Abstract
Interferon (IFN) signaling induces the expression of interferon-responsive genes and leads to the activation of pathways that are involved in the innate immune response. Ubp43 is an ISG15-specific isopeptidase, the expression of which is activated by IFN. Ubp43 knock-out mice are hypersensitive to IFN-α/β and have enhanced resistance to lethal viral and bacterial infections. Here we show that in addition to protection against foreign pathogens, Ubp43 deficiency increases the resistance to oncogenic transformation by BCR-ABL. BCR-ABL viral transduction/transplantation of wild-type bone marrow cells results in the rapid development of a chronic myeloid leukemia (CML)–like myeloproliferative disease; in contrast, a significantly increased latency of disease development is observed following BCR-ABL viral transduction/transplantation of Ubp43-deficient bone marrow cells. This resistance to leukemic development is dependent on type 1 IFN (IFN-α/β) signaling in Ubp43-deficient cells. Increased levels of type 1 IFN are also detected in the serum of CML mice. These results suggest that inhibition of Ubp43-negative effect on IFN signaling can potentiate the response to increased endogenous IFN levels in innate immune responses against cancer development, indicating that pharmacological inhibition of Ubp43 may be of benefit in cancers and others diseases in which interferon is currently prescribed.
Introduction
Innate immune responses play critical roles in the inhibition of cancer development.1 One of the key components of the innate immune response is activation of the type 1 interferon (IFN-α/β) signaling pathway.2,3 Binding of type 1 IFNs to their receptor (IFNAR) triggers the phosphorylation of receptor-associated Jak1 and Tyk2 kinases.4,5 These kinases then phosphorylate STAT1 and STAT2, leading to the activation of downstream signal transduction pathways.6–9 Furthermore, a family of suppressors of cytokine signaling (SOCS) and several protein tyrosine phosphatases negatively regulate the STAT signaling pathway.10–12 Defects in such regulators may result either in the loss of response or a hyperresponse to IFN stimulation.
Type 1 IFN signaling triggers the expression of hundreds of IFN-stimulated genes (ISGs).13,14 Among these is the ISG15 deconjugating enzyme Ubp43 (Usp18).15–19 ISG15 is a ubiquitin-like modifier whose expression and conjugation to other proteins (ISGylation) is strongly increased upon type 1 IFN stimulation.20,21 Ubp43-deficient cells accumulate higher levels of ISGylated proteins and are hypersensitive to type 1 IFN treatment, as evidenced by the enhanced and prolonged activation of STAT phosphorylation in these cells.22,23 Furthermore, Ubp43 knock-out mice show a higher resistance to viral and bacterial infection,24,25 indicating an important role for Ubp43 in the regulation of IFN signal transduction. Recently, using cells with different levels of protein ISGylation and Ubp43 expression, we demonstrated that UBP43 is a novel negative regulator of type 1 interferon signaling and this function is independent of Ubp43 isopeptidase activity against ISG15 conjugates.26,27
Type 1 IFNs suppress cell proliferation and promote apoptosis,28 as such they have been used in the clinical treatment of several cancers, including leukemia.29 A specific example is in the treatment of chronic myeloid leukemia (CML), where IFN was the primary choice before imatinib mesylate became available.30–32 In nearly all cases of CML, patients carry a reciprocal translocation between chromosomes 9 and 22.33,34 This results in a fusion protein consisting of the N-terminal portion of BCR joined to most of the ABL tyrosine kinase. The chimeric BCR-ABL tyrosine kinase is constitutively activated as a result of the oligomerization domain provided by BCR. The tyrosine kinase activity of BCR-ABL activates several signaling intermediates, such as Ras, Akt, Stat5, and p38, which in turn triggers deregulated growth and proliferation of the myeloid progenitors and promotes CML development.6,35–37
Expression of BCR-ABL in mouse bone marrow cells is sufficient for the rapid onset of a CML-like myeloproliferative disease, characterized by splenomegaly and high white blood cell counts.38–40 Given the hypersensitivity of Ubp43−/− mice to IFN and the efficacy of IFN in CML treatment, we investigated whether genetic ablation of UBP43 function resulted in delayed oncogenic transformation in vivo. Here, we report that, compared with wild-type controls, transplantation of BCR-ABL–expressing Ubp43−/− bone marrow cells into mice results in a greatly prolonged latency period in the development of the CML-like myeloproliferative disease. This resistance to leukemia development is heavily dependent upon activation of the type 1 IFN signaling pathway, suggesting that inhibition of Ubp43-negative effect on type 1 IFN signaling promotes an enhanced response to endogenous IFN levels in innate immune responses against cancer development.
Materials and methods
Animals and cells
The generation of Ubp43−/− mice and immortalized MEFs has been described previously.22,23,26 Generation and culture of KT-1 cells with stable expression of UBP43 shRNA have been described previously.26 IFN-α/β receptor R1 subunit knock-out mice (Ifnar1−/−) in the C57 background were kindly provided by Jonathan Sprent (The Scripps Research Institute)41 and bred with the Ubp43−/− mice to generate Ubp43−/−/Ifnar1−/− double knock-out mice. Wild-type C57BL/6J congenic strain C57/B6.SJL Pep3b-BoyJ mice were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were housed in a pathogen-free facility and procedures were approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute.
BCR-ABL retrovirus and retroviral transduction
293T cells were cotransfected with MCV-ecopac and either MigR1 or Mig-p210 at a 1:1 ratio by the calcium phosphate precipitation method. Media was changed 24 hours after transfection, and the retroviral supernatants were harvested the following day and filtered through a 0.45-μm filter. In most experiments, the retrovirus was used immediately to insure a high viral titer. To infect bone marrow cells, 2 mL retroviral supernatant supplemented with 8 μg/mL polybrene (Sigma, St Louis, MO) was added to 1 × 106 cells in a 6-well plate and centrifuged at 1400g for 120 minutes at 32°C. The supernatant was then removed and the cells were resuspended in the bone marrow cell culture medium. Cells were incubated overnight at 37°C in 5% CO2 before performing a second round of infection. The efficiency of retroviral transduction was determined on the basis of green fluorescence by flow cytometry 24 hours after the second round of infections.
Bone marrow transplantation
All recipient mice (6-8 weeks old) were lethally irradiated with 9 Gy in a split dose separated by at least 3 hours. Ubp43−/−, their Ubp43+/− and wild-type littermates, as well as Ubp43−/−/Ifnar1−/− donor mice were injected with 100 mg/kg body weight of 5-fluorouracil (5-FU; Sigma). Five days after 5-FU injection, bone marrow cells were harvested from these mice and spin-infected with Mig-p210 or MigR1 retrovirus as described. Twenty-four hours after the second round of infections, the bone marrow cells (4 × 105 cells) were transplanted into the recipient mice by tail vein injection. Mice were maintained in sterilized cages for 3 weeks on acidified water (pH 4.0). Upon leukemia development, moribund mice were killed.
Murine IFN injection
Where applicable, bone marrow transplant recipients were injected subcutaneously with 18 000 units mIFNβ (MP Biomedicals, Solon, OH) per day starting at day 17 (for UBP43+/+ donor cells) or day 28 (for UBP43−/− donor cells) after transplantation.
Hematological analysis
Blood (2 μL) was diluted in 78 μL Türk solution (0.01% crystal violet and 3% glacial acetic acid), and white blood cell counts were performed under microscopic observation. ACCUSTAIN Wright stain and ACCUSTAIN Giemsa stain solutions (Sigma) were used to stain peripheral blood smears as well as spleen and bone marrow cytospin slides following the 2-step staining protocol from the manufacturer. Differential counts of blood and bone marrow cells were obtained by counting 200 nucleated cells for each sample.
Histology
Spleen and liver samples were fixed with 4% paraformaldehyde in PBS overnight at room temperature and stored at 4°C. Paraffin-embedded sections were cut to a 5-μm thickness and stained with hematoxylin and eosin.
Apoptosis assay
The percentage of apoptotic cells was analyzed by fluorescence-activated cell sorting (FACS) after staining cells with the annexin V–PE apoptosis detection kit (BD Pharmingen, San Diego, CA) according to manufacturer's protocol. Flow cytometry was performed with FACSCalibur (BD immunocytometry, San Jose, CA).
Murine IFN bioassay
Serum levels of IFN were measured using an in vitro biologic assay for protection against the cytopathic effect of vesicular stomatitis virus (VSV) on murine L929 cells. Briefly, 2-fold serial dilutions were performed in 96-well plates using RPMI medium containing 2 mM glutamine, 100 units/mL penicillin/streptomycin, and 10% FBS. L929 cells (3.5 × 105) were added to each well and cultured for 24 hours at 37°C in a 5% CO2 atmosphere. VSV was then added to each well to a final MOI of 0.1. Protection against VSV-mediated cytopathic effects was then evaluated at 48 hours after addition of the virus by MTT assay.42,43 Murine IFN-β (MP Biomedicals) was used as a standard.
Isolation of RNA and Northern blot analysis
Spleen cells were harvested from the BCR-ABL–induced leukemic and MigR1 control–infected mice. RNA was isolated by the RNABee extraction kit (Tel-Test, Friendswood, TX). Total RNA (10 μg) for each sample was electrophoresed on a 1% agarose gel containing 0.22 M formaldehyde. The RNA was transferred to Hybond-N nylon membranes (Amersham, Arlington Heights, IL) and hybridized to 32P-dATP–radiolabeled probes for murine Ubp43 cDNA.
Immunoblotting
Cells were harvested in a buffer containing 50 mM Tris, pH 7.6, 3 M NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate and supplemented with protease inhibitor cocktail (Sigma) and 1 mM each of NaF and NAVO3 to inhibit protein phosphatase activity. Lysates were cleared by centrifugation at 18 000g for 10 minutes and Western blotting was performed as described previously.44 BCR-ABL was detected with the anti-ABL antibody AB-3 (Oncogene Research Products, Boston, MA). Anti-TRAIL and anti–cytochrome c antibodies are from BD Biosciences (San Jose, CA), and anti–caspase 3 antibodies are from Cell Signaling Technologies (Beverly, MA).
Statistical analyses
Statistical significance of the survival times of bone marrow transplant recipients was calculated by chi squared test using the program Prism 4 (Graphpad Software, San Diego, CA). Statistical significance of the apoptosis studies in KT-1 cells was analyzed using the 2-tailed Student t test.
Results
Ubp43-deficient cells can be used in retroviral transduction and bone marrow transplantation assays
Targeting BCR-ABL expression to the murine hematopoietic cell via the murine stem cell virus (MSCV)–based vector has been shown to cause mice to succumb rapidly to a CML-like myeloproliferative disease.39,40 We decided to investigate whether Ubp43 expression affects the development of CML using retrovirus-mediated BCR/ABL expression and bone marrow transplantation (BMT) studies. To investigate if Ubp43-deficient cells could be virally transduced and transplanted, bone marrow cells were collected from 5-FU–treated Ubp43-deficient mice and their control littermates. Since Ubp43+/+ and Ubp43+/− cells responded similarly to IFN, both cell types were used as controls based upon their availability. Two days after infection with MSCV-IRES-EGFP (MigR1) vector control virus, bone marrow cells were transplanted into γ-irradiated wild-type recipient mice. Similar percentages of retrovirally infected Ubp43+ and Ubp43− cells (EGFP+) were detected in recipient mice over a 2-month period after BMT (Figure S1, available on the Blood website; see the Supplemental Figures link at the top of the online article). These results indicate that both Ubp43+ and Ubp43− hematopoietic cells can be infected with MSCV-based retrovirus and repopulate recipient mice at equivalent efficiencies.
Murine recipients of Ubp43-deficient cells are resistant to BCR-ABL–induced CML-like disease
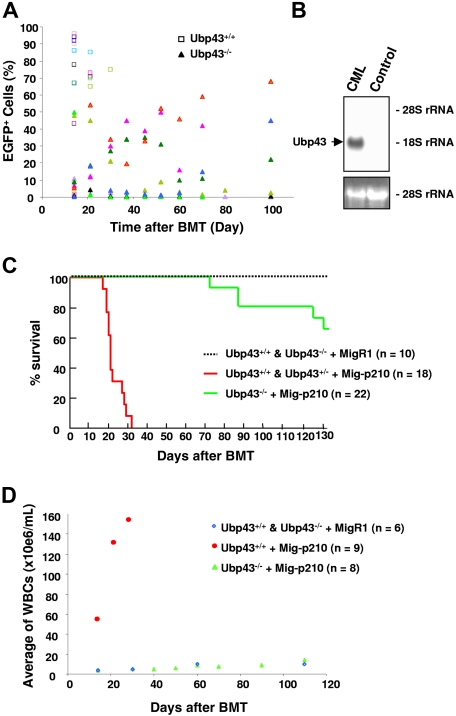
To examine the role of Ubp43 in BCR-ABL–induced leukemia development, bone marrow cells from Ubp43+/+, Ubp43+/−, and UBP43−/− mice were transduced with either MigR1 vector retrovirus or BCR-ABL–expressing retrovirus MSCV-BCR-ABLp210-IRES-EGFP (Mig-p210) and transplanted into wild-type recipient mice. Significantly, all mice that received a transplant of Mig-p210 infected Ubp43+/+ and Ubp43+/− cells developed a CML-like disease and showed a high percentage of EGFP+ cells in their peripheral blood within 28 days (Figure 1A). These mice also showed a significant increase of CD11b+/Gr-1+ myeloid cells in their bone marrow and peripheral blood (data not shown), splenomegaly, Ubp43 expression in the spleen (Figure 1B), and a fatal myeloproliferative disease by 33 days after BMT in agreement with other similar studies (Figure 1C).40,45,46 Transduction of the empty MigR1 vector control virus did not cause disease. In contrast, recipients of BCR-ABL–transduced Ubp43−/− bone marrow cells showed a substantially increased survival rate, with all mice alive at 65 days after BMT and more than 60% alive 130 days after BMT (Figure 1C). As shown in Figure 1D, the white blood cell (WBC) count in the peripheral blood also showed significant differences. In mice that received a transplant of BCR-ABL–expressing Ubp43+/+ cells, a rapid increase in WBC number was observed. In contrast, recipients of Mig-p210–transduced Ubp43−/− bone marrow cells showed comparable WBC counts with recipients of MigR1-transduced bone marrow cells. It is interesting to note that some of the mice with Mig-p210–transduced Ubp43−/− cells either temporarily or consistently showed a relatively high percentage (greater than 40%) of EGFP+ (BCR/ABL expressing) cells in their peripheral blood, but did not develop any further symptoms of a CML-like disease (Figure 1A). This suggests that the lack of Ubp43 expression suppresses the expansion of BCR-ABL–expressing cells in the peripheral blood.
Figure 1.
Significant delay of CML development with Ubp43-deficient bone marrow cells in BCR-ABL retroviral transduction/transplantation assay. (A) Percentage of BCR-ABL–expressing cells (EGFP+) in peripheral blood of mice with Mig-p210–transduced and transplanted bone marrow cells. Each color represents the result from one individual mouse. BCR/ABL was expressed in either wild-type Ubp43+/+ or Ubp43−/− bone marrow cells (triangles). (B) Ubp43 is clearly detectable in the spleen of mice that develop the CML-like disease. RNA was prepared from the spleen of a mouse that received a transplant of MigR1-transduced Ubp43+/+ bone marrow cells (control) and the spleen of a mouse with CML-like disease after transplantation with Mig-p210–infected wild-type bone marrow cells (CML). Northern blot was performed with 32P-labeled Ubp43 cDNA. Ethidium bromide–stained 28S rRNA is shown for relative RNA loading. (C) Kaplan-Meier survival curve of mice that received a transplant of Mig-p210– or MigR1-transduced Ubp43+/+, Ubp43+/−, and Ubp43−/− bone marrow cells. The result is summarized from 3 separate sets of transplantation experiments. (D) Average WBC counts of MigR1- or Mig-p210–transduced Ubp43+/+ and Ubp43−/− bone marrow cell recipients.
Pathologic changes in mice succumbing to BCR-ABL–induced leukemia
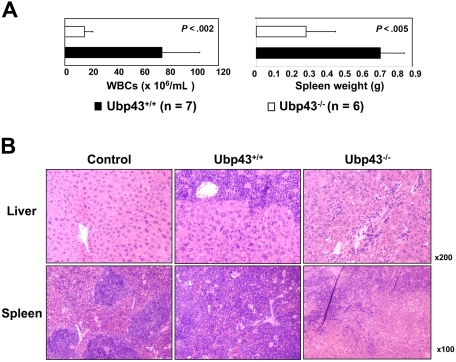
All mice that received a transplant of BCR-ABL–expressing Ubp43+ bone marrow cells developed a CML-like myeloproliferative disease. In contrast, less than 40% of mice with BCR-ABL–expressing Ubp43− cells developed a similar disease after a much longer latency period. Moreover, the severity of the CML-like disease in these 2 groups of mice also showed several differences. Mice receiving Mig-p210–transduced Ubp43+ bone marrow cells displayed the characteristic of high white blood cell counts and splenomegaly typical of this CML model (Figure 2A). In contrast, recipients of Mig-p210–transduced Ubp43− bone marrow showed much lower WBC counts and spleen weight. The leukemic cell infiltrations in the liver and spleen were also histologically examined (Figure 2B). Diseased mice with BCR-ABL–expressing Ubp43− cells were found to have significantly reduced leukemic cell infiltration of liver and spleen.
Figure 2.
Pathological analysis of diseased mice. (A) The average WBC counts and spleen weight of transplant recipients at death. The error bars represent the standard deviation. (B) Histologic analysis of spleens and livers of representative control and experimental mice that received a transplant of Mig-p210–transduced Ubp43+/+ and Ubp43−/− bone marrow cells. The tissue sections were stained by hematoxylin and eosin. Images from a Leica DMLB microscope with 10×/0.25 or 20×/0.40 objectives were captured with a Spot 2 digital camera (Diagnostic Instruments, Sterling Heights, MI) running Spot V4.64 software, and were processed with Adobe Photoshop v.5.5 (Adobe Systems, San Jose, CA).
The expression and phosphorylation of BCR-ABL are not affected by the presence of Ubp43
The BMT experiments indicated that loss of Ubp43 confers a significant survival advantage by counteracting the oncogenic properties of BCR-ABL. Since Ubp43−/− cells are hypersensitive to IFN and have increased levels of ISG15-modified proteins, we speculated that the molecular mechanism of reduced oncogenic development may be direct down-regulation of BCR-ABL expression through IFN signaling. The low infiltration of BCR-ABL–positive cells into the spleen and liver of leukemic mice with Mig-p210-Ubp43−/− cells, however, prevented elucidation of BCR-ABL levels in these mouse tissues. Instead, we performed similar retroviral transductions with MigR1 and Mig-p210 into immortalized MEFs from wild-type and Ubp43−/− mice. We also reconstituted Ubp43 expression into immortalized Ubp43−/− MEFs as an additional control (Ubp43reconst). However, there was no gross change in BCR-ABL levels and phosphorylation after 24-hour treatment with 500 U/mL IFN-β for any of the cell types, indicating that neither IFN signaling nor the absence of Ubp43 inhibited the expression and the phosphorylation of the BCR-ABL protein (Figure S2). Thus, Ubp43 inhibition of BCR-ABL driven oncogenic development occurs downstream of the BCR-ABL fusion protein.
CML mice have increased type 1 IFN in their sera
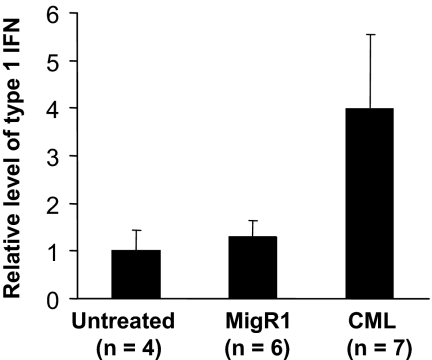
Ubp43-deficient cells are hypersensitive to IFN-α/β treatment and display a strong apoptotic response upon IFN treatment.23 It has been reported previously that human CML patients have increased production of IFN-α.47 Therefore, we hypothesized that increased type 1 IFN production during leukemogenesis and the enhanced apoptosis of Ubp43-deficient cells upon IFN stimulation could explain the resistance to CML development observed in BCR-ABL–expressing Ubp43-deficient cells. To test this hypothesis, we first analyzed the serum level of type 1 IFN. The basal level of IFN in the serum of normal healthy mice is 10 to 20 units/mL. Similar levels of IFN were detected in mice that received a transplant of retroviral vector–transduced bone marrow cells (Figure 3). However a 4-fold increase of IFN was detected in mice that have developed the CML-like myeloproliferative disease (Figure 3).
Figure 3.
Elevated type 1 IFN level is detected in the serum of mice with CML-like disease. Serum was collected from control mice, mice that received a transplant of MigR1, and BCR-ABL–induced CML mice. The concentration of type after 1 IFN in these sera was measured as described in “Materials and methods.” The relative concentrations of IFN in these sera are presented.
Increased apoptotic rate in Ubp43-deficient BCR/ABL+ cells in vivo
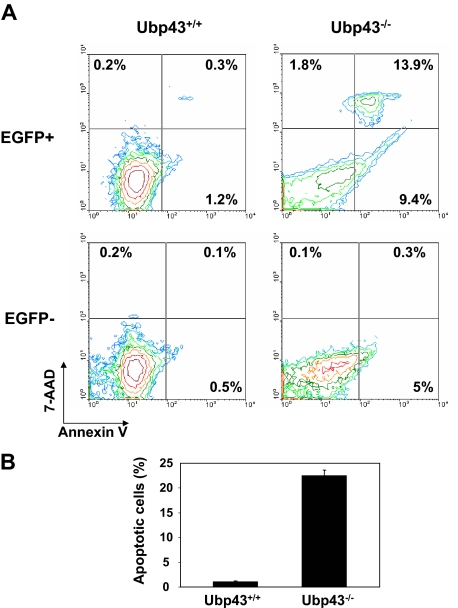
In consideration of IFN production being increased in CML mice, we examined the percentage of apoptotic cells in mice that received a transplant of BCR-ABL–expressing Ubp43+ or Ubp43− donor cells. For this analysis, we used the fact that all BCR-ABL+ cells also expressed EGFP. Three to 4 weeks after BMT, and before emergence of disease symptoms, less than 2% of early apoptotic (annexin V+/7-AAD−) and late apoptotic/necrotic (annexin V+/7-AAD+) cells were detected in the blood of mice that received a transplant of BCR-ABL–expressing wild-type donor cells (Figure 4). In contrast, more than 20% of the EGFP+ blood cell population was apoptotic in mice that received a transplant of BCR-ABL–expressing Ubp43− donor cells (Figure 4). Increased apoptosis, although to a lesser degree, was also detected in the EGFP− cells in these mice. These results support the hypothesis that the enhanced IFN sensitivity observed in Ubp43-deficient cells leads to the prevention of CML development.
Figure 4.
Increased apoptosis in Ubp43-deficient BCR-ABL+ cells in vivo. (A) Blood samples were collected from mice 3 to 4 weeks after transplantation with Mig-p210 transduced Ubp43+/+ and Ubp43−/− bone marrow cells before disease emergence. Peripheral blood cells were stained with annexin V and 7-AAD, and the percentage of apoptotic cells measured by flow cytometry. (B) The average percentages of apoptotic peripheral blood EGFP+ cells (n = 3).
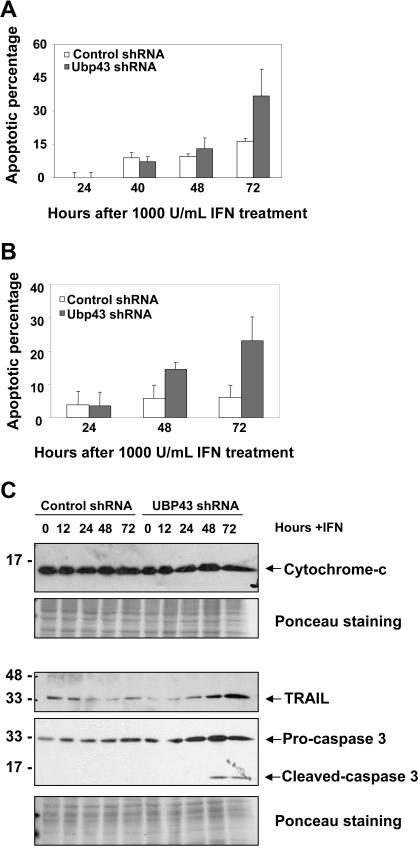
Knockdown of UBP43 sensitizes cells to the apoptotic effects of type 1 IFN
The increased apoptosis in the UBP43− cells, together with our observation of IFN up-regulation upon CML development, suggested a connection between these phenotypes and prompted us to examine whether this effect can be reproduced in the CML patient–derived KT-1 cell line.48 Pools of KT-1 cells stably expressing UBP43 shRNA show a significant increase in apoptosis upon IFNα treatment compared with cells expressing a control shRNA (Figure 5A). Furthermore, UBP43shRNA-expressing cells activated the apoptotic response even with a 10-fold reduction in the IFNα concentration, at which there is no significant increase in apoptosis for the control shRNA–expressing cells (Figure 5B). The increase in apoptosis upon IFNα treatment can be attributed to the increased expression of apoptotic genes in UBP43-deficient cells. Microarray analysis had shown several apoptosis-related genes to be up-regulated in UBP43−/− macrophages upon IFN treatment, including cytochrome c, caspase-3, and TRAIL.49 No clear increase in cytochrome c protein expression was observed in the KT-1 cell lines upon IFNα treatment (Figure 5C). However, in the UBP43 shRNA–expressing lines, there was a significant increase in the expression of TRAIL and caspase 3 proteins, as well as cleavage of caspase 3 into its active form between 24 and 48 hours after IFNα treatment, concomitant with the observed increase in apoptosis.
Figure 5.
Increased apoptosis in Ubp43 shRNA–expressing KT-1 cells treated with IFN. KT-1 cells stably transduced with the control or Ubp43 shRNA was treated with (A) 1000 units/mL or (B) 100 units/mL hIFNα, and the percentage increase in apoptotic cells (over untreated cells) was determined by annexin V/7-AAD staining at various time points. The apoptotic percentage represents the sum of early (annexin V positive) and late (annexin V/7-AAD double positive) apoptotic percentages. The results are the mean ± SD of 3 separate experiments. A comparison of the apoptotic percentage at 72 hours in control shRNA–expressing cells to that of UBP43 shRNA–expressing cells yields P values of (A) .05 and (B) .02. (C) KT-1 cells expressing the various shRNAs were treated with 1000 units/mL hIFNα. Western blots of the lysates at various time points were probed with anti–cytochrome c, anti-TRAIL, or anti–caspase 3 antibodies. Ponceau stains of the blots are shown as protein loading controls.
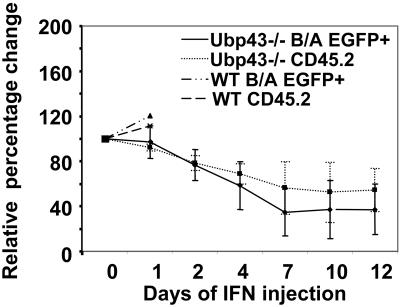
Response of Ubp43-deficient BCR/ABL+ recipient mice to IFN treatment
If the UBP43− donor cells are more sensitive to IFN-induced apoptosis as a result of the increased serum IFN levels associated with CML development, this would explain why these recipient mice show prolonged survival compared with wild-type recipients. To further test the role of IFN in disease progression in these mice, we examined the effects of daily subcutaneous injections of murine IFN into the UBP43+ or UBP43− recipient mice beginning at day 17 or day 28 after transplantation, respectively. At the starting date of injection, UBP43+ recipient mice showed approximately 50% EGFP+ cells in their peripheral blood, while UBP43− recipients showed between 15% to 45% EGFP+ cells. Wild-type recipients rapidly succumb to the BCR-ABL–induced myeloproliferative disease due to the aggressive nature of the disease and/or the lack of a sufficient response to the dosage of IFN used (Figure 6). In contrast, after 10 days of injections UBP43− recipients showed approximately 50% decrease in the initial percentage of both donor cells and donor cells expressing BCR-ABL, whereas uninjected UBP43− recipient mice over the same time period showed no such decrease (data not shown).
Figure 6.
IFN injection into Ubp43-deficient BCR-ABL+ recipient mice reduces the leukemic cell percentage. Recipients of BCR-ABL–transduced UBP43+ (n = 2) or UBP43− (n = 5) were injected subcutaneously with 18 000 units/day mIFNβ beginning at day 17 or day 28, respectively. The graph is a plot of the percentage change over time in total donor cells (CD45.2+) and BCR-ABL–positive donor cells (EGFP+) from the start date of the injection (day 0). UBP43+ recipient mice rapidly succumbed to BCR-ABL–induced leukemia, whereas UBP43− recipients showed a decrease in the donor cell numbers up to a maximum of approximately 50% at day 10.
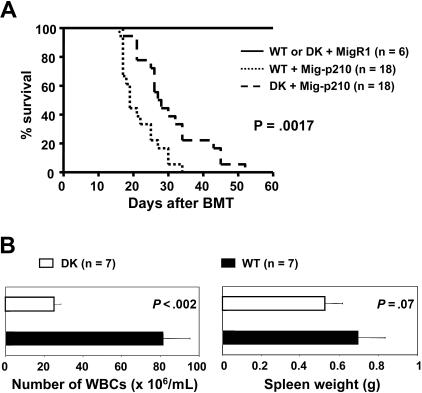
Type 1 IFN signaling is critical for the resistance to CML development in mice receiving BCR-ABL–transduced Ubp43-deficient bone marrow cells
To confirm the role of IFN in the resistance of CML development in Ubp43-deficient cells, we generated Ubp43 and IFN receptor R1 (Ifnar1) subunit double-deficient bone marrow cells by crossing Ubp43 and Ifnar1 knock-out mice. These double-deficient mice showed normal general hematopoiesis. Bone marrow cells from wild-type and double knock-out mice were used to perform the BCR-ABL retroviral transduction and bone marrow transplantation assays. Loss of type 1 IFN signaling, through loss of the IFN-α/β receptor, resulted in a reversal of the original resistance to leukemia development observed for the recipients of Mig-p210–transduced Ubp43-deficient cells (Figure 7A). The median survival times of mice with Mig-p210–transduced wild-type and Ubp43−/− cells were 19 days and 27.5 days, respectively. The CML-like pathological changes were similar in both types of mice except that substantially lower WBC counts were still observed in diseased recipients of Mig-p210-Ubp43−/−/Ifnar1−/− bone marrow cells (Figure 7B). These results demonstrate the crucial role of type 1 IFN signaling in the resistance to CML development in Ubp43-deficient bone marrow cells.
Figure 7.
IFNα/β signaling plays a critical role in resistance to leukemia development in Ubp43-deficient cells. (A) Kaplan-Meier survival curve of mice that received a transplant of MigR1- or Mig-p210–transduced wild-type bone marrow cells (WT) or Ubp43 and IFNα/β receptor subunit R1 (Ifnar1) double-deficient bone marrow cells (DK). The result is summarized from 2 separate sets of transplantation experiments. (B) WBC counts and spleen weight of moribund mice.
Discussion
Using a well-established mouse model for BCR-ABL–induced CML-like myeloproliferative disease, we have shown that Ubp43 plays an important role in regulating the latency and severity of leukemia development. Furthermore, we have detected an increase in serum levels of endogenous type I IFNs in leukemic mice. Of importance, we demonstrate that IFN signaling is critical for the inhibitory function of Ubp43 deficiency in leukemia development. This finding provides a novel molecular target to enhance innate immune responses of patients in treating various cancers that are responsive to IFN-α/β.
BCR-ABL–induced CML is a hematopoietic stem cell disease. Before the ABL tyrosine kinase–specific inhibitor imatinib became available, IFN-α had been the standard therapy choice for CML patients who were ineligible for bone marrow transplantation. Compared with IFN, imatinib is a very specific and effective drug, with fewer and less severe side effects. Of most importance, the initial response rate to imatinib is much higher than that of IFN.50 However, concerns exist over the development of imatinib drug resistance, due to amplification and mutation of BCR-ABL, and the inability of the drug to eliminate BCR-ABL+ stem cells from CML patients necessitating continued imatinib treatment even after achieving complete clinical response.51,52 In contrast, although the molecular mechanism of the IFN clinical response is not fully understood, complete remission is achievable for a significant portion of IFN-responsive patients even after stopping the administration of IFN.53,54 Recently, interferon has been reported to have higher toxicity to the more primitive CML progenitors than imatinib.55 Therefore, IFN and specific reagents that can enhance the endogenous IFN response, via the inhibition of Ubp43 or other negative regulators of IFN signaling, can be useful drugs in combination with imatinib to treat BCR-ABL–induced leukemia.
While the IFN signaling pathway is known in great detail,5 the exact molecular mechanisms whereby IFN exerts its antitumor effects have eluded researchers. Indeed, studies have shown that IFN uses a multitude of pathways that are responsible for inhibiting translation, regulating cell cycle progression, and increasing apoptosis.3,6 In vitro studies using immortalized MEFs demonstrated that IFN treatment failed to alter the expression or activation of BCR-ABL in the presence or the absence of Ubp43, suggesting that the major mechanism whereby IFN abrogates the oncogenic activity of BCR-ABL occurs further downstream. It was found that the BCR-ABL–positive cells in the peripheral blood of mice carrying Mig-p210–transduced Ubp43− bone marrow were more prone to apoptosis. The role of IFN and Ubp43 in inducing apoptosis of the leukemic cells is demonstrated both in shRNA studies of CML patient–derived cell lines (Figure 5), as well as direct injection of IFN into the recipient mice (Figure 6). Therefore, the enhanced apoptosis due to the increased IFN production during CML development may be responsible for the relatively lower WBC count, the lesser degree of splenomegaly and hepatomegaly, and the significantly extended latency of disease in mice that received a transplant of Mig-p210–transduced Ubp43−/− bone marrow cells. The results presented in Figures 4 and 6 also suggest that BCR-ABL–expressing Ubp43-deficient hematopoietic cells are more sensitive to apoptotic stimulation than non–BCR-ABL–expressing Ubp43-deficient cells. It has been reported that expression of BCR-ABL results in the increased tyrosine phosphorylation of multiple STAT family members, although the mechanism of this alteration is not clear.56–59 This increased tyrosine phosphorylation of STATs may also enhance IFN-induced JAK-STAT signaling via the complex formation of different STAT proteins and hence apoptosis. Ubp43 is a cysteine protease that catalyzes the removal of the ubiquitin-like modifier ISG15 from its conjugated targets.18,60 Protein ISGylation is strongly induced by IFN stimulation,61 as are all of the currently identified enzymes involved in ISG15 modification, including ISG15 E1–activating enzyme Ube1L, E2-conjugating enzyme UbcH8, E3 ligase Efp, and the deconjugating enzyme Ubp43.15,16,25,44,62,63 This indicates that protein ISGylation may have important functions in innate immune responses. Ubp43-deficient cells have much higher levels of ISGylated proteins than wild-type controls.18 Furthermore, Ubp43-deficient cells have an enhanced and prolonged response to type 1 IFN stimulation.23 Here, we show that Ubp43-deficient cells are resistant to BCR-ABL–induced leukemogenesis and this effect is mainly dependent on IFN signaling. We have recently demonstrated that the increased IFN sensitivity of Ubp43-deficient cells is mainly independent of its role in regulating the level of cellular protein ISGylation.26,27 Therefore, the role of protein ISGylation in cancer development and the innate immune response remains to be identified. Furthermore, besides enhancing IFN sensitivity, Ubp43 deficiency may have other currently unidentified roles in resistance of cancer development.
Supplementary Material
Acknowledgments
This work was supported by research funding from Department of Defense (DAMD17–03-1–0269) and National Institutes of Health (CA102625). The Stein Endowment Fund has partially supported the departmental molecular biology service laboratory for DNA sequencing and oligonucleotide synthesis.
We wish to thank members of Zhang lab for valuable discussions. This is manuscript 17892-MEM from The Scripps Research Institute.
Footnotes
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.Y. performed the majority of experiments, analyzed data, and wrote the paper; J.-K.L. performed a significant amount of experiments and wrote the paper; K.J.R. helped with generating UBP43 knock-out mice and edited the paper; I.S. and K.T. provided the KT-1 leukemia cell line; R.R. provided Mig-p210 construct and bone marrow transplantation method; and D.-E.Z. designed the experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong-Er Zhang, MEM-L51, The Scripps Research Institute, La Jolla, CA 92037; e-mail: dzhang@scripps.edu.
References
- 1.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Biron CA. Interferons alpha and beta as immune regulators: a new look. Immunity. 2001;14:661–664. doi: 10.1016/s1074-7613(01)00154-6. [DOI] [PubMed] [Google Scholar]
- 3.Chawla-Sarkar M, Lindner DJ, Liu YF, et al. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 5.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 6.Verma A, Platanias LC. Signaling via the interferon-alpha receptor in chronic myelogenous leukemia cells. Leuk Lymphoma. 2002;43:703–709. doi: 10.1080/10428190290016782. [DOI] [PubMed] [Google Scholar]
- 7.Fu XY. A direct signaling pathway through tyrosine kinase activation of SH2 domain-containing transcription factors. J Leukoc Biol. 1995;57:529–535. doi: 10.1002/jlb.57.4.529. [DOI] [PubMed] [Google Scholar]
- 8.Prejean C, Colamonici OR. Role of the cytoplasmic domains of the type I interferon receptor subunits in signaling. Semin Cancer Biol. 2000;10:83–92. doi: 10.1006/scbi.2000.0311. [DOI] [PubMed] [Google Scholar]
- 9.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 10.Greenhalgh CJ, Hilton DJ. Negative regulation of cytokine signaling. J Leukoc Biol. 2001;70:348–356. [PubMed] [Google Scholar]
- 11.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 12.Myers MP, Andersen JN, Cheng A, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 13.de Veer MJ, Holko M, Frevel M, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 14.Leaman DW, Chawla-Sarkar M, Jacobs B, et al. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J Interferon Cytokine Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 15.Kang D, Jiang H, Wu Q, Pestka S, Fisher PB. Cloning and characterization of human ubiquitin-processing protease-43 from terminally differentiated human melanoma cells using a rapid subtraction hybridization protocol RaSH. Gene. 2001;267:233–242. doi: 10.1016/s0378-1119(01)00384-5. [DOI] [PubMed] [Google Scholar]
- 16.Li XL, Blackford JA, Judge CS, et al. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2–5A system in attenuation of the interferon response. J Biol Chem. 2000;275:8880–8888. doi: 10.1074/jbc.275.12.8880. [DOI] [PubMed] [Google Scholar]
- 17.Liu L-Q, Ilaria R, Jr, Kingsley PD, et al. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol Cell Biol. 1999;19:3029–3038. doi: 10.1128/mcb.19.4.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 19.Schwer H, Liu LQ, Zhou L, et al. Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (Usp18). Genomics. 2000;65:44–52. doi: 10.1006/geno.2000.6148. [DOI] [PubMed] [Google Scholar]
- 20.Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 21.Dao CT, Zhang DE. ISG15: a ubiquitin-like enigma. Front Biosci. 2005;10:2701–2722. doi: 10.2741/1730. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie KJ, Malakhov MP, Hetherington CJ, et al. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 2002;16:2207–2212. doi: 10.1101/gad.1010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malakhova OA, Yan M, Malakhov MP, et al. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie KJ, Hahn CS, Kim KI, et al. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 25.Kim KI, Malakhova OA, Hoebe K, et al. Enhanced antibacterial potential in UBP43-deficient mice against salmonella typhimurium infection by up-regulating type I IFN signaling. J Immunol. 2005;175:847–854. doi: 10.4049/jimmunol.175.2.847. [DOI] [PubMed] [Google Scholar]
- 26.Malakhova OA, Kim KI, Luo JK, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim KI, Yan M, Malakhova O, et al. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol Cell Biol. 2006;26:472–479. doi: 10.1128/MCB.26.2.472-479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 29.Kirkwood J. Cancer immunotherapy: the interferon-alpha experience. Semin Oncol. 2002;29:18–26. doi: 10.1053/sonc.2002.33078. [DOI] [PubMed] [Google Scholar]
- 30.Borden EC, Lindner D, Dreicer R, Hussein M, Peereboom D. Second-generation interferons for cancer: clinical targets. Semin Cancer Biol. 2000;10:125–144. doi: 10.1006/scbi.2000.0315. [DOI] [PubMed] [Google Scholar]
- 31.Pfeffer LM, Dinarello CA, Herberman RB, et al. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 1998;58:2489–2499. [PubMed] [Google Scholar]
- 32.Talpaz M, Chernajovsky Y, Troutman-Worden K, et al. Interferon-stimulated genes in interferon-sensitive and -resistant chronic myelogenous leukemia patients. Cancer Res. 1992;52:1087–1090. [PubMed] [Google Scholar]
- 33.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining [letter]. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 34.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 35.Shet AS, Jahagirdar BN, Verfaillie CM. Chronic myelogenous leukemia: mechanisms underlying disease progression. Leukemia. 2002;16:1402–1411. doi: 10.1038/sj.leu.2402577. [DOI] [PubMed] [Google Scholar]
- 36.Van Etten RA. Mechanisms of transformation by the BCR-ABL oncogene: new perspectives in the post-imatinib era. Leuk Res. 2004;28(suppl 1):S21–S28. doi: 10.1016/j.leukres.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 38.Daley GQ, Van ER, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 39.Van Etten RA. Models of chronic myeloid leukemia. Curr Oncol Rep. 2001;3:228–237. doi: 10.1007/s11912-001-0055-y. [DOI] [PubMed] [Google Scholar]
- 40.Pear WS, Miller JP, Xu L, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 41.Muller U, Steinhoff U, Reis LF, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 42.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of radiosensitivity. Cancer Res. 1987;47:943–946. [PubMed] [Google Scholar]
- 43.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 44.Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- 46.Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szabo B, Toth FD, Kiss J, et al. Interferon production in myelo- and lymphoproliferative diseases, I: spontaneous interferon production in acute and chronic leukaemias. Acta Microbiol Hung. 1988;35:295–300. [PubMed] [Google Scholar]
- 48.Yanagisawa K, Yamauchi H, Kaneko M, et al. Suppression of cell proliferation and the expression of a bcr-abl fusion gene and apoptotic cell death in a new human chronic myelogenous leukemia cell line, KT-1, by interferon-alpha. Blood. 1998;91:641–648. [PubMed] [Google Scholar]
- 49.Zou W, Kim J-H, Handidu A, et al. Microarray analysis reveals that type I interferon strongly increases the expression of immune-response related genes in Ubp43 (Usp18) deficient macrophages. Biochem Biophys Res Commun. 2007;356:193–199. doi: 10.1016/j.bbrc.2007.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Brien SG, Guilhot F, Larson RA, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 51.Shah NP, Nicoll JM, Nagar B, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 52.Cortes J, O'Brien S, Kantarjian H. Discontinuation of imatinib therapy after achieving a molecular response [letter]. Blood. 2004;104:2204–2205. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- 53.Bonifazi F, de Vivo A, Rosti G, et al. Chronic myeloid leukemia and interferon-alpha: a study of complete cytogenetic responders. Blood. 2001;98:3074–3081. doi: 10.1182/blood.v98.10.3074. [DOI] [PubMed] [Google Scholar]
- 54.Kantarjian HM, O'Brien S, Cortes JE, et al. Complete cytogenetic and molecular responses to interferon-alpha-based therapy for chronic myelogenous leukemia are associated with excellent long-term prognosis. Cancer. 2003;97:1033–1041. doi: 10.1002/cncr.11223. [DOI] [PubMed] [Google Scholar]
- 55.Angstreich GR, Matsui W, Huff CA, et al. Effects of imatinib and interferon on primitive chronic myeloid leukaemia progenitors. Br J Haematol. 2005;130:373–381. doi: 10.1111/j.1365-2141.2005.05606.x. [DOI] [PubMed] [Google Scholar]
- 56.Carlesso N, Frank DA, Griffin JD. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J Exp Med. 1996;183:811–820. doi: 10.1084/jem.183.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ilaria RL, Jr, Van Etten RA. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271:31704–31710. doi: 10.1074/jbc.271.49.31704. [DOI] [PubMed] [Google Scholar]
- 58.Chai SK, Nichols GL, Rothman P. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol. 1997;159:4720–4728. [PubMed] [Google Scholar]
- 59.Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers CL. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13:247–254. [PubMed] [Google Scholar]
- 60.Kim KI, Zhang DE. UBP43, an ISG15-specific deconjugating enzyme: expression, purification, and enzymatic assays. Methods Enzymol. 2005;398:491–499. doi: 10.1016/S0076-6879(05)98040-3. [DOI] [PubMed] [Google Scholar]
- 61.Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. 1992;267:7806–7813. [PubMed] [Google Scholar]
- 62.Zou W, Zhang DE. The Interferon-inducible Ubiquitin-protein Isopeptide Ligase (E3) EFP Also Functions as an ISG15 E3 Ligase. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 63.Zhao C, Beaudenon SL, Kelley ML, et al. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci U S A. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.