naïve TCR repertoire (2.5 × 107; ref. 26). TCRs that are isolated from such libraries should be useful for the creation of redirected T cell populations through gene transfer of peripheral T lymphocytes (27).
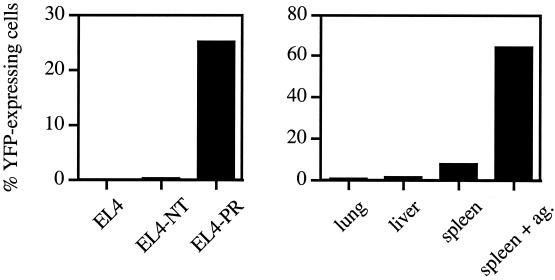
To provide an estimate of the risk of autoreactivity after the creation of cells that carry in vitro manipulated TCRs, PR-1-expressing cells were exposed to a small set of different tissue samples from H-2Db-expressing mice. In these experiments we used PR-1 TCR-expressing 34.1Lζ cells in which the CD8α coreceptor molecule was introduced, as the presence of CD8 can increase T cell sensitivity. Even though a strong T cell response is induced by splenocytes that are incubated with the A/PR8/34 influenza A CTL epitope, little activation of T cells is observed when these cells are exposed only to splenocytes, and no T cell response is induced by lung or liver tissue (Fig. 5). In addition, EL4 transfectants that produce the A/PR8/34 epitope endogenously also are recognized by PR-1-expressing T cells with no detectable crossreactivity toward the A/NT/60/68 epitope, both in the presence (Fig. 5) and the absence (data not shown) of CD8 expression.
Figure 5.
Specificity of the PR-1 TCR. (Left) PR-1-expressing CD8+ 34.1Lζ cells transduced with the NFAT-YFP construct were exposed to EL4 target cells, and cells that either endogenously produce the A/NT/60/68 (EL4NT) or A/PR8/34 (EL4PR) CTL epitope. (Right) PR-1-expressing CD8+ 34.1Lζ cells were incubated with cell suspensions from the indicated tissues or spleen cells incubated with 0.1 μM of the ASNENMETM peptide (E:T ratio of 1:50). Data shown are means of duplicates.
The introduction of in vitro generated TCRs in peripheral T cells by gene transfer without further modification likely will yield unwanted heterodimers of introduced and endogenous TCR α and β chains. Such newly generated mixed-TCR dimers may convey autoreactive behavior to these T cells. To avoid the undesirable pairing with endogenous TCR chains, CD3ζ chimeric TCRs have been produced (28). In addition, remodeling of the TCR-dimerization interface may provide an independent strategy to exclude inadvertent mixed-dimer formation. The remodeling of the structurally related Ig constant domain by Atwell et al. presents an elegant proof of principle in this regard (29).
Novel protein functions are thought to arise through natural evolution from existing molecules that bear a related structure and specificity (30). Also, for the in vitro selection of enzymes with a novel or improved catalytic activity, small-step protein evolution has proven much more successful than the fully de novo design of enzyme activities (31, 32). In line with these examples, we speculate that the selection of TCRs with novel specificities with the in vitro system outlined here is likely to be most successful for small-step specificity changes. Whereas demonstrated here for a TCR specific for an antigen of viral origin, it seems reasonable to assume that this approach will work equally well for TCRs that are specific for self antigens. This strategy may be particularly valuable for the development of a new class of high-affinity tumor/lineage-specific TCRs. In a number of systems it has been demonstrated that (central) self-tolerance results in the removal of the high-avidity T cell repertoire specific for tumor/lineage-antigens (33–36). However, high-avidity T cells that recognize structurally related peptide antigens are retained within the T cell repertoire (K. E. de Visser, H.W.H.G.K., A. M. Kruisbeck, and T.N.M.S., unpublished data). The retroviral TCR display system as outlined here provides a unique opportunity to redirect the TCR carried by such T cells toward recognition of self antigens. The creation of collections of high-affinity TCRs that target lineage antigens for which self-tolerance precludes their in vivo production may thereby now be feasible.
Acknowledgments
The authors thank A. Kruisbeek for the 34.1L T cell line, T. Kitamura for the pMX retroviral vector, G. Nolan for the Phoenix-A cell line, M. Toebes for preparation of MHC tetramers, M. Wolkers for generation of the EL4PR and EL4NT cell lines and useful suggestions, and E. Noteboom and A. Pfauth for expert technical assistance. This research was supported by the Dutch Cancer Society Grants (NKI 97-1442 and NKI 99-2036).
Abbreviations
- TCR
T cell receptor
- CDR
complementarity determining region
- APC
allophycocyanin
- NFAT
nuclear factor of activated T cells
- NP
nucleoprotein
- GFP/YFP
green/yellow fluorescent protein
- CTL
cytotoxic T lymphocyte
References
- 1.Sprent J, Kishimoto H. Ann NY Acad Sci. 1998;841:236–245. doi: 10.1111/j.1749-6632.1998.tb10933.x. [DOI] [PubMed] [Google Scholar]
- 2.Stockinger B. Adv Immunol. 1999;71:229–265. doi: 10.1016/s0065-2776(08)60404-6. [DOI] [PubMed] [Google Scholar]
- 3.Clackson T, Hoogenboom H R, Griffiths A D, Winter G. Nature (London) 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 4.Weidanz J A, Card K F, Edwards A, Perlstein E, Wong H C. J Immunol Methods. 1998;221:59–76. doi: 10.1016/s0022-1759(98)00153-7. [DOI] [PubMed] [Google Scholar]
- 5.Kieke M C, Shusta E V, Boder E T, Teyton L, Wittrup K D, Kranz D M. Proc Natl Acad Sci USA. 1999;96:5651–5656. doi: 10.1073/pnas.96.10.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holler P D, Holman P O, Shusta E V, O'Herrin S, Wittrup K D, Kranz D M. Proc Natl Acad Sci USA. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. . (First Published April 25, 2000; 10.1073/pnas.080078297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monks C R, Freiberg B A, Kupfer H, Sciaky N, Kupfer A. Nature (London) 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 8.Grakoui A, Bromley S K, Sumen C, Davis M M, Shaw A S, Allen P M, Dustin M L. Science. 1999;285:221–227. [PubMed] [Google Scholar]
- 9.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 10.Haanen J B, Toebes M, Cordaro T A, Wolkers M C, Kruisbeek A M, Schumacher T N. Eur J Immunol. 1999;29:1168–1174. doi: 10.1002/(SICI)1521-4141(199904)29:04<1168::AID-IMMU1168>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Oosterwegel M, Timmerman J, Leiden J, Clevers H. Dev Immunol. 1992;3:1–11. doi: 10.1155/1992/27903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorer P A. Br J Cancer. 1950;4:372–379. doi: 10.1038/bjc.1950.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanenberg H, Xiao X L, Dilloo D, Hashino K, Kato I, Williams D A. Nat Med. 1996;2:876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
- 14.Crameri A, Cwirla S, Stemmer W P. Nat Med. 1996;2:100–102. doi: 10.1038/nm0196-100. [DOI] [PubMed] [Google Scholar]
- 15.Hooijberg E, Bakker A Q, Ruizendaal J J, Spits H. Blood. 2000;96:459–466. [PubMed] [Google Scholar]
- 16.Moskophidis D, Kioussis D. J Exp Med. 1998;188:223–232. doi: 10.1084/jem.188.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Townsend A R, Gotch F M, Davey J. Cell. 1985;42:457–467. doi: 10.1016/0092-8674(85)90103-5. [DOI] [PubMed] [Google Scholar]
- 18.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 19.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 20.Ding Y H, Smith K J, Garboczi D N, Utz U, Biddison W E, Wiley D C. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 21.Teng M K, Smolyar A, Tse A G, Liu J H, Liu J, Hussey R E, Nathenson S G, Chang H C, Reinherz E L, Wang J H. Curr Biol. 1998;8:409–412. doi: 10.1016/s0960-9822(98)70160-5. [DOI] [PubMed] [Google Scholar]
- 22.Fiering S, Northrop J P, Nolan G P, Mattila P S, Crabtree G R, Herzenberg L A. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 23.Kersh G J, Kersh E N, Fremont D H, Allen P M. Immunity. 1998;9:817–826. doi: 10.1016/s1074-7613(00)80647-0. [DOI] [PubMed] [Google Scholar]
- 24.Busch D H, Pamer E G. J Exp Med. 1999;189:701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savage P A, Boniface J J, Davis M M. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 26.Arstila T P, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 27.Clay T M, Custer M C, Sachs J, Hwu P, Rosenberg S A, Nishimura M I. J Immunol. 1999;163:507–513. [PubMed] [Google Scholar]
- 28.Willemsen R A, Weijtens M E, Ronteltap C, Eshhar Z, Gratama J W, Chames P, Bolhuis R L. Gene Ther. 2000;7:1369–1377. doi: 10.1038/sj.gt.3301253. [DOI] [PubMed] [Google Scholar]
- 29.Atwell S, Ridgway J B, Wells J A, Carter P. J Mol Biol. 1997;270:26–35. doi: 10.1006/jmbi.1997.1116. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S, Greene E A, Pietrokovski S, Bork P, Attwood T K, Hood L. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 31.Blacklow S C, Liu K D, Knowles J R. Biochemistry. 1991;30:8470–8476. doi: 10.1021/bi00098a026. [DOI] [PubMed] [Google Scholar]
- 32.Altamirano M M, Blackburn J M, Fersht A R. Nature (London) 2000;403:617–622. doi: 10.1038/35001001. [DOI] [PubMed] [Google Scholar]
- 33.Kappler J W, Roehm N, Marrack P. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 34.Kisielow P, Bluthmann H, Staerz U D, Steinmetz M, von Boehmer H. Nature (London) 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 35.Sha W C, Nelson C A, Newberry R D, Kranz D M, Russell J H, Loh D Y. Nature (London) 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- 36.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel R M. Nature (London) 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 37.Whelan J A, Dunbar P R, Price D A, Purbhoo M A, Lechner F, Ogg G S, Griffiths G, Phillips R E, Cerundolo V, Sewell A K. J Immunol. 1999;163:4342–4348. [PubMed] [Google Scholar]
- 38.Karttunen J, Shastri N. Proc Natl Acad Sci USA. 1991;88:3972–3976. doi: 10.1073/pnas.88.9.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ljunggren H G, Stam N J, Ohlen C, Neefjes J J, Hoglund P, Heemels M T, Bastin J, Schumacher T N, Townsend A, Karre K, et al. Nature (London) 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]