Abstract
HSPA2 (formerly HSP70.2) is a testis-specific member of the HSP70 family, known to play a critical role in the completion of meiosis during male germ cells differentiation. Although abundantly present in post-meiotic cells, its function during spermiogenesis remained obscure. Here, using a global proteomic approach to identify genome-organizing proteins in condensing spermatids, we discovered an unexpected role for HSPA2, which acquires new functions and becomes tightly associated with major spermatid DNA-packaging proteins, Transition Proteins4 (TP) 1 and 2. Hence, HSPA2 is identified here as the first TP chaperone and these data shed a new light on the yet totally unknown process of genome condensing structures assembly in spermatids.
Although several waves of genome-wide reorganization have long been known to occur during male germ cells post-meiotic maturation, the underlying mechanisms still remain obscure. The most spectacular of these reorganizations are those associated with the replacement of histones by transition proteins (TPs) and of TPs by protamines (1–4). In somatic cells, specific sets of chaperones control the assembly of nucleosomes containing canonical and variant histones and hence are considered as major regulators of the establishment of differential genome organization (5). In differentiating spermatids, despite the fundamental role of TPs and protamines in genome organization, nothing is known on the chaperone system escorting these proteins and mediating their assembly into new DNA packaging structures. In general, very little is known on chaperones specific for histone and non-histone genome organizers, which are in action during spermiogenesis. The only characterized testis-specific histone chaperone is tNASP which binds the male germ cell linker histone, Hlt (6). Here, by analyzing the nature of proteins soluble in acids present in condensing spermatids, we have discovered an unexpected property of the testis-specific chaperone HSPA2, which sheds lights on potential mechanisms controlling the post-meiotic genome reorganization. HSPA2 had already been shown to possess an essential and specific role in male germ cell meiosis (7–10). However, because of a meiotic blockade and massive apoptosis of spermatocytes in HSPA2 knockout mice, the post-meiotic function of this protein has remained obscure. Here, HSPA2 is identified as the first chaperone of transition proteins, TP1 and TP2. Our data therefore strongly suggest that, after meiosis, HSPA2 acquires new functions and contributes to the dramatic spermatid-specific genome wide reorganisation.
EXPERIMENTAL PROCEDURES
Detailed methods are available in supplementary information.
Germinal cells fractionation
The fraction composed of condensing spermatids (steps 12–16) used for mass spectrometry analysis was obtained as described in (11).
Other spermatogenic cell fractions were obtained using the Bellvé sedimentation method (12,13). Cell fractions were enriched at 80% in each major stages (Spc, spermatocytes; R, round spermatids; RE, mix of round and elongating spermatids; EC, mix of elongating and condensing spermatids).
Acid extraction
Proteins were extracted by direct sonication of spermatogenic cell nuclei in sulphuric acid (0,2 N). The quality of extraction was controlled by SDS-PAGE gel stained by Coomassie.
Immunoprecipitation
25–40 μg of acid soluble proteins were incubated with 1–2 μg of anti HSPA2 or anti HA (Santa Cruz) antibodies in the appropriate buffer (see Supp. Information). Protein G coupled to dynabeads (Invitrogen) were used following the manufacturer’s instructions. Bound proteins were washed, edited in loading buffer and analysed by SDS-PAGE and silver staining (SilverQuest, Invitrogen), or western blots using standard procedures.
Chromatin fibres extraction
Nuclei of pooled fractions obtained after fractionation were submitted to micrococcal nuclease digestion in the appropriate conditions (see Supp Methods), Nuclei were broken by osmotic shock by re-suspension in H2O, TSA 300nM and antiprotease cocktail Complete (Roche). Micrococcal nuclease released proteins were analysed by SDS-PAGE, using core histones amounts as internal loading control.
Immunofluorescence on germ cells
Nuclei were prepared as described in “Chromatin fibres” section, and fixed on Superfrost slides. Detection details are available in Supp Methods, Confocal images were taken using a CLSM microscope (Zeiss) with slices of 0.7–1 μm.
RESULTS
Mass spectrometry analysis of acid soluble proteins
Many genome-organizing proteins including histones and testis-specific DNA-packaging proteins, such as TPs and protamines, are highly-basic and can be purified thanks to their solubility in acid. We reasoned that a global identification of acid soluble proteins extracted from condensing spermatids would allow us to gain an insight into the nature of genome-organizing proteins in these cells. Accordingly, nuclei from a population of sonication resistant mouse spermatids, composed mostly of condensing spermatids at step 12 to 16, were used to extract proteins soluble in 0,2 N sulfuric acid. A proleomic approach for the global identification of these proteins was undertaken. The identified proteins are listed in supplementary table 1. As expected, many DNA packaging proteins are on the list. Among the linker histones, the newly identified Hlt2 (14) was present and, within the high mobility group proteins, testis-specific as well as yet uncharacterized members could be identified. In addition to canonical core histones, many variants were found including new H2A and H2B variants, which we called H2AL1, H2AL2 and H2BL1, and characterized elsewhere (Govin et al., submitted). Other proteins non-related to histones were also found.
We then examined the isoelectric points (pI) of all the proteins found soluble in H2SO4 and plotted the pI values for each of these proteins along a pI scale (Fig. 1). This representation revealed the presence of two distinct groups of proteins. The first group contained, as expected, basic and mostly DNA-packaging structural proteins (Fig. 1, black arrows). Surprisingly the other group was mostly composed of acidic proteins with a pI between 4,5 and 6 (Fig. 1 grey arrows). An analysis of the nature of these acidic proteins revealed that known chaperones are among them, Based on these data, we hypothesized that a tight association between these acidic proteins, mainly the chaperones, and their basic partners may have induced their solubility in H2SO4. Accordingly, we had here an interesting opportunity to uncover new functions for some of these chaperones in spermatids by identifying their partners.
Fig. 1. Presence of acidic proteins in nuclear acid extracts from condensing mouse spermatids.
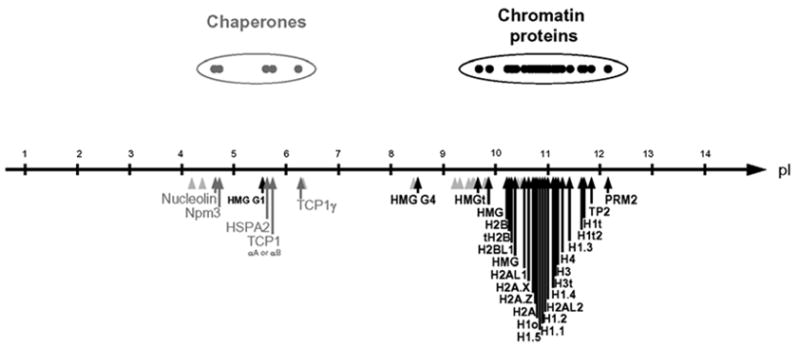
A global identification of proteins present in acid extracts from condensing spermatid nuclei was carried out, and the values of their isoelectric points (pI, supplementary table 1) were plotted along a pI scale. Black arrows indicate pI of chromatin proteins, grey arrows for chaperones; and grey arrowheads for miscellaneous proteins.
Stage-specific HSPA2 acid solubilization
Among the identified chaperones, HSPA2, a testis-specific member of the HSP70 family (15), appeared to us as an interesting candidate chaperone. Indeed, the data presented above provided an excellent opportunity to investigate its functions in post-meiotic cells where its role had remained obscure.
Spermatogenic cells were fractionated and pools of cells enriched in spermatocytes (Spc), round spermatids (R), a mix of round and elongated spermatids (RE), and a mix of elongating and condensing spermatids (EC) were obtained and used to confirmed that HSPA2 is expressed in meiotic as well as in post-meiotic cells (Fig. 2A). Our proteomic data suggested that HSPA2 becomes acid soluble owing to its tight association with DNA-packaging basic proteins. To test this hypothesis, fractionated spermatogenic cells obtained as above were submitted to acid extraction. Figure 2B shows the results of two independent experiments. Very interestingly, a fraction of HSPA2 becomes acid soluble only at late stages of spermiogenesis, after the accumulation of transition protein 2 (TP2). In the same extracts we did not find any evidence for the selective solubility of HSPA1B/HSP70 (Fig. 2B, Exp 2 and not shown). Because of high sequence identity between HSPA2 and HSPA1B, this experiment ruled out the possibility that the stage-specific acid solubility of HSPA2 could be due to non-specific binding of basic proteins to HSPA2 during the extraction procedure.
Fig. 2. Selective acid solubility of HSPA2 in elongating/condensing spermatids.
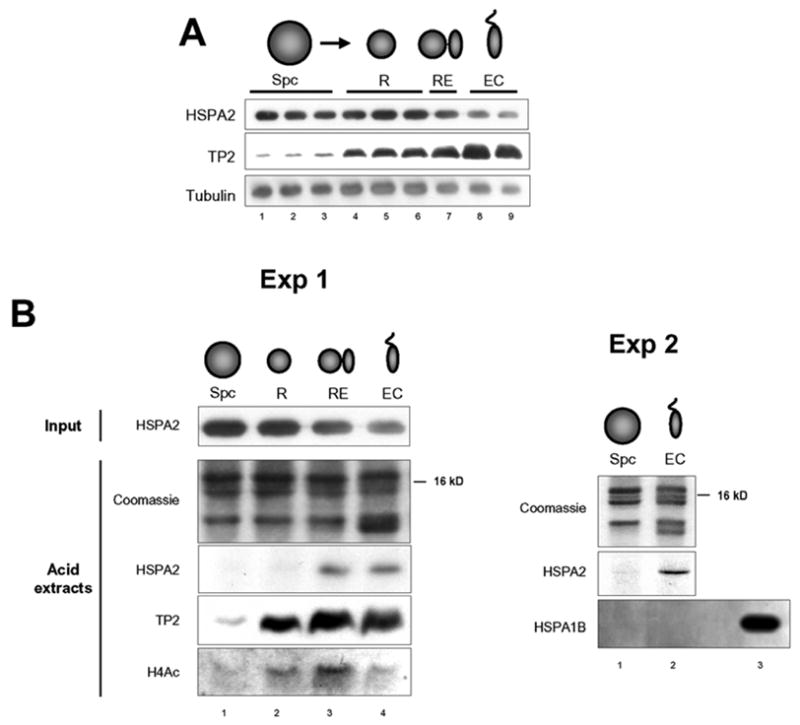
A) Equivalent amounts of total cell extracts from fractionated spermatogenic cells were used to visualize the presence of HSPA2, TP2 and tublin (Tub) in cells at different stages. Fractions are as follows: Spc, spermatocytes; R, round spermatids; RE, round and elongating spermatids; EC, elongating and condensing spermatids. Bars indicate non-pooled fractions obtained during the fractionation procedure.
B) Nuclei from the indicated pooled fractions were purified and used for an acid extraction. The presence of HSPA2 before acid extraction is shown in the input HSPA2 panel. After acid extraction proteins were recovered and solubilized in an appropriate buffer (material and methods). The presence of HSPA2, TP2 and acetylated histone H4 was monitored by the indicated antibodies. The coomassie stained gel (acid extracts) shows the histone part of the extracted proteins (Exp. 1).
In another independent experiment (Exp. 2), acid extraction was performed in a pooled spermatocyte fraction (Spc) and a pooled elongating/condensing spermatids fraction (EC). The selective solubility of HSPA2 in pooled spermatids was monitored as above. An immunodetection of HSPA1B was also performed in these fractions. As a control, the blot also contained the total nuclear extracts from the EC cells before acid extraction (lane 3).
HSPA2 function switch during spermiogenesis
Our data suggested that HSPA2’s function changes during spermatogenesis, and that it becomes specifically associated with spermatid-specific DNA-packaging proteins during the late stages of spermiogenesis. According to this hypothesis, we predicted a recruitment of HSPA2 to DNA-organizing structures as spermiogenesis proceeds. Proteins preferentially released by micrococcal nuclease from post-meiotic cell nuclei (EC) compared to a spermatocyte pool (Spc), were identified by a proteomic approach. A silver stain analysis of these proteins showed that at least two major proteins are preferentially released from the nuclei of spermatids compared to spermatocytes (Fig. 3A, arrows). Interestingly, in band N°1, in addition to HSPA2, mass spectrometry identified HSPA1L/HSC70t, another testis-specific member of the HSP/HSC70 family. Band N°2 contained disulfide isomerase A3 (PDA3).
Fig. 3. Stage-specific redistribution of HSPA2.

A) Stage-specific micrococcal nuclease-dependent release of HSPA2 from spermatogenic cell nuclei. Nuclei were prepared from pooled fractions of spermatocytes (Spc) and elongating and condensing spermatids (EC) and submitted to extensive micrococcal nuclease digestion. Material released through the action of the enzyme was visualized after silver staining of a gel. The two indicated bands were cut on another gel and identified by mass spectrometry. Band N°1 contained HSPA2 and HSPA1L/HSC70t and band N°2 contained disulfide isomerase A3 (PDA3).
B) Stage-specific intranuclear redistribution of HSPA2 during spermiogenesis.
Suspension of spermatogenic cells were used to prepare nuclei which were then subjected to co-immunolocalization of HSPA2 and acetylated lysines (monoclonal antibody detecting essentially histones in the nucleus). Images were acquired using a confocal microscope. Four representative situations are shown, R, round spermatids; E, elongating spermatids and C, condensing spermatids.
These experiments were performed on purified nuclear suspensions, because the cytoplasmic HSPA2 signal interfered with a clear detection of nuclear HSPA2, which was the object of this study. Some of the remaining cytoplasmic HSPA2 can be observed in the C panel (arrow head). Scale bar represents 2 μm.
We also tried to visualize the stage-specific HSPA2 redistribution in situ. A wave of histone acetylation is known to occur prior to histone replacement during spermatid elongation (2,3). In order to have an insight into the relationship between HSPA2 localization and histone replacement, both histone acetylation and HSPA2 were detected in spermatogenic cells, using respectively a monoclonal mouse anti-acetyl-lysine antibody (16) and a rabbit polyclonal antibody specific for HSPA2, Figure 3B shows examples of three critical periods during spermiogenesis as a function of changes occurring in global histone acetylation. In round spermatids (R), HSPA2 is almost homogenously distributed in the nucleus and, as previously shown, histones are found underacetylated in these cells (17). Interestingly, the wave of histone acetylation observed at stage VIII spermatids (17), is associated with a dramatic re-distribution of HSPA2, which becomes concentrated in a cap-like structure penetrating into the inner side of the nucleus. Co-localization with DAPI DNA staining clearly shows that these regions are DNA-containing sub-acrosomal domains and interestingly also correspond to the domain previously shown to accumulate the spermatid-specific linker Hlt2 at stage VIII (14). Moreover, the absence of hyperacetylated histones and accumulation of Hlt2 in this region indicate that this particular zone is probably one of the first concerned by the replacement of somatic-type histones in the nucleus of condensing spermatids. Figure 3B also shows an example of a more advanced spermatid (C), where most of the histone replacements have already occurred. It is interesting to note that the global disappearance of histone acetylation nicely matches the extension of HSPA2 localization into various regions and its concentration in the nucleus.
Acid-resistant association between TP1/TP2 and HSPA2
In order to visualize the binding of proteins to HSPA2 in condensing spermatids, which supposedly mediates the latter’s acid solubility, acid extracts from round and elongating/condensing pools of fractionated spermatogenic cells were used to immunoprecipitate HSPA2. The HSPA2 antibody-did not immunoprecipitate any histone, showing that if there is any HSPA2-histone interaction, it does not resist acid extraction (lane 3). The silver stained profile of the immunoprecipitated proteins, as well as other data shown here (mainly the timing of TP accumulation and HSPA2 acid-solubility) suggest that HSPA2 can form an acid-resistant complex with TP1 and TP2. In order to confirm this hypothesis, acid extracts from condensing spermatids were immunoprecipitated either with an irrelevant antibody (Ir) or the anti-HSPA2. Western blots confirmed the existence of a specific acid resistant complex between TP1/TP2 and HSPA2 in elongating and condensing spermatids. It is interesting to mention that, HSPA2 immunoprecipitation, after sonication of elongating-condensing spermatids in a buffer containing increasing amounts of salt, also allowed to pull-down DNA fragments containing TP1 and TP2 (not shown). This confirms the association of HSPA2 with genomic regions where TP1/2 assembly takes place.
DISCUSSION
HSPA2, although sharing important sequence identity with other members of the HSP70 family, shows unique functions in spermatogenic cells. Indeed, the knockout approach revealed the occurrence of specific meiotic defects in male germ cells despite of the presence of another testis-specific member, HSPA1L/HSP70t, and the somatic type HSP70 in these cells (8). Male germ cells lacking HSPA2 arrest in prophase of meiosis I and pachytene spermatocytes undergo massive apoptosis (8). In these cells, HSPA2 was shown to be associated with the synaptonemal complex and to play a role in desynapsis (7–9). The protein also appears to play a role in the assembly of the Cdc2-cyclin B1 complex during meiosis (10). Although the amount of HSPA2 in post-meiotic cells is equivalent to that in meiotic cells, no function has been attributed to this chaperone in spermatids. Here we show that a fraction of HSPA2 becomes soluble in acid only in elongating spermatids. Specific association of HSPA2 with spermatid basic proteins, including TP1 and TP2, accounts for this selective solubility of HSPA2 when extracted from elongating and condensing spermatids.
There is a considerable lack of information on the chaperone systems involved in the assembly of spermatid-specific DNA packaging structures, Chaperones such as nucleolin or NPM3, known to be expressed in spermatogenic cells (18–20), have also been found among the acid soluble proteins identified here (Fig. 1 and supplementary table 1), but no information on their chaperone activity during spermiogenesis is available.
This report provides the first evidence for such an activity and moreover reveals a unique physiological situation where the activity of a chaperone, HSPA2, evolves in a differentiation-dependent manner. The detailed analysis of the nuclear distribution of HSPA2 in spermatids, with respect to histone acetylation, confirms the existence of a specific nuclear sub-acrosomal compartment in elongating spermatids, where transitions in genome reorganizing structures seem to start. Interestingly, Davidson and colleagues had already noticed that Hlt2, a new spermatid-specific linker histone, accumulates in a highly polar manner at the apical pole of stage V-VIII spermatids under the acrosome compartment, before its spreading whithin the nucleus in stage X spermatids (14), We observe a specific accumulation of HSPA2 in the same region at approximately the same stage (VIII). Moreover, this particular mobilization of HSPA2 perfectly correlates with the disappearance of histone acetylation. Like Hlt2 (14), HSPA2 accumulation spreads to various nuclear regions at later stages. Since histone acetylation is thought to be linked to their removal (2,3), our data on the intranuclear distribution of HSPA2 strongly suggest that its spreading within the nucleus follows histone removal and the assembly of new spermatid-specific structures. This hypothesis is further supported by the fact that, at these stages, specific acid resistant complexes are formed, containing HSPA2 and TP1 and TP2.
Based on all these data we propose here that, after the completion of meiosis, HSPA2 acquires a new function as a chaperone of spermatid-specific DNA packaging proteins, and hence could be considered as the first identified factor controlling the histone to TP transition.
Supplementary Material
Acid soluble proteins identified from elongating/condensing spermatids. Acid soluble proteins from elongating/condensing proteins were extracted in sulfuric acid at 0.2N, and identified by mass spectrometry. A complete list of the identified proteins, including accession numbers, isoelectric points (see Figure 1), molecular weights and identified peptides is presented.
Fig. 4. Stage-specific acid resistant HSPA2-TP1/TP2 complex formation.
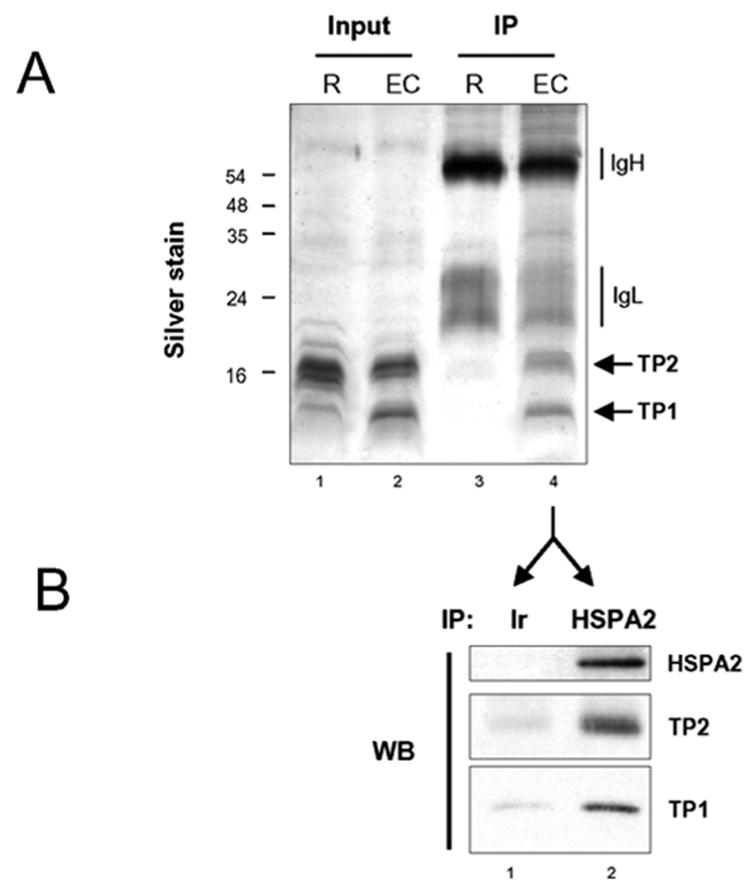
A) Acid extracts from pooled fractions containing round spermatids (R) or pooled fractions containing elongating and condensing spermatids (EC), were immunoprecipitated using the anti-HSPA2 antibody. A fraction of input and immunoprecipitated materials was visualized after silver staining of a gel. Expected TP1 and TP2 positions are indicated (arrows).
B) Acid extracts from elongating and condensing spermatids (EC), as shown in A, were immunoprecipitated with equivalent amounts of an irrelevant antibody (Ir, anti-HA) or an anti-HSPA2, The presence of HSPA2, TP1 and TP2 was monitored by a western blot on the immunoprecipitated materials.
Acknowledgments
We are grateful to Drs. Steve Kistler and Minoru Yoshida for the generous gifts of anti-TP1, anti-TP2 and anti-acetylated lysines antibodies, respectively. We wish to thank Sandrine Curtet-Benitski and Isabelle Galvez, for expert technical assistance, S.K. laboratory is supported by grants from the Regulome consortium (ANR-05-BLAN-0396-04) and the CLARA (EpiMed and EpiPro programs). E.M. Eddy laboratory is supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
The abbreviations used are
- TP
transition protein
- HSP
heat shock protein
- Spc
spermatocyte
- R
round spermatid
- RE
round and elongating spermatids
- EC
elongating and condensing spermatids
- SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- TSA
tricostatine A
References
- 1.Lewis JD, Song Y, de Jong ME, Bagha SM, Ausio J. Chromosoma. 2003;111:473–482. doi: 10.1007/s00412-002-0226-0. [DOI] [PubMed] [Google Scholar]
- 2.Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. Eur J Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 3.Caron C, Govin J, Rousseaux S, Khochbin S. Prog Mol Subcell Biol. 2005;38:65–89. doi: 10.1007/3-540-27310-7_3. [DOI] [PubMed] [Google Scholar]
- 4.Kimmins S, Sassone-Corsi P. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 5.Polo SE, Almouzni G. Curr Opin Genet Dev. 2006;16:104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Alekseev OM, Widgren EE, Richardson RT, O’Rand MG. J Biol Chem. 2005;280:2904–2911. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- 7.Allen JW, Dix DJ, Collins BW, Merrick BA, He C, Selkirk JK, Poorman-Allen P, Dresser ME, Eddy EM. Chromosoma. 1996;104:414–421. doi: 10.1007/BF00352265. [DOI] [PubMed] [Google Scholar]
- 8.Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, Poorman-Allen P, Goulding EH, Eddy EM. Proc Natl Acad Sci USA. 1996;93:3264–3268. doi: 10.1073/pnas.93.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dix DJ, Allen JW, Collins BW, Poorman-Allen P, Mori C, Blizard DR, Brown PR, Goulding EH, Strong BD, Eddy EM. Development. 1997;124:4595–4603. doi: 10.1242/dev.124.22.4595. [DOI] [PubMed] [Google Scholar]
- 10.Zhu D, Dix DJ, Eddy EM. Development. 1997;124:3007–3014. doi: 10.1242/dev.124.15.3007. [DOI] [PubMed] [Google Scholar]
- 11.Marushige Y, Marnshige K. Biochim Biophys Acta. 1983;761:48–57. doi: 10.1016/0304-4165(83)90361-6. [DOI] [PubMed] [Google Scholar]
- 12.Bellve AR. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- 13.Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S. Mol Cell Biol. 2003;23:5354–5365. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Proc Natl Acad Set US A. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eddy EM. Rev Reprod. 1999;4:23–30. doi: 10.1530/ror.0.0040023. [DOI] [PubMed] [Google Scholar]
- 16.Komatsu Y, Yukutake Y, Yoshida M. J Immunol Methods. 2003;272:161–175. doi: 10.1016/s0022-1759(02)00500-8. [DOI] [PubMed] [Google Scholar]
- 17.Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S. Eur J Cell Biol. 2000;79:950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- 18.Biggiogera M, Kaufmann SH, Shaper JH, Gas N, Amalric F, Fakan S. Chromosoma. 1991;100:162–172. doi: 10.1007/BF00337245. [DOI] [PubMed] [Google Scholar]
- 19.Shackleford GM, Ganguly A, MacArthur CA. BMC Genomics. 2001;2:8. doi: 10.1186/1471-2164-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eirin-Lopez JM, Frehlick LJ, Ausio J. Genetics. 2006 doi: 10.1534/genetics.106.058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosario MO, Perkins SL, O’Brien DA, Allen RL, Eddy EM. Dev Biol. 1992;150:1–11. doi: 10.1016/0012-1606(92)90002-x. [DOI] [PubMed] [Google Scholar]
- 22.Zhao M, Shirley CR, Yu YE, Mohapatra B, Zhang Y, Unni E, Deng JM, Arango NA, Terry NH, Weil MM, Russell LD, Behringer RR, Meistrich ML. Mol Cell Biol. 2001;21:7243–7255. doi: 10.1128/MCB.21.21.7243-7255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Acid soluble proteins identified from elongating/condensing spermatids. Acid soluble proteins from elongating/condensing proteins were extracted in sulfuric acid at 0.2N, and identified by mass spectrometry. A complete list of the identified proteins, including accession numbers, isoelectric points (see Figure 1), molecular weights and identified peptides is presented.


