Abstract
The translocated intimin receptor (Tir) is a key virulence factor of enteropathogenic Escherichia coli and related bacteria. During infection, Tir is translocated via a type III secretion system into host intestinal epithelial cells, where it inserts into the target cell membrane and acts as a receptor for the bacterial adhesin intimin. The effects of phosphorylation by cAMP-dependent kinase at two serine residues (Ser-434 and Ser-463) within the C-terminal domain of Tir, which may be involved in mediating structural/electrostatic changes in the protein to promote membrane insertion or intermolecular interactions, have previously been investigated. This study has focused on defining the conformation of Tir in solution and assessing any conformational changes associated with serine phosphorylation at positions 434/463. In addition to phosphorylated protein, combinations of Ala (unphosphorylatable) and Asp (phosphate-mimic) mutations of Ser-434 and Ser-463 have been generated, and a range of techniques (sodium dodecyl sulfate polyacrylamide gel electrophoresis, circular dichroism spectroscopy, analytical ultracentrifugation) used to further dissect the structural role and functional implications of changes in residue size/charge at these positions. The results have shown that under physiological NaCl concentrations, Tir is a monomer and adopts a highly elongated state in solution, consistent with a natively unfolded conformation. Despite this, perturbations in the structure in response to buffer conditions and the nature of the residues at positions 434 and 463 are apparent, and may be functionally relevant.
INTRODUCTION
Infection with the gram-negative bacterium enteropathogenic Escherichia coli (EPEC) is a major cause of infantile diarrhea in the developing world (1,2). EPEC infection is characterized by destruction of host-cell intestinal microvilli, actin rearrangement within epithelial cells, and the formation of a raised platform or pedestal at the site of bacterial attachment (3). EPEC utilizes a type III secretion system to deliver a series of protein effectors directly into target epithelial cells. The translocated intimin receptor (Tir), a 56-kDa protein essential for EPEC virulence, becomes inserted into the plasma membrane of host epithelial cells after translocation through the type III secretion system (4). After membrane insertion in vivo, Tir adopts a structure comprising N- and C-terminal domains on the cytosolic face of the membrane, linked via two transmembrane helices to an extracellullar domain that mediates intimin binding (5,6). Recently, the protein, which can be expressed and purified in a soluble form, has been shown to insert directly into membranes from solution in vitro, adopting a topology as found in vivo (7).
Tyrosine phosphorylation plays an important role in EPEC Tir function, with modification of Tyr-474 essential for recruitment of the Nck adaptor molecule, and actin nucleating machinery, which results in pedestal formation (8,9). A role for phosphorylation of Tyr-454 in recruitment of cytoskeletal proteins and pedestal formation has also been proposed (10). EPEC Tir resolves on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels at a higher molecular mass (∼78 kDa) than would be predicted from its sequence, which has been interpreted as the protein retaining some residual structure under these conditions (4). On translocation into mammalian cells, Tir undergoes changes in apparent molecular mass resulting in protein isolated from infected cell membranes running at ∼90 kDa on SDS-PAGE gels (4). These observations have led to the use of anomalous shifts on SDS-PAGE gels as probes for the conformational state of Tir in solution, revealing that changes in apparent molecular mass are mediated by serine phosphorylation (11) and can be mimicked in vitro by phosphorylation of Ser-434 and Ser-463 residues with cAMP-dependent protein kinase A (PKA) (12). Tir is also a substrate for PKA in vivo (11,12). As phosphorylation of tyrosine residues within Tir has no effect on its apparent molecular mass (11,12), the observed shifts cannot be fully accounted for by electrostatic effects of charge alteration on the protein (as discussed in Hawrani et al. (12)). Although Tir has recently been shown to insert directly into membranes in the absence of modification or accessory factors in vitro, it remains an attractive proposal that serine phosphorylation may act as a trigger or facilitator of Tir insertion into membranes in vivo. This could be mediated either directly, through conformation/electrostatic changes promoting insertion, or indirectly, by assisting interactions with accessory factors.
One concern with the use of phosphorylated proteins for molecular/structural analysis is sample homogeneity, especially where multiple phosphorylation sites exist. The mutation of serine residues to Asp or Glu to mimic the effects of phosphorylation has become a well established technique (13,14). Of the amino acids, Asp and Glu best retain the shape and charge of phosphorylated serine residues. Using mutagenesis rather than enzymatic phosphorylation ensures homogeneity in the sample and allows the effects of individual phosphorylation events to be studied. When combined with alanine mutants (which are not phosphorylatable) and phosphorylation assays of the native protein (15), a detailed picture of the effects of phosphorylation can be obtained.
One interpretation of the anomalous migration of Tir in SDS-PAGE gels is that the protein may adopt a natively unfolded/disordered state in solution. This would be consistent with recent studies showing that natively unfolded proteins give apparent molecular masses 1.2–1.8 times higher on SDS-PAGE gels than would be expected from their sequence or measured by mass spectrometry (16). Recently, a number of studies have used circular dichroism (CD, a measure of the secondary structure content in proteins (16)), gel filtration (shape of protein, i.e., asymmetry/elongation, expressed in terms of hydrodynamic (Stokes) radius (Rs) (17)), and small-angle scattering (radius of gyration (16,18,19)) to characterize natively unfolded proteins. For example, the value of Rs in relation to the mass of the protein is usually determined by gel filtration or dynamic light scattering, and appears to be one of the major parameters indicative of the protein being intrinsically unfolded/disordered (16,20).
Analytical ultracentrifugation (AUC) has recently been used for mass determination of natively unfolded proteins (21,22). Sedimentation equilibrium studies gave an accurate value of protein molecular weight that was unaffected by its shape. The sedimentation velocity approach, which is a hydrodynamic technique, contains information about both protein mass and shape. Ordinarily, one-dimension sedimentation velocity data treatment gives a value for the protein mass that is affected by its shape (usually the mass is underestimated for elongated particles, especially if a mixture of species is present). Recently, a method for two-dimension treatment of sedimentation velocity has been proposed (23), which allows for determination of protein molecular mass and shape (expressed in terms of friction coefficient (f/f0) or Rs) independently. Therefore, this approach can be particularly useful in characterizing natively unfolded proteins adopting extended conformations in solution.
In this study, a set of mutants has been generated to probe the effects of serine phosphorylation at positions 434 and 463 on the solution structure of EPEC Tir. Within a C-terminal domain construct (cTir), both Ser-434 and Ser-463 have been mutated to Ala or Asp in all combinations. Additionally, a double Asp mutant (Ser-434-Asp/Ser-463-Asp) has been generated in the full-length protein (FL-Tir). These mutants are potentially useful tools for further study of Tir function where fine control of the phosphorylation state of these residues is important (for instance, studies of protein/membrane interaction). However, to be useful tools, the mutations should mimic the effects of phosphorylation on the structure of Tir. A combination of SDS-PAGE and phosphorylation assays (to track changes in apparent molecular mass), CD spectroscopy, and AUC (sedimentation velocity method) has been used to characterize the conformations of cTir and FL-Tir in solution and investigate structural changes after modification at the Ser-434 and Ser-463 positions. The results reveal that both FL-Tir and its C-terminal domain are monomeric and adopt highly elongated conformations at physiological conditions. This fact, combined with the limited amount of secondary structure observed for these proteins, suggests that they adopt natively unfolded states in solution, which may be important for function.
MATERIALS AND METHODS
Site-directed mutagenesis
Ala and Asp mutants were generated in cTir by overlap-extension polymerase chain reaction (PCR) (24). Template plasmids (pET27b) containing the wild-type and S434A mutant cTir constructs were provided by Prof. B. Kenny (Newcastle University, Newcastle upon Tyne, UK). Primers were designed to span the regions encoding residues Ser-434 and Ser-463 and incorporate the appropriate mutation (base changes underlined): Ser-434-Asp, 5′-GAATAGACGTGATGATCAGGGGAGTGTT-3′ (Forward), 5′-AACACTCCCCTGATCATCACGTCTATTC-3′ (Reverse); Ser-463-Ala, 5′-GGCTCGGAATGCTCTATCGGCTCATCAG-3′ (Forward), 5′-CTGATGAGCCGATAGAGCATTCCGAGCC-3′ (Reverse); Ser-463-Asp, 5′-GGCTCGGAATGATCTATCGGCTCATCAG-3′ (Forward), 5′-CTGATGAGCCGATAGATCATTCCGAGCC-3′ (Reverse). Two PCR reactions were used to generate the upstream (T7 promoter primer and reverse primer encoding the mutation) and downstream (forward primer encoding the mutation and T7 terminator primer) regions of the coding sequence, with a third reaction used to generate the full length sequence (T7 promoter and T7 terminator primers, relevant products above as the template). Where generation of a second mutation was required, the appropriate template DNA was used. Full-length PCR products were cloned into pGEM-T (Promega, Madison, WI), and mutations were confirmed by sequencing. Coding regions were cut from pGEM-T with BamH1 and Nhe1, and ligated into pET27b, predigested with the same enzymes.
The Ser-434-Asp/Ser-463-Asp FL-Tir double mutant was generated in two steps using the Quikchange kit (Stratagene, La Jolla, CA), following standard procedures. The primers used were the same as above. First, the S434D mutant was generated; this construct was then used as template DNA for the second reaction, generating the S463D mutation. The mutations were confirmed by DNA sequencing.
Protein expression and purification
pET27b plasmids encoding cTir and FL-Tir proteins were transformed into E. coli BL21 (DE3) cells for overexpression. Recombinant proteins were expressed and purified as described previously (7,12), but with the addition of a final gel filtration step (HiLoad S75 16/60 column equilibrated in 20 mM Tris and 150 mM NaCl, pH 7.5) to ensure high purity. All protein concentrations were determined by absorbance at 280 nm using a theoretically derived extinction coefficient based on protein sequence.
Phosphorylation assays
Phosphorylated forms of cTir and FL-Tir were generated by incubating 0.3 mg of the substrate with 100 μM ATP and 3000 units of PKA (New England Biolabs, Beverly, MA) in 1× PKA reaction buffer (New England Biolabs) at 30°C for 4 h. Control experiments (not shown) were the same, but with water substituted for PKA. Proteins were visualized using either 12% (cTir) or 8% (FL-Tir) SDS-PAGE.
Circular dichroism spectroscopy
CD spectra were collected using a JASCO-810 spectropolarimeter (Jasco, Tokyo, Japan) fitted with a Peltier temperature controller. Spectra of native and mutant cTir and FL-Tir were collected from 190 to 250 nm, using a 0.1-mm-pathlength cuvette, at 25°C in 20 mM sodium phosphate and 150 mM NaCl, pH 7.5. The concentration of each stock protein solution (∼10 mg/ml) was determined by absorbance at 280 nm followed by dilution to 0.2 mg/ml immediately before the experiments. Final spectra were generated as averages of five repeat scans, with appropriate protein-free buffer spectra subtracted. The data were plotted without smoothing using Excel. The data were truncated to 190 nm on the wavelength axis corresponding to the value where the high-tension voltage rose above 600 V (the accepted measure of the signal/noise ratio for this machine, ensuring accurate measurements). The spectra were analyzed with programs available at the DICHROWEB web server (http://www.cryst.bbk.ac.uk/cdweb/html/) (25). Secondary-structure estimations were obtained using the Provencher and Glockner method (26).
Analytical ultracentrifugation
Sedimentation velocity (SV) experiments were carried out in a Beckman Coulter (Palo Alto, CA) ProteomeLab XL-I analytical ultracentrifuge using both absorbance at 280 nm and interference optics. All AUC runs were carried out at the rotation speed of 48,000 rpm and experimental temperature 4°C. The sample volume was 400 μl for the SV experiments, and the sample concentrations ranged between 0.2 and 1.8 mg/ml. The partial specific volumes (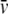 ) for the protein was calculated from the protein amino acid sequence, using the program SEDNTERP (27), and extrapolated to the experimental temperature following the method described in Durchschlag (28). The density and viscosity of the buffer (20 mM Tris pH 7.5, 50/150 mM NaCl) at the experimental temperature was also calculated using SEDNTERP.
) for the protein was calculated from the protein amino acid sequence, using the program SEDNTERP (27), and extrapolated to the experimental temperature following the method described in Durchschlag (28). The density and viscosity of the buffer (20 mM Tris pH 7.5, 50/150 mM NaCl) at the experimental temperature was also calculated using SEDNTERP.
The distributions of sedimenting material were modeled as a distribution of Lamm equation solutions (29), where the measured boundary a(r,t) was modeled as an integral over differential concentration distributions c(s).
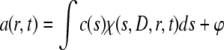 |
(1) |
where ϕ is the noise component, r is the distance from the center of rotation, and t is time. The expression χ(s, D, r, t) denotes the solution of the Lamm equation (30) for a single species by finite-element methods (31). Eq. 1 is solved numerically by discretization into a grid of 200 sedimentation coefficients for both absorbance and interference data and the best-fit concentrations for each plausible species are calculated according to a linear least-squares fit (as implemented in the program SEDFIT (http://www.analyticalultracentrifugation.com)). The sedimentation velocity profiles were fitted using a maximum entropy regularization parameter of p = 0.95. The weight-average sedimentation coefficient was calculated by integrating the differential sedimentation coefficient distribution (32):
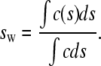 |
(2) |
Sedimentation coefficients were extrapolated to zero concentration and converted to standard conditions, i.e., those that would be measured at 20°C in water. The diffusion coefficient (D) corresponding to each sedimentation coefficient value was estimated from a weight-average frictional ratio (f/f0)w (33):
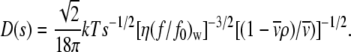 |
(3) |
The integration of the mass distribution c(M) was made similarly to as in Eq. 2 to determine the weight-average molecular mass of solute.
The two-dimensional “size-and-shape” distribution model (c(s,f/f0)) was applied to determine, from sedimentation velocity data, precisely an accurate value for the molecular mass (23). In this model, a differential distribution of sedimentation coefficients and frictional ratios (fr, i.e., f/f0) is defined as
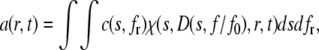 |
(4) |
where all symbols are the same as in Eq. 1. In this model, sedimentation coefficient and friction ratio (i.e., mass) for sedimenting species are fitted as independent variables. The numerical solutions of Eq. 4 were obtained in a discrete grid of 50 sedimentation coefficients and 10 values of f/f0.
PONDR prediction for cTir/FL-Tir
Protein sequences were submitted to the PONDR web engine (www.pondr.com) using the neural network predictor VL-XT (34). Access to PONDR was provided by Molecular Kinetics (Indianapolis, IN).
Stokes radius calculations for cTir/FL-Tir
The theoretical Stokes radius (Rs) of the native ( ) and fully unfolded (
) and fully unfolded ( ) proteins were determined as described in Uversky (17):
) proteins were determined as described in Uversky (17):
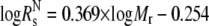 |
(5) |
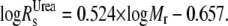 |
(6) |
RESULTS
Phosphorylation of residues Ser-434 and Ser-463 in the EPEC Tir virulence protein by cAMP-dependent kinase (PKA) have been observed both in vitro and in vivo and are potentially important events for protein function (12). However, the detailed effects of these phosphorylation events on both protein structure and function remain to be fully determined. In an attempt to further characterize the effects of modification at these two sites, and generate tools for further study, a series of Ala and Asp mutations have been constructed and characterized in vitro using a number of techniques.
Changes in apparent molecular mass on SDS-PAGE
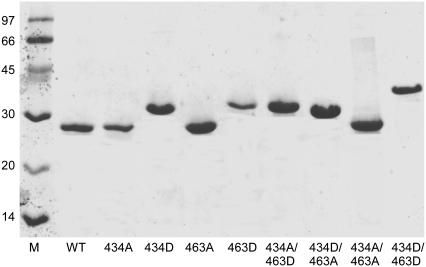
SDS-PAGE has previously been used as a tool for visualizing conformational dynamics within the cTir and FL-Tir proteins on phosphorylation (5,11,12). Fig. 1 shows an SDS-PAGE gel of unphosphorylated native and mutant cTir proteins revealing the Ser-434-Asp, Ser-463-Asp, and Ser-434-Asp/Ser-463-Asp mutations result in significant increases in apparent molecular mass above that expected for a single residue change. As compared to the wild-type protein, the increases in apparent molecular mass are ∼4 kDa for Ser-434-Asp, ∼6 kDa for Ser-463-Asp, and ∼12 kDa for the Ser-434-Asp/Ser-463-Asp double mutant (judged from the positions of the bands on the gel compared to the size marker). The shifts for the single point mutations differ, suggesting that they are not solely due to a change in the protein's charge. These shifts correlate well with previous in vitro and in vivo phosphorylation assays with both cTir and FL-Tir, where progressive stepwise increases in apparent molecular mass have been attributed to successive phosphorylation of Ser-434 and Ser-463 (11,12). The Ser-434-Ala/Ser-463-Asp double mutant resolves at a higher molecular weight than Ser-434-Asp/Ser-463-Ala, consistent with the pattern observed for the native/Asp mutants. The combined Ser-434-Asp/Ser-463-Asp mutant shows the greatest shift, closely approaching the change observed for the fully PKA-phosphorylated native cTir (below and Hawrani et al. (12)).
FIGURE 1.
12% SDS-PAGE gel of cTir WT and Ala/Asp mutant proteins. WT, wild-type protein. Ala and Asp residues are referred to by their residue number and single-letter code. Protein (8 ng) was loaded in each lane and samples were visualized by Coomassie blue staining.
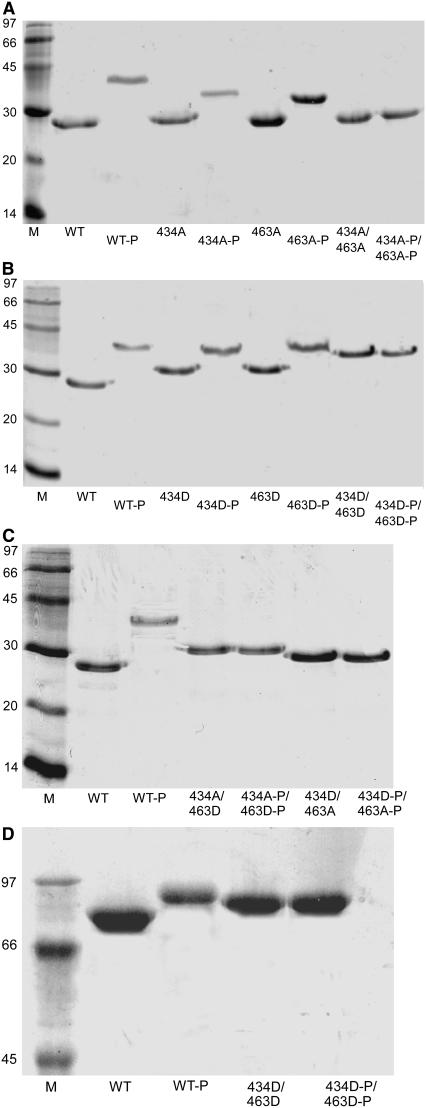
The effect of the Ser-434-Asp/Ser-463-Asp mutant on the resolving properties of FL-Tir on SDS-PAGE is shown in Fig. 2 D. As for the equivalent cTir mutant, the gel reveals a significant apparent molecular weight shift compared to the native protein.
FIGURE 2.
SDS-PAGE gels showing the effects of phosphorylation on (A) the Ala mutants, (B) the Asp mutants, and (C) the Ala/Asp combinations in cTir and (D) FL-Tir. cTir samples were run on 12% gels, FL-Tir samples on 8% gels. WT, wild-type protein. Ala and Asp residues are referred to by their single-letter code. Phosphorylation assays were performed as described in Materials and Methods. cTir (8 ng) and FL-Tir (10 ng)were loaded in each lane as appropriate and samples were visualized by Coomassie blue staining.
Phosphorylation assays
The effects of PKA-mediated phosphorylation on wild-type and mutant cTir (detectable using the gel-shift analysis) are shown in Fig. 2, and reveal that the modifications only involve residues Ser-434 and Ser-463. In any of the cTir mutant combinations where both residues are nonphosphorylatable, no additional apparent molecular mass shifts are observed on SDS-PAGE gels. Where a single serine remains, incubation with PKA results in apparent molecular mass shifts, showing that this residue has been modified. This confirms that each serine residue is independently available for modification, and mutation of one site does not preclude phosphorylation of the second. Consistent with the observed effects of the Asp mutants in isolation, in the constructs where one of the Ser residues is replaced by an Asp, the apparent molecular mass of the PKA-modified proteins on SDS-PAGE gels are higher than for the equivalent Ala substitutions.
Fig. 2 D shows the resolving properties of native and phosphorylated FL-Tir in addition to the Ser-434-Asp/Ser-463-Asp mutant. The FL-Tir Ser-434-Asp/Ser-463-Asp mutant displays resolving properties similar to those of the phosphorylated wild-type protein, confirming, at least in this assay, that the Ser-434-Asp/Ser-463-Asp mutant is an effective phosphorylation mimic. Attempts to phosphorylate the Ser-434-Asp/Ser-463-Asp mutant also suggest that there are no further sites for PKA phosphorylation in FL-Tir (identifiable using this assay).
CD spectroscopy
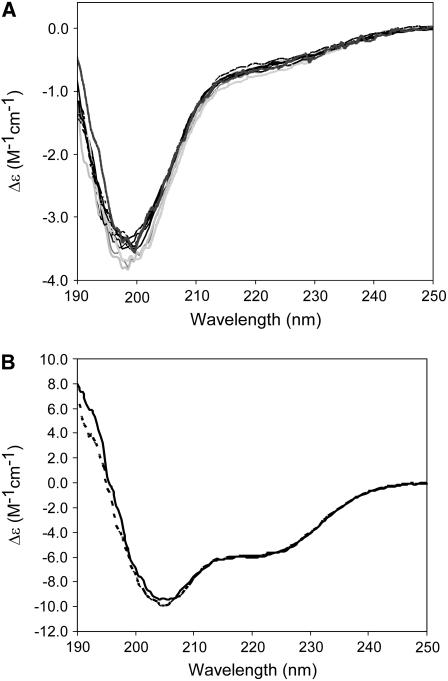
Far-ultraviolet (UV) CD spectra of the cTir and FL-Tir proteins are shown in Fig. 3, A and B, respectively. For each of the cTir mutants, although essentially the same trace was obtained, two distinct classes are apparent. For three of the proteins (Ser-434-Ala, Ser-434-Asp/Ser-463-Ala, and Ser-434-Asp/Ser-463-Asp), there is a slight shift to shorter wavelength and a deepening of the trough at just under 200 nm (see Fig. 3 A). Deconvolution of the spectra into secondary-structure components reveals that the proteins largely adopt an “unordered” structure in solution (66–74%, see Table 1) and that none of the mutations described result in a significant change in the secondary structure composition. Attempts to investigate the proteins' tertiary structure by near-UV CD revealed there was no significant spectroscopic signal in the 250–350 nm range, even at high protein concentrations. These results are consistent with previously published observations of native and phosphorylated cTir, where it was predicted that cTir would adopt a partially disordered or loosely packed structure (12).
FIGURE 3.
Far-UV circular dichroism. (A) Wavelength scans of cTir WT and mutants. The data display two overall patterns, as discussed in the text. Traces for WT, Ser-434-Asp, Ser-463-Ala, Ser-463-Asp, Ser-434-Ala/Ser-463-Ala, and Ser-434-Ala/Ser-463-Asp are solid; Ser-434-Ala, Ser-434-Asp/Ser-463-Ala, and Ser-434-Asp/Ser-463-Asp are shaded. (B) Wavelength scans for full-length protein (solid line, WT; dashed line, Ser-434-Asp/Ser-463-Asp).
TABLE 1.
Secondary structure components of cTir and FL-Tir as determined by deconvolution of CD spectra
| α-Helix | β-Strand | Turn | Unordered | |||||
|---|---|---|---|---|---|---|---|---|
| % | n* | % | n* | % | n* | % | n* | |
| cTir WT (150 mM NaCl) | 9.9 | 16 | 8.2 | 14 | 10.3 | 17 | 71.8 | 118 |
| cTir WT (50 mM NaCl) | 10.4 | 17 | 7.1 | 12 | 9.5 | 16 | 73.0 | 120 |
| cTir WT-P | 13.4 | 22 | 9.4 | 16 | 9.2 | 15 | 68.0 | 112 |
| cTir 434A | 7.2 | 12 | 8.1 | 14 | 14.2 | 23 | 70.5 | 116 |
| cTir 434D | 10.1 | 17 | 7.9 | 13 | 12.6 | 21 | 69.4 | 114 |
| cTir 463A | 8.7 | 14 | 7.2 | 12 | 10.6 | 17 | 73.5 | 122 |
| cTir 463D | 13.3 | 22 | 9.7 | 16 | 10.8 | 18 | 66.2 | 109 |
| cTir 434A/463D | 14.4 | 24 | 8.3 | 14 | 10.5 | 17 | 66.8 | 110 |
| cTir 434D/434A | 9.8 | 16 | 8.7 | 14 | 11.2 | 18 | 70.3 | 117 |
| cTir 434A/463A | 8.8 | 15 | 7.4 | 12 | 10.2 | 17 | 74.0 | 121 |
| cTir 434D/463D | 13.6 | 22 | 8.9 | 15 | 9.6 | 16 | 67.9 | 112 |
| FL-Tir WT | 20.9 | 115 | 24.3 | 134 | 22.0 | 121 | 32.2 | 188 |
| FL-Tir WT (50 mM NaCl)† | 25.8 | 142 | 23.3 | 117 | 21.5 | 107 | 29.0 | 159 |
| FL-Tir WT-P | 24.4 | 134 | 24.8 | 136 | 23.1 | 127 | 27.7 | 153 |
| FL-Tir 434D/463D | 25.1 | 138 | 23.9 | 131 | 22.5 | 124 | 28.5 | 154 |
Values represent the average of separate calculations using CONTIN, CDSSTR, and SELCON3.
The number of residues adopting this secondary structure in the protein (based on a total of 165 residues in cTir and 550 residues in FL-Tir).
Reference 12.
Deconvolution of the CD spectra for the native and Ser-434-Asp/Ser-463-Asp FL-Tir is also presented in Table 1 (spectra in Fig. 3 B). As for the comparison of the cTir wild-type (WT) and mutant proteins, these two spectra essentially overlay, indicating a very similar overall secondary structure composition. However, it is interesting to note that the shift to the left and deepening of the trough in the trace of the Ser-434-Asp/Ser-463-Asp mutant at ∼205 nm compared to the native protein is consistent with the observed shift between the spectra of native and phosphorylated FL-Tir (12). This suggests that the Ser-434-Asp/Ser-463-Asp mutant may be mimicking any subtle changes made by phosphorylation in the secondary-structure content of FL-Tir. No significant near-UV CD spectra could be observed for FL-Tir at 10 mg/ml (180 μM); previously, Tir was unstable at the concentrations required for this technique (12).
Hydrodynamic studies of cTir/FL-Tir by sedimentation velocity
SV provides a qualitative measure for protein size and shape. Recent advances in SV data treatment enable accurate determination of molecular mass (to include the association state of macromolecules), where it was previously necessary to use sedimentation equilibrium experiments (33,35).
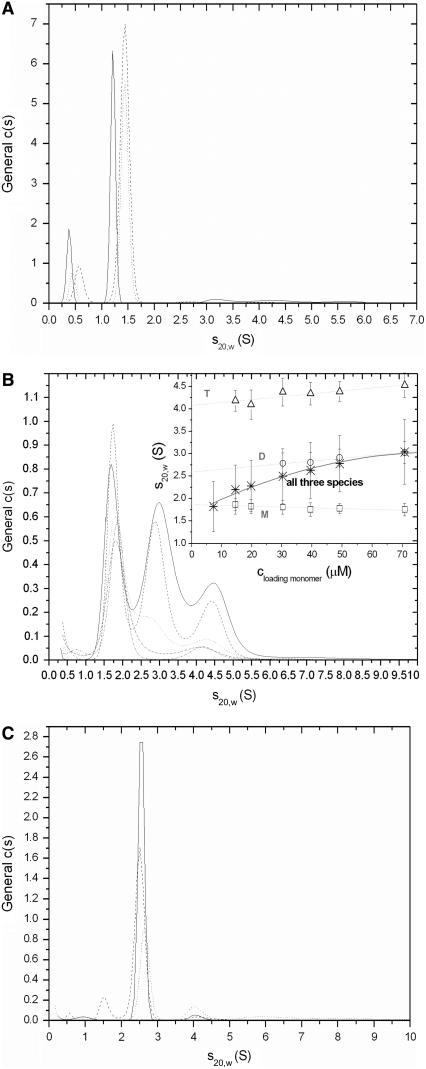
Each of the cTir mutants and the WT have been examined for heterogeneity using the continuous c(s) size-distribution approach. One major dominant species was evident as a single peak centered around an apparent sedimentation coefficient (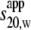 ) of 1.5 S for all the cTir mutants (in 150 mM NaCl). The peak corresponding to the major species of the cTir WT samples was displaced toward lower s-values (1.2 S). The integration of this peak gave a weight-average sedimentation coefficient (extrapolated to zero concentration) for cTir mutants that varied between 1.46 S for the Ser-434-Ala/Ser-463-Ala mutant and 1.52 S for the Ser-434-Asp mutant (Fig. 4 A and Table 2). All measured values of s are significantly less than those calculated for a sphere of the same molecular mass with typical hydration (2.46 S), which implies substantial elongation of cTir in solution.
) of 1.5 S for all the cTir mutants (in 150 mM NaCl). The peak corresponding to the major species of the cTir WT samples was displaced toward lower s-values (1.2 S). The integration of this peak gave a weight-average sedimentation coefficient (extrapolated to zero concentration) for cTir mutants that varied between 1.46 S for the Ser-434-Ala/Ser-463-Ala mutant and 1.52 S for the Ser-434-Asp mutant (Fig. 4 A and Table 2). All measured values of s are significantly less than those calculated for a sphere of the same molecular mass with typical hydration (2.46 S), which implies substantial elongation of cTir in solution.
FIGURE 4.
General size distribution (c(s)) analysis of the SV data converted to standard conditions for (A) cTir WT in 150 mM NaCl, c = 0.92 mg/ml (solid line), Ser-434-Asp/Ser-463-Asp mutant, c = 1 mg/ml (dashed line), and phosphorylated sample, c = 0.92 mg/ml (dotted line); (B) cTir in 50 mM NaCl, with sample concentrations ranging from 0.3 mg/ml to 1.4 mg/ml. (Inset) Linear extrapolation to zero concentrations sw for each detected species (monomer (□), dimer (○), tetramer (▵), dashed lines) and weight-average s for all three species together (*, solid line); (C) FL-Tir, with samples and symbols as in A. The concentrations were 1.2 mg/ml for WT; 0.87 mg/ml for Ser-434-Asp/Ser-463-Asp mutant, and 0.85 mg/ml for phosphorylated protein.
TABLE 2.
Hydrodynamic parameters for cTir/FL-Tir derived from AUC experiments (interference data)
| Sedimentation coefficient (S)
|
Mass (kDa)
|
Hydrodynamic radius (Å)
|
|||||
|---|---|---|---|---|---|---|---|
| s1 | s2 | s3 | M1 | M2 | M3 | Rs | |
| cTir WT (150 mM NaCl) | 1.20 ± 0.01 | — | — | 19.6 ± 0.4 | — | — | 38.8 ± 0.2 |
| cTir WT (50 mM NaCl) | 1.78 ± 0.10 | 2.61 ± 0.14 | 4.22 ± 0.1 | 18.5 ± 0.5 | 39.7 ± 2.1 | 68.2 ± 2.2 | — |
| cTir WT-P | 1.47 ± 0.01 | — | — | 19.3 ± 0.4 | — | — | 35.4 ± 0.5 |
| cTir 434A | 1.48 ± 0.08 | — | — | 20.3 ± 0.4 | — | — | 37.4 ± 0.3 |
| cTir 434D | 1.52 ± 0.03 | — | — | 19.5 ± 0.4 | — | — | 35.0 ± 0.9 |
| cTir 463A | 1.48 ± 0.02 | — | — | 20.5 ± 0.5 | — | — | 37.0 ± 0.7 |
| cTir 463D | 1.49 ± 0.04 | — | — | 18.1 ± 0.4 | — | — | 33.8 ± 0.4 |
| cTir 434A/463D | 1.48 ± 0.01 | — | — | 20.5 ± 0.2 | — | — | 37.3 ± 0.4 |
| cTir 434D/463A | 1.50 ± 0.01 | — | — | 19.9 ± 0.2 | — | — | 36.2 ± 0.3 |
| cTir 434A/463A | 1.46 ± 0.02 | — | — | 17.7 ± 0.7 | — | — | 33.8 ± 0.8 |
| cTir 434D/463D | 1.47 ± 0.02 | — | — | 19.5 ± 0.5 | — | — | 34.7 ± 1.2 |
| FL-Tir WT (150 mM NaCl) | 2.65 ± 0.01 | — | — | 60.5 ± 2.1 | — | — | 57.0 ± 0.6 |
| FL-Tir WT (50 mM NaCl) | 2.67 ± 0.02 | 58.4 ± 2.6 | 54.3 ± 2.9 | ||||
| FL-Tir WT-P | 2.68 ± 0.03 | — | — | 58.5 ± 2.5 | — | — | 56.2 ± 2.4 |
| FL-Tir 434D/463D | 2.65 ± 0.03 | — | — | 61.9 ± 1.4 | — | — | 62.7 ± 0.7 |
The c(s) distribution of cTir WT in 50 mM NaCl had three easily distinguishable peaks centered at 1.7 S, 2.65 S, and 4.25 S (Fig. 4 B). Importantly, the peak positions are almost independent of protein concentration, suggesting that the timescale of exchange between oligomeric species is slow compared to the duration of the SV experiment (32). The overall integration of c(s) suggests that the oligomerization of cTir WT in 50 mM NaCl is an equilibrium association process, since the value of weight-average sedimentation coefficient for the whole system increases with sample concentration (Fig. 4 B, inset). Linear extrapolation to zero concentration gives sedimentation coefficient values under standard conditions of 1.78 ± 0.1 S for the monomeric species, 2.61 ± 0.14 S for dimers, and 4.22 ± 0.09 S for tetramers. Interestingly, the sedimentation coefficient for the cTir monomer in 50 mM NaCl is much higher than the s value for the same sample in 150 mM NaCl. Conversion of the c(s) distribution to the c(M) distribution (data not shown) followed by peak integration yields a molecular mass of 18.5 ± 0.5 kDa for monomers, 39.7 ± 2.6 kDa for dimers, and 68.2 ± 2.2 kDa for tetramers, which is in reasonable agreement with that predicted from the amino acid composition of the protein (19.8 kDa for monomers, 39.6 kDa for dimers, and 79.2 kDa for tetramers, Table 2).
For some cTir mutants, minor species of lower molecular weight were observed on the continuous c(s)/c(M) distribution, resulting in inaccurate determination of molecular mass after peak integration. To compensate for this, a two-dimensional distribution “mass-and-shape” model was applied to provide shape-independent estimation of the molecular mass from the SV data (23). This two-dimensional distribution c(s,fr) with subsequent peak integration also includes the determination of the Stokes radius (Rs) of the particles from SV profiles. The theoretical calculations of Rs (see Materials and Methods) for cTir in a folded and unfolded state based on its molecular mass gave the values of 21.4 Å and 39.3 Å, respectively. The experimentally determined values of Rs for cTir WT and all mutants ranged between 33.8 and 38.8 Å (see Table 2), again suggesting that they adopt an unfolded conformation in solution, consistent with the CD data. The most extended conformation was observed for cTir-WT in 150 mM NaCl, whereas the most “compact” samples were the S434A/S463A and S43D/S463D mutants; the phosphorylated protein also seems to be more compact than wild-type cTir (35.4 Å compared to 38.8 Å).
FL-Tir forms an essentially homogeneous monomeric species in solution, as determined by AUC, with an s value of 2.65 (Fig. 4 C, Table 2) and a molecular mass of 60.5 kDa ± 2.1 kDa. A small peak at ∼4 S that may represent a dimeric species (or other form of aggregate) did not exceed 3% in solution. There were no significant changes observed in the hydrodynamic properties of FL-Tir after phosphorylation and no significant difference was observed between FL-Tir and the Ser-434-Asp/Ser-463-Asp mutant. However, the phosphorylated sample seems to possess a slightly more compact conformation, as revealed by its sedimentation coefficient and Stokes radius. The calculated Stokes radius for FL-Tir was 32.1 Å for the folded and 69.8 Å for the unfolded states. The experimentally determined radius for native FL-Tir was 57 Å; the phosphorylated FL-Tir has an almost identical Stokes radius (56.2 Å). The Ser-434-Asp/Ser-463-Asp mutant was slightly more extended (62.7 Å).
DISCUSSION
The aim of this study was to investigate the solution structure of EPEC Tir and determine whether the effects of PKA-mediated phosphorylation in the protein can be mimicked by site-directed mutagenesis. Both cTir and FL-Tir are shown to adopt natively unfolded conformations in solution. The importance of natively unfolded proteins in mediating protein-protein interactions is an emerging theme in molecular recognition. Within this context, SDS-PAGE, CD spectroscopy, and AUC have been used to characterize the solution structure of Tir constructs in which Ser-434 and Ser-463 have been mutated to Ala and Asp, and these have been compared to both the native protein and the protein phosphorylated at these positions. Implications for the function of Tir are discussed.
cTir and FL-Tir are natively unfolded proteins in solution
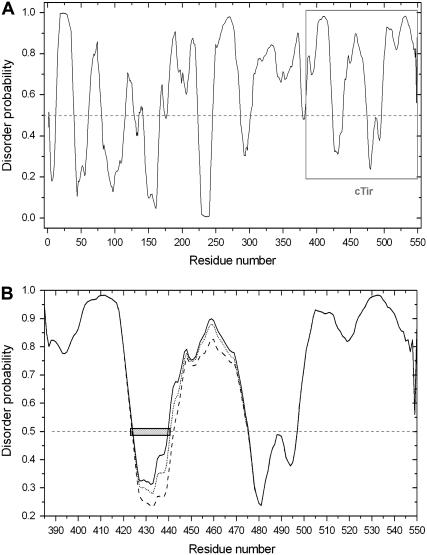
Analysis of the structure of cTir and FL-Tir by CD spectrometry and AUC demonstrates experimentally that they adopt extended conformations in solution, with little tertiary structure. A low degree of compactness (i.e., high hydrodynamic radius of molecule) is an important characteristic in the definition of a natively unfolded state in solution (17). The experimentally determined hydrodynamic radii of cTir and FL-Tir in solution have been compared with both 1), proteins previously demonstrated to be natively unfolded; and 2), proteins unfolded in 8 M urea (Table 3). The high similarity between the mass/hydrodynamic radius ratio observed for cTir and FL-Tir with these proteins provides further evidence that they adopt an essentially unfolded state in solution. Further, bioinformatic analysis of the amino acid composition of cTir reveals that the protein contains 31% “disorder-promoting” residues (such as Pro, Glu, Ser, and Gln (17,36) and only 11% “order-promoting” residues (Tyr, Phe, Trp, Ile, and Leu; Cys is absent). The same analysis for FL-Tir reveals 28% disorder-promoting and 14% order-promoting residues. This reflects the percentage of disordered structure predicted by PONDR (34), 78.4% for cTir and 67.6% for FL-Tir (Fig. 5 A). It is also worth noting that the amino acid sequence of FL-Tir contains four proline-rich clusters (either P-X-X-P or P-P/G sequences), which have a high propensity to form PPII-type helical structures (37).
TABLE 3.
Hydrodynamic characteristics of cTir/FL-Tir compared to other natively unfolded/urea-unfolded proteins
| Mass (kDa) | Hydrodynamic radius (Å) | Reference | ||
|---|---|---|---|---|
| cTir WT (150 mM NaCl) | 19.8 | 38.8 ± 0.2 | ||
| Natively unfolded proteins | Stathmin | 17 | 33 | (42) |
| CFos-AD domain, 216-380 fragment | 17.3 | 35 | (43) | |
| Calf thymus histone | 19.8 | 36.7 | (44) | |
| PPI-1 | 20.8 | 32.3 | (45) | |
| Securin | 22.2 | 39.7 | (21) | |
| DARPP-32 | 23.1 | 34 | (46) | |
| β-Casein | 24 | 41.7 | (44) | |
| 8 M urea unfolded proteins without cross-links | Myoglobin | 16.9 | 35.1 | (47) |
| β-Lactoglobulin | 18.5 | 37.8 | (47) | |
| Chymotrypsinogen | 25.7 | 45 | (47) | |
| FL-Tir WT | 59.2 | 57.0 ± 0.6 | ||
| Natively unfolded proteins | Taka-amylase A, reduced | 52.5 | 43.1 | (44) |
| GARP1 | 64.5 | 65 | (48) | |
| SdrD protein B1-B5 fragment | 64.8 | 54.7 | (49) | |
| 8 M urea unfolded proteins without cross-links | Serum albumin | 66.3 | 74 | (47) |
FIGURE 5.
PONDR prediction for intrinsic disorder from amino acid sequences (A) FL-Tir, (B) cTir WT (solid line), Ser-434-Asp/Ser-463-Asp (dashed line), and Ser-434-Ala/Ser-463-Ala (dotted line). The predicted MoRF sequence, as discussed in the text, is shown as a hatch-marked box.
Before adopting its final conformation as an integral transmembrane receptor, Tir is 1), bound by a molecular chaperone (CesT) after overexpression in the bacteria (which protects the protein from degradation and may be involved in delivery of Tir to the type III secretion system); and 2), passes through the “needle” of the type III translocon in an at least partially unfolded state. The protein is then predicted to be released into the cytoplasm, where it inserts into the membrane. It would seem likely that the expressed protein described here represents this preinsertion form, and this is natively unfolded, but soluble in solution. The importance of natively unfolded proteins in promoting efficient interactions with other molecules (for example, in regulation and cell signaling) has recently been investigated (16,20,36,38,39). It is an attractive hypothesis that the natively unfolded state adopted by FL-Tir in solution is important for the structural transition into an integral membrane protein by providing an appropriate interaction surface or promoting penetration of the lipid bilayer due to exposed hydrophobic residues.
Bioinformatic analysis of the predicted regions of disorder in Tir (as generated by PONDR (Fig. 5)) reveals the presence of a MoRF-like sequence (molecular recognition feature (40,41)) centered around residue Ser-434. MoRFs are defined as short, loosely structured regions in proteins that are surrounded by largely disordered sequences (41), and have been implicated in molecular recognition events, particularly protein/protein interactions associated with cell signaling and regulation. Upon binding their partners, these MoRFs undergo disorder-order transitions. Additionally, it has been shown that a significant proportion of MoRFs contain a predicted phosphorylation site, and phosphorylation may be a common mechanism to modulate MoRF binding activity (41). The identification of a MoRF centered on the 434 region of Tir supports a role for this region in regulating intermolecular interactions with an as yet unidentified binding partner, and suggests that phosphorylation of this residue may modulate binding affinity.
Oligomerization of cTir in solution
Previously, when Tir proteins were prepared in 50 mM NaCl, it was shown that cTir existed in a monomer-dimer equilibrium (by AUC and native gels), with phosphorylation affecting the oligomerization state of the protein; the implications for Tir function were discussed (12). In this study, protein preparations and experiments were conducted at more physiological NaCl concentrations (150 mM in all buffers, also with an additional gel filtration purification step where a single peak was observed). At these salt concentrations, the AUC experiments reveal that there is no change in oligomeric state 1), after cTir phosphorylation or 2), for any of the cTir variant proteins. Also, only single bands are seen on native gels for all samples (data not shown). If cTir samples are prepared in only 50 mM NaCl, then evidence of changes to compactness and oligomeric state (dimer and possibly tetramer) are seen using AUC, but only a single band is apparent on native gels (in contrast to the two bands observed in Hawrani et al. (12) (data not shown)). The most extended conformation of cTir-WT was observed at approximately physiological conditions (pH 7.5, 150 mM NaCl), whereas in low salt concentrations (50 mM NaCl), the protein was significantly more compact, giving a friction coefficient of 1.21 (averaged value for all species present in solution), indicative of a more globular state. These differences in the compactness of cTir and its oligomerization state in response to NaCl concentration are not distinguishable by AUC in the context of the full-length protein in solution, as the distribution of FL-Tir purified in 50 mM NaCl is virtually identical to that of protein prepared in 150 mM NaCl (data not shown).
Considering the two studies together, it would appear that isolated cTir is able to oligomerize, and the oligomerization state of the protein can be perturbed on phosphorylation, but this is dependent on salt concentration. It may be that preparation at higher salt concentrations (as predominant in this study) is important for ensuring/retaining a monomer conformation, as addition of NaCl to 150 mM from samples initially prepared in 50 mM, before running native gels, maintains the monomer/dimer equilibrium of cTir (12). However, in 150 mM NaCl buffer, a monomer conformation was more readily observed after incubation with PKA (12). These effects suggest that subtle changes in the composition of the cell medium may affect the structural state of Tir, and this may have implications for the mechanism of host-cell membrane insertion, or downstream signaling.
How does phosphorylation or phosphorylation-mimicking mutations affect the natively unfolded state of cTir/FL-Tir?
All of the eight cTir variants produced as part of this study, and both the native and phosphorylated proteins, appear to adopt a largely unfolded or loosely packed domain structure in solution. Deconvolution of the CD spectra for the proteins shows a range of 66.2–74% “unordered” structure and modeling of the AUC data reveals a frictional coefficient that, when considering the molecular weight of these proteins, translates to a very elongated state that cannot be correlated with a folded domain structure. Also, the proteins all display similar conformational stability (as assayed by tryptic digest (1 ng/μl trypsin) followed by SDS-PAGE analysis (data not shown)), with a single major breakdown product stable for ∼15 min but degraded after 60 min. The extent of digestion is essentially the same at each time point for all constructs. Despite their unfolded state, the proteins appear stable in solution to high concentrations and show no sign of aggregation up to at least 10 mg/ml (monodisperse as observed by dynamic light scattering, single peak on a gel filtration column (well after the void volume), single band on native-PAGE gels (not shown)), the proteins also retain the ability to be phosphorylated by PKA and display changes in apparent molecular mass on SDS-PAGE gels, similar to the full-length protein.
Bioinformatic analysis suggests that the phosphorylation-mimicking mutations are on average slightly order-promoting (Fig. 5 B). This agrees with the experimental data (for instance, the value of the sedimentation coefficient and deconvolution of the CD spectra for the Ser-434-Asp mutant suggests a more folded and compact structure). Interestingly, the first site (Ser-434) for PKA phosphorylation spanning residues Ala-424 to Ser-440 is situated in a region that has MoRF-like properties, as discussed above, whereas the second site, Ser-463, is situated in a disordered region (Thr-441 to Asp-475 (Fig. 5 B)). However, it is apparent from the computational and experimental analysis that neither phosphorylation nor the phosphorylation-mimicking mutations dramatically affect the structure of the cTir domain in solution. It may be that cTir requires either the remaining domains of the protein, or a combination of the rest of the protein with the host-cell membrane or other factors to refold to a compact state, if indeed it does fold in the context of the intact protein. Although cTir itself has some affinity for membranes in vitro (7), it is not known whether this forms part of the insertion mechanism; there is no evidence to suggest that cTir can refold on a membrane alone (there is no change in the CD spectra of cTir on incubation with small unilamellar vesicles mimicking the lipid composition of the host-cell membrane (data not shown)).
As for the cTir domain, AUC reveals that FL-Tir does not undergo any major structural rearrangements after modification of Ser-434/Ser-463 by phosphorylation or mutagenesis. This shows that the subtle changes observed by CD are not linked to large-scale changes in tertiary structure. Phosphorylation does appear to stabilize a higher-molecular-weight component of FL-Tir in solution (peak at ∼4 S, 10% rather than 3% in the native protein, Fig. 3 C), but it is not known at present whether this is an oligomeric state of the protein or nonspecific aggregation. In either case, this component is a minor species in solution. The conformational stability of FL-Tir does not appear to be significantly affected by modification at residues 434/463, as determined by tryptic digest (1 ng/μl trypsin) (data not shown)).
In summary, this study aimed to determine the effects of site-directed mutagenesis at previously identified serine phosphorylation sites on the structure of the EPEC virulence protein Tir. It has been shown that both the isolated cTir domain and the FL-Tir protein appear to be natively unfolded in solution, and modification of the targeted residues has little effect on the structures (be it enzymatic phosphorylation or mutagenesis) at physiological NaCl concentrations. It still remains plausible that phosphorylation of these residues has a role in the mechanism of membrane insertion or promoting structural rearrangements postinsertion through either charge effects, recognition of accessory factors (or both), or other, as yet unidentified, downstream signaling activities. The recognition of Tir adopting a natively unfolded state in solution has importance for directing further structural studies. At present, low-resolution, solution-based approaches appear to be the most appropriate way to investigate the structure of Tir, both in the aqueous phase and inserted into the membrane.
Acknowledgments
The authors thank Brendan Kenny for discussion, Dave Scott (University of Nottingham, Nottingham, UK) for initial AUC data, and Duncan McNulty for technical help.
This work was funded by the Royal Society (United Kingdom) through the award of a University Research Fellowship to M.J.B. M.J.B. and A.S.S. also thank the University of Newcastle for financial support.
Editor: Jane Clarke.
References
- 1.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kenny, B. 2002. Enteropathogenic Escherichia coli (EPEC)–a crafty subversive little bug. Microbiology. 148:1967–1978. [DOI] [PubMed] [Google Scholar]
- 3.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA. 97:8799–8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 91:511–520. [DOI] [PubMed] [Google Scholar]
- 5.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229–1241. [DOI] [PubMed] [Google Scholar]
- 6.de Grado, M., A. Abe, A. Gauthier, O. Steele-Mortimer, R. DeVinney, and B. B. Finlay. 1999. Identification of the intimin-binding domain of Tir of enteropathogenic Escherichia coli. Cell. Microbiol. 1:7–17. [DOI] [PubMed] [Google Scholar]
- 7.Race, P. R., J. H. Lakey, and M. J. Banfield. 2006. Insertion of the enteropathogenic Escherichia coli Tir virulence protein into membranes in vitro. J. Biol. Chem. 281:7842–7849. [DOI] [PubMed] [Google Scholar]
- 8.Campellone, K. G., A. Giese, D. J. Tipper, and J. M. Leong. 2002. A tyrosine-phosphorylated 12-amino-acid sequence of enteropathogenic Escherichia coli Tir binds the host adaptor protein Nck and is required for Nck localization to actin pedestals. Mol. Microbiol. 43:1227–1241. [DOI] [PubMed] [Google Scholar]
- 9.Gruenheid, S., R. DeVinney, F. Bladt, D. Goosney, S. Gelkop, G. D. Gish, T. Pawson, and B. B. Finlay. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856–859. [DOI] [PubMed] [Google Scholar]
- 10.Campellone, K. G., and J. M. Leong. 2005. Nck-independent actin assembly is mediated by two phosphorylated tyrosines within enteropathogenic Escherichia coli Tir. Mol. Microbiol. 56:416–432. [DOI] [PubMed] [Google Scholar]
- 11.Warawa, J., and B. Kenny. 2001. Phosphoserine modification of the enteropathogenic Escherichia coli Tir molecule is required to trigger conformational changes in Tir and efficient pedestal elongation. Mol. Microbiol. 42:1269–1280. [DOI] [PubMed] [Google Scholar]
- 12.Hawrani, A., C. E. Dempsey, M. J. Banfield, D. J. Scott, A. R. Clarke, and B. Kenny. 2003. Effect of protein kinase A-mediated phosphorylation on the structure and association properties of the enteropathogenic Escherichia coli Tir virulence protein. J. Biol. Chem. 278:25839–25846. [DOI] [PubMed] [Google Scholar]
- 13.Huffine, M. E., and J. M. Scholtz. 1996. Energetic implications for protein phosphorylation. Conformational stability of HPr variants that mimic phosphorylated forms. J. Biol. Chem. 271:28898–28902. [DOI] [PubMed] [Google Scholar]
- 14.Jang, Y. J., S. Ma, Y. Terada, and R. L. Erikson. 2002. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 277:44115–44120. [DOI] [PubMed] [Google Scholar]
- 15.Leger, J., M. Kempf, G. Lee, and R. Brandt. 1997. Conversion of serine to aspartate imitates phosphorylation-induced changes in the structure and function of microtubule-associated protein τ. J. Biol. Chem. 272:8441–8446. [DOI] [PubMed] [Google Scholar]
- 16.Receveur-Brechot, V., J. M. Bourhis, V. N. Uversky, B. Canard, and S. Longhi. 2006. Assessing protein disorder and induced folding. Proteins. 62:24–45. [DOI] [PubMed] [Google Scholar]
- 17.Uversky, V. N. 2002. What does it mean to be natively unfolded? Eur. J. Biochem. 269:2–12. [DOI] [PubMed] [Google Scholar]
- 18.Uversky, V. N., J. R. Gillespie, I. S. Millett, A. V. Khodyakova, R. N. Vasilenko, A. M. Vasiliev, I. L. Rodionov, G. D. Kozlovskaya, D. A. Dolgikh, A. L. Fink, S. Doniach, E. A. Permyakov, and V. M. Abramov. 2000. Zn(2+)-mediated structure formation and compaction of the “natively unfolded” human prothymosin α. Biochem. Biophys. Res. Commun. 267:663–668. [DOI] [PubMed] [Google Scholar]
- 19.Li, J., V. N. Uversky, and A. L. Fink. 2002. Conformational behavior of human α-synuclein is modulated by familial Parkinson's disease point mutations A30P and A53T. Neurotoxicology. 23:553–567. [DOI] [PubMed] [Google Scholar]
- 20.Uversky, V. N. 2002. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11:739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Puig, N., D. B. Veprintsev, and A. R. Fersht. 2005. Human full-length Securin is a natively unfolded protein. Protein Sci. 14:1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batra-Safferling, R., K. Abarca-Heidemann, H. G. Korschen, C. Tziatzios, M. Stoldt, I. Budyak, D. Willbold, H. Schwalbe, J. Klein-Seetharaman, and U. B. Kaupp. 2006. Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J. Biol. Chem. 281:1449–1460. [DOI] [PubMed] [Google Scholar]
- 23.Brown, P. H., and P. Schuck. 2006. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90:4651–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular Cloning. A Laboratory Manual, 3rd ed. Coldspring Harbor Laboratory Press, Woodbury, NY.
- 25.Whitmore, L., and B. A. Wallace. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32:W668–W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provencher, S. W., and J. Glockner. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 20:33–37. [DOI] [PubMed] [Google Scholar]
- 27.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. Pelletier. 1992. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Redwood Press, Melksham, UK.
- 28.Durchschlag, H. 1986. Specific volumes of macromolecules and some other molecules of biological interest. In Thermodynamic Data for Biochemistry and Biotechnology. H.-J. Hinz, editor. Springer-Verlag, Berlin. 45–128.
- 29.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamm, O. 1929. Die Differentialgleichung der Ultrazentrifugierung. Ark. Mat. Astr. Fys. 21B:1–4. [Google Scholar]
- 31.Schuck, P. 1998. Sedimentation analysis of noninteracting and self-associating solutes using numerical solutions to the Lamm equation. Biophys. J. 75:1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuck, P. 2003. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320:104–124. [DOI] [PubMed] [Google Scholar]
- 33.Dam, J., and P. Schuck. 2004. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 384:185–212. [DOI] [PubMed] [Google Scholar]
- 34.Romero, P., Z. Obradovic, X. Li, E. C. Garner, C. J. Brown, and A. K. Dunker. 2001. Sequence complexity of disordered protein. Proteins. 42:38–48. [DOI] [PubMed] [Google Scholar]
- 35.Lebowitz, J., M. S. Lewis, and P. Schuck. 2002. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11:2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tompa, P. 2002. Intrinsically unstructured proteins. Trends Biochem. Sci. 27:527–533. [DOI] [PubMed] [Google Scholar]
- 37.Stapley, B. J., and T. P. Creamer. 1999. A survey of left-handed polyproline II helices. Protein Sci. 8:587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fink, A. L. 2005. Natively unfolded proteins. Curr. Opin. Struct. Biol. 15:35–41. [DOI] [PubMed] [Google Scholar]
- 39.Uversky, V. N., C. J. Oldfield, and A. K. Dunker. 2005. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 18:343–384. [DOI] [PubMed] [Google Scholar]
- 40.Oldfield, C. J., Y. Cheng, M. S. Cortese, P. Romero, V. N. Uversky, and A. K. Dunker. 2005. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry. 44:12454–12470. [DOI] [PubMed] [Google Scholar]
- 41.Mohan, A., C. J. Oldfield, P. Radivojac, V. Vacic, M. S. Cortese, A. K. Dunker, and V. N. Uversky. 2006. Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362:1043–1059. [DOI] [PubMed] [Google Scholar]
- 42.Belmont, L., T. Mitchison, and H. W. Deacon. 1996. Catastrophic revelations about Op18/stathmin. Trends Biochem. Sci. 21:197–198. [PubMed] [Google Scholar]
- 43.Campbell, K. M., A. R. Terrell, P. J. Laybourn, and K. J. Lumb. 2000. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry. 39:2708–2713. [DOI] [PubMed] [Google Scholar]
- 44.Ptitsyn, O. 1995. Molten globule and protein folding. Adv. Protein Chem. 47:83–229. [DOI] [PubMed] [Google Scholar]
- 45.Nimmo, G. A., and P. Cohen. 1978. The regulation of glycogen metabolism. Purification and characterisation of protein phosphatase inhibitor-1 from rabbit skeletal muscle. Eur. J. Biochem. 87:341–351. [DOI] [PubMed] [Google Scholar]
- 46.Hemmings, H. C., Jr., A. C. Nairn, D. W. Aswad, and P. Greengard. 1984. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Purification and characterization of the phosphoprotein from bovine caudate nucleus. J. Neurosci. 4:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uversky, V. N. 1993. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 32:13288–13298. [DOI] [PubMed] [Google Scholar]
- 48.Wiesner, B., J. Weiner, R. Middendorff, V. Hagen, U. B. Kaupp, and I. Weyand. 1998. Cyclic nucleotide-gated channels on the flagellum control Ca2+ entry into sperm. J. Cell Biol. 142:473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Josefsson, E., D. O'Connell, T. J. Foster, I. Durussel, and J. A. Cox. 1998. The binding of calcium to the B-repeat segment of SdrD, a cell surface protein of Staphylococcus aureus. J. Biol. Chem. 273:31145–31152. [DOI] [PubMed] [Google Scholar]