Abstract
Self-assembled DNA delivery systems based on cationic lipids are simple to produce and weakly hazardous in comparison with viral vectors, but possess a significant toxicity at high doses. Phospholipids are in contrast intrinsically safe; yet their association with DNA is problematic because of unfavorable electrostatic interactions. We achieve the phospholipid-DNA complexation through the like-charge attraction induced by cations. Monovalent cations are inappropriate due to their poor binding affinity with lipids as inferred from electrophoretic mobility, whereas x-ray diffractions reveal that with multivalent cations, DNA is complexed within an inverted hexagonal liquid-crystalline phase. Coarse-grained Monte Carlo simulations confirm the self-assembly of a DNA rod wrapped into a lipid layer with cations in between acting as molecular glue. Transfection experiments performed with Ca2+ and La3+ demonstrate efficiencies surpassing those obtained with optimized cationic DOTAP-based systems, while preserving the viability of cells. Inspired by bacteriophages that resort to polycations to compact their genetic materials, complexes assembled with tetravalent spermine achieve unprecedented transfection efficiencies for phospholipids. Influence of complex growth time, lipid/DNA mass ratio, and ion concentration are examined. These complexes may initiate new developments for nontoxic gene delivery and fundamental studies of biological self-assembly.
INTRODUCTION
Gene therapy suffers from a lack of efficient and nontoxic delivery systems (1). DNA is traditionally delivered by either viral or nonviral vector-mediated systems (2). Viral methods are by far the most efficient in terms of delivery and expression, owing to the highly evolved and specialized components of natural viruses (3,4). They present, however, a number of restrictions due to the induced toxicity and immunogenicity, the limited size of DNA that can be carried, the lack of specificity, and the high cost of production (5). Considerably easier to prepare and safer to use, artificial viruses are made of self-assembling complexes of DNA with positively charged molecules such as lipids, polymers, peptides, or combinations thereof; yet two major difficulties have limited their success so far in clinical applications (5–8): i), low transfection efficiencies (TE) in comparison with that of their viral counterpart, and ii), problems of carrier toxicity, elimination, and biodegradability, especially occurring at high injected doses when increasing the total amount of delivered DNA.
A particular emphasis has been given to lipids because they are the main constituents of cell and organelle membranes for all living organisms; accordingly, numerous cationic lipids have been synthesized to date for the delivery of nucleic acids into cultured cells (9–11), as well as in clinical trials (12–14). Despite a few successful and promising attempts in vivo, the toxicity arising from these synthetic materials has been hampering their use in the pharmaceutical industry. Strong administered doses and high lipid charges are generally more toxic to a variety of cell types including cancer cell lines. Cell shrinking, inflammatory reactions, and immunotoxicity are among the many harmful effects associated with cationic lipids (15).
Unlike positively charged lipids that are only found in extremely small amounts in certain tissues (16), phospholipids, either anionic or zwitterionic, are ubiquitous in cell membranes and may thereby constitute better candidates for lipid-based delivery systems. The challenge is to achieve the complexation between nucleic acids and phospholipids, given their low affinity arising from unfavorable electrostatic interactions. Several x-ray diffraction studies have revealed ordered phases in which DNA was complexed within liquid-crystalline structures of phospholipids through the mediation of divalent cations (17–20). Interestingly, the geometry of these complexes was identical to that observed with cationic lipids, that is, lamellar and inverted hexagonal structures (21). These findings support the capability of phospholipids to compact nucleic acids under certain conditions. Very recently, phosphatidylcholine lipids could transfer plasmid DNA into mouse fibroblasts in the presence of bivalent metal cations. Unfortunately, the transfection efficiency achieved was too low for practical applications (22).
In this work, we report highly efficient phospholipid-DNA complexes assembled by using multivalent cations including trivalent and tetravalent cations. We also attempt to shed light on the mechanisms underlying the self-assembly process, as well as to identify the key parameters necessary to achieve high delivery performances. We first investigate the binding properties of multivalent cations with phospholipid membranes and correlate them with the complexation of DNA. X-ray diffractions and Monte Carlo simulations enable us to get some insight into the assembly of complexes at a molecular level. We next monitor the transfection efficiencies of complexes made of various phospholipids and multivalent cations, and compare them to a cationic mixture of lipids. The cytotoxicity on cultured cells is also assessed. Then, we seek the factors affecting the formation of complexes, and analyze their influence on the transfection performances. We finish with a summary on the supramolecular structure of complexes, the key parameters for gene delivery, and on further possible improvements.
Self-assembly of phospholipid-DNA complexes
Ion adsorption at the surface of biological membranes contributes to many cellular processes such as cell adhesion, ion transport, or binding of ligands. Previous studies, involving techniques such as electrophoretic mobility, NMR, titration calorimetry, and molecular dynamics simulations, reported a good affinity between phospholipid membranes and multivalent cations (23–25). Table 1 summarizes the binding properties of some inorganic cations with 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE). The binding constant K measures how strong the affinity of ions with the lipid membrane is, whereas the density of binding sites N represents the maximum number of ions that can be adsorbed per unit area. These values were obtained by fitting the ion concentration dependence of membrane surface potentials deduced from electrophoresis data, to the Gouy-Chapman-Stern (GCS) theory (25). Note that as the valence and the size of ions increase, the GCS theory deviates from the experiment, and in particular, we could not estimate accurately the binding parameters of large polycations such as spermine on phospholipid membranes. A direct consequence of cation binding is the increase of the membrane charge density—calculated from electrophoresis data through the Grahame equation—with ion concentration, as illustrated on Fig. 1 A. At physiological pH, DOPE had a slightly negative intrinsic membrane charge density  Due to a low density of binding sites, K+ did not alter the charge density of DOPE even at high concentrations, whereas Ca2+ and La3+ could make it strongly positive. Thanks to a 100-fold larger K, the charge density increased much more rapidly upon the addition of La3+ than with Ca2+. Moreover, we can see that the binding model behind the GCS theory accounts well for the concentration dependence of the charge density and this agreement supports the accuracy of the values listed in Table 1.
Due to a low density of binding sites, K+ did not alter the charge density of DOPE even at high concentrations, whereas Ca2+ and La3+ could make it strongly positive. Thanks to a 100-fold larger K, the charge density increased much more rapidly upon the addition of La3+ than with Ca2+. Moreover, we can see that the binding model behind the GCS theory accounts well for the concentration dependence of the charge density and this agreement supports the accuracy of the values listed in Table 1.
TABLE 1.
Intrinsic binding constants (K) and density of binding sites (N) describing the adsorption of inorganic cations to zwitterionic DOPE membranes
| Ion | K (M−1) | N (nm−2) |
|---|---|---|
| K+ | ∼0 | <0.02 |
| Mg2+ | 4.4 ± 1.4 | 0.29 ± 0.09 |
| Ca2+ | 4.0 ± 1.1 | 0.63 ± 0.15 |
| La3+ | 450 ± 30 | 0.32 ± 0.01 |
These parameters were inferred from electrophoresis data fitted with the GCS theory.
FIGURE 1.
Ion binding and self-assembly structure for zwitterionic phospholipid-DNA complexes. (A) surface charge density of DOPE membrane as a function of the ion concentration in solution for three cations of different valence: La3+ (•), Ca2+ (▪), and K+ (▴). The values were calculated from experimental electrophoresis data with the Grahame equation. The solid line represents the prediction from the Gouy-Chapman-Stern (GCS) theory by using the parameters given in Table 1. (B) Small angle x-ray scattering patterns of DOPE-DNA complexes at ion concentrations of 200 mM and L/D = 7. The integers in parentheses are Miller indices.
Because multivalent cations bind to lipid membranes and reverse their charge density, DNA can be complexed into a liquid-crystalline structure determined by small angle x-ray scattering. Fig. 1 B shows x-ray patterns measured on mixtures of DOPE, DNA, and cations. Upon the use of 200 mM of monovalent K+, the sample did not precipitate and no ordered structure was visible within the detection range of our instrument, suggesting that DNA complexation did not occur consistently with the poor binding properties of K+. In contrast, the complexation of DOPE-DNA by 200 mM of Ca2+ and La3+ gave rise clearly to the coexistence of two hexagonal phases: one denoted HII and corresponding to a phase of pure DOPE (26), and a second one labeled  and referring to a phase where DNA strands are surrounded by a lipid monolayer and arranged on a hexagonal lattice (17,21,27). In the case of Ca2+, the unit cell sizes could be inferred from the positions of Bragg peaks as
and referring to a phase where DNA strands are surrounded by a lipid monolayer and arranged on a hexagonal lattice (17,21,27). In the case of Ca2+, the unit cell sizes could be inferred from the positions of Bragg peaks as  and
and  this latter spacing corresponding well to the thickness of one DOPE membrane bilayer (∼45 Å) plus the diameter of one DNA chain (∼20 Å) and two layers of ∼2 Å each to accommodate the bridging cations. We found for La3+,
this latter spacing corresponding well to the thickness of one DOPE membrane bilayer (∼45 Å) plus the diameter of one DNA chain (∼20 Å) and two layers of ∼2 Å each to accommodate the bridging cations. We found for La3+,  and
and  which are close to the values obtained for Ca2+. Furthermore, there are some evidences that the complexed lipids were in liquid state: first, DOPE has a gel-to-liquid phase transition temperature of −16°C, and it was reported by other groups that the complexation with DNA and cations increases the transition temperature by no more than 10°C (28). Also, our differential scanning calorimetry measurements from 0°C to 60°C revealed no phase transition, indicating the complexed lipid chains were already melted.
which are close to the values obtained for Ca2+. Furthermore, there are some evidences that the complexed lipids were in liquid state: first, DOPE has a gel-to-liquid phase transition temperature of −16°C, and it was reported by other groups that the complexation with DNA and cations increases the transition temperature by no more than 10°C (28). Also, our differential scanning calorimetry measurements from 0°C to 60°C revealed no phase transition, indicating the complexed lipid chains were already melted.
Although x-ray diffractions experiments give valuable information on liquid-crystalline structures, we cannot be sure about the detailed assembly. To support the above results, we carried out Monte Carlo simulations with a simple coarse-grained water-free model of lipids, which was reportedly proven to yield a self-assembly of membrane bilayer (29). The model was only modified to set a low bending modulus to membranes, so as to get mechanical properties close to those of DOPE. At the initial state of simulations, zwitterionic lipids and divalent cations were distributed randomly around a negatively charged DNA rod (Fig. 2 A). After the equilibrium was reached (Fig. 2 B), the cations coated the DNA uniformly, and the assembly was wrapped up in a lipid bilayer, consistently with the structure suggested by x-ray experiments. In absence of cations, no stable configuration could be obtained as the repulsion between DNA and the negative part of zwitterionic lipid heads prevents their association. With cations, the repulsive electrostatic forces are screened—or even become attractive—and short-range interactions ensure stable binding. It should be noted here that since only one DNA rod was introduced in the simulations, the lipids formed a bilayer to protect the exposed hydrophobic tails from the bulk aqueous environment. In reality, in presence of several DNA molecules, only one lipid monolayer should coat each DNA.
FIGURE 2.
Monte Carlo simulations of the self-assembly of zwitterionic lipids (hydrophilic part in dark gray and hydrophobic one in light gray) and DNA (central disk) with divalent cations (small light gray spheres). (A) Initial and (B) equilibrium configurations.
Gene transfection and toxicity
We assessed the transfection capabilities of phospholipid-DNA complexes on two different human tumor cell lines, one from glioblastoma (U87), the other from hepatoma (HepG2). For the sake of clarity and to rule out a possible bias introduced by the physiological state of cells, the transfection efficiencies, quantified by luminescence assay, were normalized to the values obtained under the same experimental conditions with a binary mixture of 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and DOPE at a ratio of 60:40%. This lipid mixture is a commonly used cationic lipid transfectant and good performances have been reported in the literature (30). Fig. 3, A–B, shows the relative transfection efficiencies for DOPE-DNA complexes. The lipid/DNA mass ratio (L/D) and the growth time of complexes before transfection (τg) were both maintained at 7 and 2 h, respectively. By using Ca2+, we successfully transfected U87 and HepG2 cell lines, and the level of luciferase activity was comparable to that achieved with cationic DOTAP/DOPE for ion concentrations of 200 mM and 50 mM, respectively (Fig. 3 A). In the absence of lipids, the relative TE did not go beyond ∼0.03 for U87 and ∼0.08 for HepG2. Fig. 3 B depicts the performances obtained on U87 with DOPE-DNA complexed by cations of different valences. As expected, lipids mixed with K+ failed because DNA was not even complexed. With multivalent cations, the efficiencies increased in the following order: Mg2+ < Ca2+ < La3+, [La3+] = 100 μM giving the best level at ∼2.6-fold higher than cationic DOTAP/DOPE. It is worth noting that the efficiency curve for La3+ is shifted toward lower ion concentrations by three orders of magnitude with respect to that of Ca2+, which is ascribed to the much higher binding constant of the trivalent cations. The presence of a peak in the transfection efficiency curve is attributed to the interactions between lipid membrane and DNA: at low cation concentrations, only a few DNA molecules are complexed, and the total number of delivered DNA is therefore low. At high ion concentrations, DNA molecules are believed to be strongly bound to lipids and cannot be released properly in the endosome toward the cell nucleus. The optimal ion concentration is cell dependent (Fig. 3 A), and this can be seen as a valuable advantage for in vivo cell targeting, ensuring that one cell line is efficiently transfected whereas the others remain unaffected.
FIGURE 3.
Relative transfection efficiencies of DOPE (A) with Ca2+ on U87 (red line) and HepG2 (blue line) cells, and (B) on U87 cells with different cations: La3+ (▪), Ca2+ (•), Mg2+ (▴), and K+ (▾). Inset is a close-up view at ion concentrations comprised between 100 and 400 mM. In all experiments, L/D = 7 and  (C) Cell proliferation assays for varying amounts of DNA to transfect. Complexes are made of DOPE and
(C) Cell proliferation assays for varying amounts of DNA to transfect. Complexes are made of DOPE and  The tested cell lines are U87 and HepG2, the incubation time 48 h, and the lipid/DNA mass ratio maintained at L/D = 7.
The tested cell lines are U87 and HepG2, the incubation time 48 h, and the lipid/DNA mass ratio maintained at L/D = 7.
We monitored the toxicity induced by the complexes by performing proliferation assays on the transfected cells. To keep levels of transfection optimal, we varied the amount of delivered DNA while maintaining the L/D ratio constant. We retained the combination of DOPE with 100 μM of La3+, and the corresponding proliferation data of transfected U87 and HepG2 cell lines are shown on Fig. 3 C. With 0.2 μg of DNA considered as a nominal value, the graph shows that even with 10-fold more DNA to transfect, 89% and 79% of U87 and HepG2 cells, respectively, were still viable after 48 h of culture. Other assays involving 200 mM of Ca2+ for the preparation of complexes resulted in lower viabilities, emphasizing the importance of ion concentration to the toxicity. In a pharmaceutical context, the use of cations having a high affinity with lipids and DNA such as La3+ is beneficial for reduced toxicity, because it allows the preparation of phospholipid-DNA complexes with small amounts of cations (several orders of magnitude between Ca2+ and La3+). Thus, the intracellular ionic environment is less affected and the cell viability is preserved.
Phospholipid-DNA complexation is not limited to inorganic ions. Tetravalent polycation spermine (spr4+) is found in a wide variety of organisms and tissues, and is in particular exploited by bacteriophage T4 and herpes simplex virion as a compacting agent to accommodate DNA in their tiny viral capsid (31,32). In vitro, spermine is also known to precipitate pure DNA or in a mononucleosome core particle state (33–35). We assembled phospholipid-DNA complexes that are inspired by these above-mentioned natural viruses, by substituting inorganic cations with spermine. We found qualitatively from electrophoresis data that the binding properties of spermine were not satisfactory with DOPE, but were good enough to reverse the membrane charge density of anionic phospholipids such as 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA). Fig. 4 A shows the very high efficiency exhibited by the combination DOPA-spr4+ on U87 cells: an ∼18-fold higher expression of luciferase than that achieved with cationic DOTAP/DOPE, for just 500 μM of spermine. This outcome was confirmed under fluorescence microscope by expressing green fluorescent protein instead of luciferase in cells (Fig. 4 B). Even in the presence of serum and transfecting directly in supplemented culture medium, the luminescence assay yielded better readings because the complexes were in contact with cells for longer durations (data not shown). In absence of DOPA or cations, the level of transfection was negligible. Note that the optimal experimental conditions, namely the lipid/DNA mass ratio and the growth time before transfection, differed from the case with DOPE.
FIGURE 4.
Transfection of U87 with DOPA and spermine (spr4+). (A) Relative transfection efficiency as a function of spermine concentration. Inset is the efficiency obtained in absence of lipids. L/D = 20 and  (B) Fluorescence image of U87 cells transfected with plasmid DNA encoding GFP, by DOTAP/DOPE (top) and
(B) Fluorescence image of U87 cells transfected with plasmid DNA encoding GFP, by DOTAP/DOPE (top) and  L/D = 20,
L/D = 20,  (bottom), after 3 days of culture.
(bottom), after 3 days of culture.
Factors affecting the delivery
The factors conditioning the delivery are exemplified with natural phospholipids extracted from egg yolk, on U87 cell line: they were egg phosphatidylethanolamine (egg PE), egg phosphatidylcholine (egg PC), and egg phosphatidic acid (egg PA). Egg PE achieved a good relative TE of ∼2.2 at  as shown by Fig. 5 A. In contrast, egg PC, yet also a mixture of zwitterionic lipids, did not exceed 0.09, which is barely better than in absence of lipids. At an intermediary level, anionic egg PA stood with a relative TE approaching 0.25. These results suggest that the biophysical properties of lipids, especially the membrane bending modulus that was much higher for PC (≈ 10 kT, k the Boltzmann constant and T the temperature) than for PE (≈ kT) membrane, and the intrinsic charge density play a critical role in the delivery process. Similar conclusions were drawn from experiments on cationic lipids (9,30).
as shown by Fig. 5 A. In contrast, egg PC, yet also a mixture of zwitterionic lipids, did not exceed 0.09, which is barely better than in absence of lipids. At an intermediary level, anionic egg PA stood with a relative TE approaching 0.25. These results suggest that the biophysical properties of lipids, especially the membrane bending modulus that was much higher for PC (≈ 10 kT, k the Boltzmann constant and T the temperature) than for PE (≈ kT) membrane, and the intrinsic charge density play a critical role in the delivery process. Similar conclusions were drawn from experiments on cationic lipids (9,30).
FIGURE 5.
Gene transfections with natural phospholipids and Ca2+ ions. (A) Relative transfection efficiency with varying concentrations of Ca2+ on U87 cells: egg PE (black ▪), egg PA (gray •), egg PC (◊). L/D = 7 and  (B) Transfection efficiency of egg PE in relative light units per mg of proteins versus growth time of lipoplexes before transfection τg (bars, left axis), and their corresponding size (♦, right axis). L/D = 7 and
(B) Transfection efficiency of egg PE in relative light units per mg of proteins versus growth time of lipoplexes before transfection τg (bars, left axis), and their corresponding size (♦, right axis). L/D = 7 and  (C) Relative transfection efficiency of egg PE as a function of both L/D and
(C) Relative transfection efficiency of egg PE as a function of both L/D and  at fixed
at fixed 
Like cationic lipid systems (36), the size of complexes before transfection was found to be determinant as depicted on Fig. 5 B. Size and transfection efficiency were strongly correlated through the growth time before transfection τg. Naked egg PE particles had a size distribution centered at ∼96 nm. Within the first seconds after complex formation, the size jumped to 300 nm and reached ∼425 nm after 15 min. The transfection efficiency was the best when complexes were administered to cells at this very moment. It is worth noting that the optimal growth time could vary strongly from one lipid-ion combination to another, ranging from 15 min for egg PE-Ca2+ to 4 h for DOPA-spr4+. As egg PE-DNA complexes continued to self-assemble, their size increased to ∼650 nm after 4 h. In terms of transfection, the latter were 10-fold less efficient than their 425-nm counterparts. This is easily understandable as small complexes do not carry enough DNA to transfect and large complexes are too big to be assimilated into cells by endocytosis and/or cell-complex fusion.
Fig. 5 C gives the dependencies of the lipid/DNA mass ratio and the ion concentration for egg PE/Ca2+. The transfection efficiency peaked at L/D = 10 and  and besides this, the efficiency dropped steeply yielding low values. Both parameters set the overall charge of phospholipid-DNA complexes: if too low, the uptake to cells is inefficient; if too high, the complexes are trapped inside the endosome and DNA cannot migrate to the nucleus. In principle, L/D, the ion concentration, and τg should all be adjusted simultaneously to achieve optimal delivery to a given cell line. This may appear troublesome when one wants to transfect a large variety of cells. However, gene therapy requires only well-specified cells to be transfected and the delivery into nontargeted cells can lead to a fatal outcome. The narrow margins of operation may therefore ensure further safety for in vivo applications.
and besides this, the efficiency dropped steeply yielding low values. Both parameters set the overall charge of phospholipid-DNA complexes: if too low, the uptake to cells is inefficient; if too high, the complexes are trapped inside the endosome and DNA cannot migrate to the nucleus. In principle, L/D, the ion concentration, and τg should all be adjusted simultaneously to achieve optimal delivery to a given cell line. This may appear troublesome when one wants to transfect a large variety of cells. However, gene therapy requires only well-specified cells to be transfected and the delivery into nontargeted cells can lead to a fatal outcome. The narrow margins of operation may therefore ensure further safety for in vivo applications.
MATERIALS AND METHODS
Phospholipid-DNA complex preparation
Lipids purchased from Avanti Polar Lipids (Alabaster, AL) and dissolved in chloroform:methanol 2:1 (v/v) were dried by rotary evaporation under vacuum at 42°C. They were placed afterwards in a vacuum chamber overnight so as to remove the last traces of organic solvent. Millipore (Billerica, MA) sterilized water was added to form liposomes in solution at 0.5 mg/mL for transfection, and 5 mg/mL for x-ray diffraction. After overnight incubation at room temperature, the liposomes were tip sonicated for 10 min, and extruded through a 0.2-μm polycarbonate filter. Lipids and DNA first, then cations in chloride form (KCl, CaCl2…) were mixed together in desired ratios and incubated at room temperature during a period τg (growth time) before transfection. For x-ray experiments, λ-DNA (Roche Applied Science, Singapore) was precipitated and mixed thoroughly with concentrated lipids by gentle stirring; after addition of ions, the complexes were allowed to form for several days at 4°C. The liposomal transfectant of reference was prepared from a binary mixture of 60% DOTAP and 40% DOPE in mol. DNA was complexed in Millipore sterilized water at a DOTAP-to-DNA charge ratio of 3, and the growth time before transfection was limited to 30 min. These conditions were reportedly optimal for gene delivery (30). The size of complexes was determined by dynamic light scattering using a ZetaSizer Nano system from Malvern Instruments (Malvern, UK).
Ion binding assessment
Liposomes with no DNA were prepared as described above in buffer solution of 2 mM Tris pH 7.0, and ions were added up to the desired concentration. After overnight equilibration, ζ-potentials were measured with a ZetaPALS system from Brookhaven Instruments (Holtsville, NY). The surface potentials were obtained by solving numerically the Poisson-Boltzmann equation, considering a thickness of shearing layer of 1 Å. The binding constants K and the densities of binding sites N were inferred from least-square minimization between the Gouy-Chapman-Stern theory and the computed surface potentials (25). The charge densities shown on Fig. 1 A were calculated through the Grahame equation,
 |
where σ is the charge density, ɛ and ɛ0 the dielectric constant and the permittivity of free space, respectively, k the Boltzmann constant, T the temperature, ni(∞) the bulk ion concentrations, zi the ion valences, e the elementary charge, and ψs the surface potential of liposomes. The sum is performed over all ions in solution.
X-ray diffraction
The small angle x-ray scattering setup comprised the SAXSess small angle x-ray scattering instrument from Anton Paar GmbH (Graz, Austria) and the PW3830 laboratory x-ray generator (40 kV, 50 mA) with a long-fine focus sealed x-ray tube (CuKα wavelength of  ) from PANalytical (Almelo, The Netherlands). Detection was performed with the two-dimensional imaging-plate reader Cyclone by Perkin Elmer (Wellesley, MA). The sample was sandwiched in a Kapton cell and measurements were performed in the slit configuration with a solid sample holder for 20 min. The data were collected up to a q-value of 4 nm−1, where
) from PANalytical (Almelo, The Netherlands). Detection was performed with the two-dimensional imaging-plate reader Cyclone by Perkin Elmer (Wellesley, MA). The sample was sandwiched in a Kapton cell and measurements were performed in the slit configuration with a solid sample holder for 20 min. The data were collected up to a q-value of 4 nm−1, where  is the length of the scattering vector and θ is the scattering angle, i.e., the angle of observation. Data was corrected from instrumental broadening effects (“slit-smearing”) with the GIFT software and fitted with a sum of Lorentzian functions. Differential scanning calorimetry was performed with a Q 100 calorimeter (TA Instruments, New Castle, DE) for temperatures ramped up from 0°C to 60°C at a rate of 3°C/min.
is the length of the scattering vector and θ is the scattering angle, i.e., the angle of observation. Data was corrected from instrumental broadening effects (“slit-smearing”) with the GIFT software and fitted with a sum of Lorentzian functions. Differential scanning calorimetry was performed with a Q 100 calorimeter (TA Instruments, New Castle, DE) for temperatures ramped up from 0°C to 60°C at a rate of 3°C/min.
Monte Carlo simulations
The model used was based on a coarse-grained water-free membrane model, which has been proven to produce self-assembled lipid bilayers (29,37). The lipids consisted of one hydrophilic and two hydrophobic spherical particles with short-range pair interactions between them. Their diameter was set to 6.3 Å and the Lennard-Jones pair potentials taken as in Farago (29) except for  and
and  so as to lower the bending modulus of lipid membrane. The hydrophilic spheres carried one dipole (moment 5 e.Å) oriented along the molecule to account for the zwitterionic charge of lipids. The cations were modeled as charged spheres of 2 Å in diameter interacting with other cations and lipids through a purely repulsive 12th-order Lennard-Jones potential, plus the Coulomb potential. An infinitely long rigid rod with a uniform axial charge
so as to lower the bending modulus of lipid membrane. The hydrophilic spheres carried one dipole (moment 5 e.Å) oriented along the molecule to account for the zwitterionic charge of lipids. The cations were modeled as charged spheres of 2 Å in diameter interacting with other cations and lipids through a purely repulsive 12th-order Lennard-Jones potential, plus the Coulomb potential. An infinitely long rigid rod with a uniform axial charge  and a diameter
and a diameter  was employed for DNA. It interacted with all other particles through the following pair potential,
was employed for DNA. It interacted with all other particles through the following pair potential,
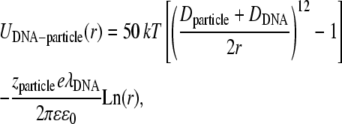 |
where Dparticle and zparticle stand for the diameter and the valence of the particle in consideration, and r the distance particle-DNA. Monte Carlo simulations were used to generate the temporal evolution of the lipids, while the position of the rod was fixed. Each Monte Carlo step consisted of an attempt to translate the particles and rotate the lipids around their midatom. The size of the simulation box was 80 Å and 171 lipids were simulated.
Cell transfection
Human glioblastoma U87 and human hepatoma HepG2 were cultured at 37°C in supplemented cell medium (Dulbecco's modified eagle's medium (DMEM) with 10% fetal bovine serum, penicillin-streptomycin, and L-glutamine) in a humidified atmosphere containing 5% CO2. The day before transfection, cells were seeded in 48-well plates so that they were 70 ∼ 80% confluent at the time of transfection. For each well, 200 ng of plasmid DNA encoding for a modified firefly luciferase (Photinus Pyralis) under the control of a strong viral promoter (SV40) (pGL3 promoter, Promega, Madison, WI) was used. Fig. 4 B involved 200 ng of plasmid DNA encoding green fluorescent protein. The cells were incubated for 4 h with 250 μL of OptiMEM reduced serum medium (Invitrogen, Carlsbad, CA) and 50 μL of phospholipid-DNA complexes, then with supplemented cell medium for an additional 24 h. Gene expression was measured with the Luciferase Assay System from Promega, and luminescence quantified by a Berthold Lumat LB 9507 luminometer (Bad Wildbad, Germany). The light output readings, in relative light units (RLU), were normalized to the weight of total cellular protein determined using Bio-Rad protein assay dye reagent (Bio-Rad, Hercules, CA). The relative transfection efficiency was obtained by further normalizing to the transfection efficiency of cationic DOTAP/DOPE under the same experimental conditions. All experiments were performed in triplicate.
Cytotoxicity
The cells of interest were seeded in 96-well plates, 1 day before transfection. Phospholipid-DNA complex solution (20 μL) containing 80 ng of plasmid DNA was added to each well filled with 100 μL of OptiMEM, and incubated for 4 h. The wells were subsequently refilled with supplemented medium and the plate stored for 48 h in the incubator. Cell proliferation was evaluated using the CellTiter 96 assay from Promega with 3 h of incubation, and the quantity of living cells in culture was determined by absorbance at 490 nm.
CONCLUDING REMARKS
In spite of unfavorable electrostatic interactions, phospholipids and DNA can be assembled into an ordered liquid-crystalline structure upon the mediation of multivalent cations. The process takes advantage of the like-charge attraction arising from the counterions that screen repulsive electrostatic forces between lipids and DNA, and allow short-ranged attractive forces to come into play (38). The underlying physics, involving electrodynamic theories, is still far from being understood, and phospholipid-DNA complexes may constitute a valuable platform for fundamental studies on biological self-assembly.
The internal structure of complexes consists of cylindrical DNA strands coated by lipid monolayers with ions in between, and arranged on a two-dimensional hexagonal lattice ( phase) as depicted by Fig. 6. Upon mixing of the components, the complexes spontaneously grow in size until equilibration.
phase) as depicted by Fig. 6. Upon mixing of the components, the complexes spontaneously grow in size until equilibration.
FIGURE 6.
Schematic of inverted micellar columns of lipids, cations, and DNA in  phase. The blue helixes stand for DNA, the yellow spheres are cations, the red and gray objects represent the hydrophilic and hydrophobic parts of lipids, respectively.
phase. The blue helixes stand for DNA, the yellow spheres are cations, the red and gray objects represent the hydrophilic and hydrophobic parts of lipids, respectively.
The efficacy in gene delivery with phospholipid-DNA complexes relies on critical factors:
The growth time before transfection τg sets the size of complexes when administered to cells, and conditions the complex uptake.
The ion concentration and the lipid/DNA mass ratio L/D set the overall charge of complexes, which is connected to the complex uptake and to the DNA release from endosome.
The concentration and valence of ions control the like-charge attraction and therefore the DNA complexation.
The lipids themselves take part in the complex uptake and the induced toxicity.
Comprehensive literature already exists on lipid research unlike other chemical reagents such as dendrimers and prophyrin derivatives, for which a great deal of work remains to be done before these are used clinically. Elaborate strategies originally developed for liposomal drug and gene delivery are readily applicable to phospholipid-DNA complexes: specific cell targeting by antibody conjugation (39,40) or colloidal stabilization by polymer grafting (41) are among the numerous techniques devoted to lipids to achieve artificial delivery systems with virus-like functions.
Acknowledgments
The authors thank Oded Farago and Suren Tatulian for their helpful advice, Juergen Pipper for the discussions, Adeline Goh for proofreading, and Jackie Ying and Shu Wang for their support. The x-ray experiments were carried out with the support of Nanyang Technological University.
This work was funded by the Agency for Science, Technology and Research (A*STAR) in Singapore.
Editor: Jonathan B. Chaires.
References
- 1.Ferber, D. 2001. Gene therapy: safer and virus-free? Science. 294:1638–1642. [DOI] [PubMed] [Google Scholar]
- 2.Gardlík, R., R. Pálffy, J. Hodosy, J. Lukács, J. Turňa, and P. Celec. 2005. Vectors and delivery systems in gene therapy. Med. Sci. Monit. 11:110–121. [PubMed] [Google Scholar]
- 3.Kootstra, N. A., and I. M. Verma. 2003. Gene therapy with viral vectors. Annu. Rev. Pharmacol. Toxicol. 43:413–439. [DOI] [PubMed] [Google Scholar]
- 4.Lundstrom, K. 2003. Latest development in viral vectors for gene therapy. Trends Biotechnol. 21:117–122. [DOI] [PubMed] [Google Scholar]
- 5.Luo, D., and M. W. Saltzman. 2000. Synthetic DNA delivery systems. Nat. Biotechnol. 18:33–37. [DOI] [PubMed] [Google Scholar]
- 6.Allen, T. M., and P. R. Cullis. 2004. Drug delivery systems: entering the mainstream. Science. 303:1818–1822. [DOI] [PubMed] [Google Scholar]
- 7.Mastrobattista, E., M. A. E. M. van der Aa, W. E. Hennink, and D. J. A. Crommelin. 2006. Artificial viruses: a nanotechnological approach to gene delivery. Nat. Rev. Drug Discov. 5:115–121. [DOI] [PubMed] [Google Scholar]
- 8.Li, S., and L. Huang. 2000. Nonviral gene therapy: promises and challenges. Gene Ther. 7:31–34. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad, A., H. M. Evans, K. Ewert, C. X. George, C. E. Samuel, and C. R. Safinya. 2005. New multivalent cationic lipids reveal bell curve for transfection efficiency versus membrane charge density: lipid-DNA complexes for gene delivery. J. Gene Med. 7:739–748. [DOI] [PubMed] [Google Scholar]
- 10.Ewert, K. K., H. M. Evans, A. Zidovska, N. F. Bouxsein, A. Ahmad, and C. R. Safinya. 2006. A columnar phase of dendritic lipid-based cationic liposome-DNA complexes for gene delivery: hexagonally ordered cylindrical micelles embedded in a DNA honeycomb lattice. J. Am. Chem. Soc. 128:3998–4006. [DOI] [PubMed] [Google Scholar]
- 11.Felgner, P. L., T. R. Gadek, M. Holm, R. Roman, H. W. Chan, M. Wenz, J. P. Northrop, G. M. Ringold, and M. Danielsen. 1987. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA. 84:7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergen, M., R. Chen, and R. Gonzalez. 2003. Efficacy and safety of HLA-B7/beta-2 microglobulin plasmid DNA/lipid complex (Allovectin-7 (R)) in patients with metastatic melanoma. Expert Opin. Biol. Ther. 3:377–384. [DOI] [PubMed] [Google Scholar]
- 13.Ewert, K., A. Ahmad, H. M. Evans, and C. R. Safinya. 2005. Cationic lipid-DNA complexes for non-viral gene therapy: relating supramolecular structures to cellular pathways. Expert Opin. Biol. Ther. 5:33–53. [DOI] [PubMed] [Google Scholar]
- 14.Stopeck, A. T., A. Jones, E. M. Hersh, J. A. Thompson, D. M. Finucane, J. C. Gutheil, and R. Gonzalez. 2001. Phase II study of direct intralesional gene transfer of allovectin-7, an HLA-B7/beta-2 microglobulin DNA-liposome complex, in patients with metastatic melanoma. Clin. Cancer Res. 7:2285–2291. [PubMed] [Google Scholar]
- 15.Dass, C. R. 2004. Lipoplex-mediated delivery of nucleic acids: factors affecting in vivo transfection. J. Mol. Med. 82:579–591. [DOI] [PubMed] [Google Scholar]
- 16.Hikita, T., K. Tadano-Aritomi, N. Iida-Tanaka, S. B. Levery, I. Ishizuka, and S. Hakamori. 2002. Cationic glycosphingolipids in neuronal tissues and their possible biological significance. Neurochem. Res. 27:575–581. [DOI] [PubMed] [Google Scholar]
- 17.Francescangeli, O., M. Pisani, V. Stanić, P. Bruni, and T. M. Weiss. 2004. Evidence of an inverted hexagonal phase in self-assembled phospholipid-DNA-metal complexes. Europhys. Lett. 67:669–675. [Google Scholar]
- 18.Francescangeli, O., V. Stanić, L. Gobbi, P. Bruni, M. Iacussi, G. Tosi, and S. Bernstorff. 2003. Structure of self-assembled liposome-DNA-metal complexes. Phys. Rev. E. 67:011904. [DOI] [PubMed] [Google Scholar]
- 19.Gromelski, S., and G. Brezesinski. 2006. DNA condensation and interaction with zwitterionic phospholipids mediated by divalent cations. Langmuir. 22:6293–6301. [DOI] [PubMed] [Google Scholar]
- 20.Liang, H., D. Harries, and G. C. L. Wong. 2005. Polymorphism of DNA-anionic liposome complexes reveals hierarchy of ion-mediated interactions. Proc. Natl. Acad. Sci. USA. 102:11173–11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Safinya, C. R. 2001. Structures of lipid-DNA complexes: supramolecular assembly and gene delivery. Curr. Opin. Struct. Biol. 11:440–448. [DOI] [PubMed] [Google Scholar]
- 22.Bruni, P., M. Pisani, A. Amici, C. Marchini, M. Montani, and O. Francescangeli. 2006. Self-assembled ternary complexes of neutral liposomes, deoxyribonucleic acid, and bivalent metal cations. Promising vectors for gene transfer? Appl. Phys. Lett. 88:073901. [Google Scholar]
- 23.Böckmann, R. A., and H. Grubmüller. 2004. Multistep binding of divalent cations to phospholipid bilayers: a molecular dynamics study. Angew. Chem. Int. Ed. Engl. 43:1021–1024. [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin, A., C. Grathwohl, and S. McLaughlin. 1978. The adsorption of divalent cations to phosphatidylcholine bilayer membranes. Biochim. Biophys. Acta. 513:338–357. [DOI] [PubMed] [Google Scholar]
- 25.Tatulian, S. A. 1999. Surface electrostatics of biological membranes and ion binding. In Surface Chemistry and Electrochemistry of Membranes. T. S. Sørensen, editor. Marcel Dekker, New York. 871–922.
- 26.Turner, D. C., and S. M. Gruner. 1992. X-ray diffraction reconstruction of the inverted hexagonal (HII) phase in lipid-water systems. Biochemistry. 31:1340–1355. [DOI] [PubMed] [Google Scholar]
- 27.Koltover, I., T. Salditt, J. O. Rädler, and C. R. Safinya. 1998. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 281:78–81. [DOI] [PubMed] [Google Scholar]
- 28.Pisani, M., P. Bruni, G. Caracciolo, R. Caminiti, and O. Francescangeli. 2006. Structure and phase behavior of self-assembled DPPC-DNA-metal cation complexes. J. Phys. Chem. B. 110:13203–13211. [DOI] [PubMed] [Google Scholar]
- 29.Farago, O. 2003. “Water-free” computer model for fluid bilayer membranes. J. Chem. Phys. 119:596–605. [Google Scholar]
- 30.Lin, A. J., N. L. Slack, A. Ahmad, C. X. George, C. E. Samuel, and C. R. Safinya. 2003. Three-dimensional imaging of lipid gene-carriers: membrane charge density controls universal transfection behavior in lamellar cationic liposome-DNA complexes. Biophys. J. 84:3307–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson, W., and B. Roizman. 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA. 68:2818–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabor, C. W., and H. Tabor. 1984. Polyamines. Annu. Rev. Biochem. 53:749–790. [DOI] [PubMed] [Google Scholar]
- 33.Raspaud, E., I. Chaperon, A. Leforestier, and F. Livolant. 1999. Spermine-induced aggregation of DNA, nucleosome and chromatin. Biophys. J. 77:1547–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raspaud, E., D. Durand, and F. Livolant. 2005. Interhelical spacing in liquid crystalline spermine and spermidine-DNA precipitates. Biophys. J. 88:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloomfield, V. A. 1997. DNA condensation by multivalent cations. Biopolymers. 44:269–282. [DOI] [PubMed] [Google Scholar]
- 36.Ross, P. C., and S. W. Hui. 1999. Lipoplex size is a major determinant of in vitro lipofection efficiency. Gene Ther. 6:651–659. [DOI] [PubMed] [Google Scholar]
- 37.Farago, O., N. Grønbech-Jensen, and P. Pincus. 2006. Mesoscale computer modeling of lipid-DNA complexes for gene therapy. Phys. Rev. Lett. 96:018102. [DOI] [PubMed] [Google Scholar]
- 38.Grosberg, A. Y., T. T. Nguyen, and B. I. Shklovskii. 2002. Colloquium: the physics of charge inversion in chemical and biological systems. Rev. Mod. Phys. 74:329–345. [Google Scholar]
- 39.Lee, H. Y., H. S. Jung, K. Fujikawa, J. W. Park, J. M. Kim, T. Yukimasa, H. Sugihara, and T. Kawai. 2005. New antibody immobilization method via functional liposome layer for specific protein assays. Biosens. Bioelec. 21:833–838. [DOI] [PubMed] [Google Scholar]
- 40.Mason, J. T., L. Xu, Z.-M. Sheng, and T. J. O'Leary. 2006. A liposome-PCR assay for the ultrasensitive detection of biological toxins. Nat. Biotechnol. 24:555–557. [DOI] [PubMed] [Google Scholar]
- 41.Allen, C., N. Dos Santos, R. Gallagher, G. N. C. Chiu, Y. Shu, W. M. Li, S. A. Johnstone, A. S. Janoff, L. D. Mayer, M. S. Webb, and M. B. Bally. 2002. Controlling the physical behavior and biological performance of liposome formulations through use of surface grafted poly(ethylene glycol). Biosci. Rep. 22:225–249. [DOI] [PubMed] [Google Scholar]








