Abstract
This paper describes a Grid-aware image reviewing system (GridIMAGE) that allows practitioners to (a) select images from multiple geographically distributed digital imaging and communication in medicine (DICOM) servers, (b) send those images to a specified group of human readers and computer-assisted detection (CAD) algorithms, and (c) obtain and compare interpretations from human readers and CAD algorithms. The currently implemented system was developed using the National Cancer Institute caGrid infrastructure and is designed to support the identification of lung nodules on thoracic computed tomography. However, the infrastructure is general and can support any type of distributed review. caGrid data and analytical services are used to link DICOM image databases and CAD systems and to interact with human readers. Moreover, the service-oriented and distributed structure of the GridIMAGE framework enables a flexible system, which can be deployed in an institution (linking multiple DICOM servers and CAD algorithms) and in a Grid environment (linking the resources of collaborating research groups). GridIMAGE provides a framework that allows practitioners to obtain interpretations from one or more human readers or CAD algorithms. It also provides a mechanism to allow cooperative imaging groups to systematically perform image interpretation tasks associated with research protocols.
Key words: Grid computing, grid architectures, caBIG™, caGrid, computer-assisted detection, digital imaging and communications in medicine
INTRODUCTION
This paper describes a Grid-aware image reviewing system (GridIMAGE) that allows users to (a) select images from multiple geographically distributed digital imaging and communications in medicine (DICOM) servers, (b) send those images to a particular collection of human readers and computer-assisted detection (CAD) algorithms,1 and (c) obtain and compare interpretations from human readers and CAD algorithms. Several current challenges motivated the development of GridIMAGE. The first involves the need for medical professionals to obtain external consultation. Depending on the circumstance, a radiologist, clinician, or other professional might seek opinions from one or more radiologists and/or direction from one or more different CAD algorithms. Another challenge involves the growing need for support for protocol-driven image interpretations carried out by research studies. In this case, a protocol would be likely to specify that specific images must be interpreted by specific sets of human readers or by specific sets of CAD algorithms. The final challenge is to create new pathways that allow access to multiple CAD algorithms and data sets and thereby facilitate novel and complex analytic approaches. In comparative analysis, for example, an investigator can evaluate multiple groups of image data sets (obtained on different groups of subjects and by different research teams) using a CAD method to search for patterns across subject groups, investigate differences, and assess the efficacy of different imaging instruments. In a multianalysis scenario, on the other hand, the investigator applies multiple CAD methods to a data set. This function can be useful for mining patterns in a given data set and examining the effectiveness of different algorithms.
The effective application of decentralized human review and CAD in a distributed environment requires solutions to additional challenges associated with accessing data and application software across geographically distributed locations. Image data sets are usually captured and stored at diverse locations in a multi-institutional setting. Human readers may be drawn from geographically distributed institutions, and different CAD methods and systems may be installed at different institutions.
Although a centralized system could be used in some cases to host all data and all CAD algorithms, logistic, financial, licensing, and regulatory constraints are likely to make a centralized approach difficult. Moreover, even if data and/or CAD algorithms were centralized, practitioners and human readers would still most likely be geographically separated.
We present here a novel application of Grid1 computing using the National Cancer Institute (NCI) caGrid infrastructure2 to support access to distributed CAD technology, distributed human readers, and image data sets for medical imaging research. GridIMAGE enables researchers and clinicians to share data sets and CAD analytical resources and allows human readers to view and manually specify and annotate regions of interest. To our knowledge, this is the first report of a Grid-enabled CAD system and provides a prototype for future Grid-based teleradiology systems that integrate human and CAD image interpretation.
The service-oriented and distributed structure of the GridIMAGE framework facilitates a flexible, scalable, and extensible system. Although this paper focuses on the Grid aspects of GridIMAGE, the system can easily be deployed within an institution as an enterprise system to facilitate access to multiple DICOM servers and CAD systems in that institution through a unified infrastructure. The advantages of GridIMAGE deployment in a single institution is that the system can be easily extended; new DICOM servers and CAD systems (or other image analysis systems) can be added without needing to restructure the entire system. Because there is no need for a centralized server, the image and analytic resources (eg, DICOM storage systems, machines that host CAD systems) can be placed in different buildings in the institution, thus reducing the need to relocate the existing servers to a new building as more resources are added. Moreover, when cross-institutional collaborative projects are initiated, collaborating researchers can easily link distributed image and analysis resources in compliance with institutional security and access control policies.
The current implementation of GridIMAGE is designed to carry out pulmonary nodule detection for thoracic computerized tomography (CT) images. In this implementation, the practitioner queries a set of image databases and selects studies, series, or images to be interpreted. For each image selected, the practitioner can specify one or more radiologists and the CAD algorithms to be used to identify candidate nodules. Pulmonary nodule candidates are identified as 3-dimensional structures and are typically defined using parameters that specify a boundary and/or centroid.
A graphical user interface (GUI) has been developed to display CAD findings on CT images as overlays. Using the GUI, the practitioner can (a) query, select, and preview the subsets of distributed lung CT databases to be included in a study; (b) select the CAD algorithms and human readers to be involved in the analysis; and (c) visualize the human annotations and mark-ups and those produced by the CAD algorithms. Because human mark-ups are captured based on the reader’s availability, the system was designed to handle the asynchronous nature of this workflow.
All components of the software infrastructure described in this paper, with the exception of the vendor-supplied proprietary CAD algorithms, will be made available publicly in late fall 2006 at the Ohio State University Biomedical Informatics Department web page.3
BACKGROUND
The GridIMAGE application has been implemented using two Grid middleware infrastructures: the caBIG™ caGrid infrastructure2 and the Mobius framework.4 After a brief overview of these systems, we will describe the design and implementation of the GridIMAGE application.
Grid Computing and Medical Imaging
Grid computing refers to hardware platforms and software systems (middleware, tools, and applications) that create an infrastructure to support remote and secure access to disparate resources.5–7 Using Grid-enabled software built on open protocols and standard frameworks, users and institutions can share resources, including software, applications, data, and compute and storage platforms, giving them virtually unlimited capacity. Different Grid-enabled middleware infrastructures are being developed to address the challenges associated with sharing, transferring, and processing images.8–10 The approach taken by Espert et al.11 has created virtual repositories of distributed storage sites and provides support for global querying, autoencryption, and pseudoanonymization. The MammoGrid project is developing a CAD system for the detection of microcalcifications and masses. This system will be integrated as a local node of the European Grid.10 In the GRID Platform for Computer Assisted Library for Mammography project, a CAD system developed for the detection of microcalcifications and masses, will be made available to hospitals via the Grid.12
GridIMAGE differs from these applications in several ways: (a) It is being developed using the caGrid infrastructure of the NCI-funded caBIG™ effort and will therefore be implemented as a suite of Grid services, utilizing common functions provided by caGRID, such as security.13 (b) It enables human diagnosis, as well as computer-assisted analysis, and provides support for multiple mark-ups from radiologists at remote locations. (c) It facilitates access to distributed image data storage and CAD resources. (d) Images can be queried and retrieved from geographically disparate picture archiving and communication systems (PACS). (e) Distributed CAD analysis can be carried out for lung nodule detection. (f) It enables the review of CAD analysis results and mark-ups from a single thin client.
caBIGTM Grid Technology: caGrid
The NCI initiated the cancer Biomedical Informatics Grid (caBIG™) program in 2003 to provide broad and scalable solutions to the information management, integration, analysis, and sharing needs of basic and clinical cancer research projects (including single investigator-initiated studies, collaborative projects, and multi-institutional coordinated efforts). Through a community effort, caBIG™ continues to develop a common set of applications, standards for cancer data models and vocabularies, and a standard informatics infrastructure to support development and deployment of applications and resources in a multi-institutional environment. To enable development and deployment of caBIG™-compliant resources, a Grid middleware infrastructure, referred to as caGrid,2 is being developed. caGrid version 0.5 was released to the caBIG™ community in September 2005, and version 1.0 was under development for release at the end of 2006.
The caGrid infrastructure is designed as a service-oriented architecture. caGrid data and analytical services expose an object-oriented view of backend data and analytical resources. An important aspect of caGrid is that it employs a model- and metadata-driven approach and utilizes what are referred to as strongly typed services to ensure syntactic and semantic interoperability among services in the environment. The structure and semantic definitions of data objects, which are consumed or produced by a caGrid service, are drawn from common data elements and controlled vocabularies registered and published in the caGrid system. It is also required that all caGrid services be strongly typed services. This means that data elements are exchanged between two Grid endpoints (eg, clients and services or two services) as XML documents conforming to XML schemas published in the system. This enables syntactic interoperability and is the basis of the concept of strongly typed Grid services. An XML schema represents the structure of the data object that is exchanged between two Grid endpoints. When an XML document is received by a client or a service, the document is converted into a Java object, which is an instance of the corresponding data object. Similarly, when sending a data element to a Grid end-point, the corresponding Java object is converted into an XML document conforming to the respective XML schema. Conversion between XML documents and Java objects simplifies programmatic interaction with data by client-side programs and server-side backend systems.
The caGrid infrastructure builds on tools developed at the NCI and on Grid middleware systems, such as the Globus Toolkit 4.0,14 the Mobius Global Model Exchange (GME) services,4 and the NCI cancer Data Standards Repository and Enterprise Vocabulary Services.15 Using these tools and middleware systems, caGrid provides common services to support core functions, including secure communication, authentication and authorization, service invocation, and support for common data elements; controlled vocabularies; and management of XML schemas to facilitate semantic and syntactic interoperability. It also provides toolkits for development and deployment of secure, strongly typed data and analytic services and application program interfaces for building client applications that can interact with these services.
Among the various caGrid services and toolkits, the Introduce toolkit, the Dorian security infrastructure, and the Mobius GME service were employed in implementing GridIMAGE. Introduce provides an easy-to-use environment for development and deployment of caGrid services.16 It is designed to simplify the creation of secure and strongly typed caGrid services for domain scientists interested in creating caBIG™-compliant Grid applications. It hides the complex steps needed to generate a suitable service layout, create configurable service properties, and register service metadata and to configure the service for secure access and deploy it for execution. Dorian is the security component of caGrid that enables the federation of user identities across the collaborative environment.13 It provides support for users to authenticate to the Grid environment using their institutions’ authentication mechanism.
Mobius
The Mobius GME service4 provides support for distributed and coordinated creation, registration, and management of XML schemas. The GME is employed in caGrid to facilitate strongly typed services. Mobius is a service-based framework designed to support distributed and coordinated data and metadata management within the Grid.4,17 It provides a suite of services for distributed creation, versioning, management of data models and data instances, on-demand creation of XML databases, federation of existing databases as XML data sources, and query of data in a distributed environment. Mobius services employ XML schemas to represent data definitions and XML documents to represent and exchange data instances. Mobius client interfaces enable clients to interact with Mobius data services over a network and submit, query, and retrieve data instances using well-defined interfaces and protocols. XML schemas are managed by the Mobius GME service. The Mobius Mako service provides support for on-demand creation of databases conforming to GME-registered XML schemas and federation of existing databases in a distributed setting. In GridIMAGE, the Mako service is used to provide support for storage and management of results from CAD services.
METHODS
The Ohio State University Institutional Review Board has granted an exemption to this study for the use of 100 existing, deidentified thoracic CT image sets collected at Baltimore VA Hospital with patient consent forms. These images were used for the development and testing of GridIMAGE.
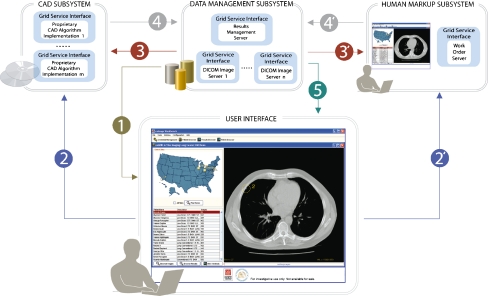
The goal of the GridIMAGE system is to support access to and review of distributed image data sets and enable invocation of remote automated and manual analysis services. This is facilitated by the architecture design shown in Figure 1. The system architecture includes three subsystems and a suite of GUIs.
Fig 1.
Schematic representation of GridIMAGE showing the sequence of interaction among different subsystems. The data management subsystem stores and manages image data sets and analysis results. The CAD subsystem enables access to distributed CAD applications. The human mark-up subsystem allows review and analysis of image data by humans. A suite of GUIs enables clients to log on to the environment, interact with services in each subsystem, submit requests for data query, apply CAD processing, provide human mark-up, and review the analysis results. Steps 1 through 5 show the sequence of interactions when submitting images for CAD processing and reviewing results. Steps 2′ through 5′ show the sequence of interactions when marking the images.
The data management subsystem consists of two services: DICOM image servers and results management service (RMS), for storing and managing image data sets and analysis results. The CAD subsystem enables access to distributed CAD applications and is made up of the CAD application service (CAS). The human mark-up subsystem allows review and analysis of image data by humans. It includes the work order service (WOS) to manage image analysis requests from clients. The suite of GUI software enables clients to log on to the environment, interact with services in each subsystem, submit requests for data query, apply CAD processing, provide human mark-up, and review the analysis results.
The primary use case that has motivated the design of GridIMAGE focuses upon the distributed analysis of an image data set on the Grid and involves two types of actors: practitioners (who may be involved in patient care or research) and human readers. In this use case, a practitioner wishes to access a data set and perform multiple-algorithm analysis on the data set.
-
Step 1
After signing on to the system through the GUI using unique credentials, the practitioner selects one or more image servers to query for available data sets. The results of this query are returned to the client as a list of data sets.
-
Step 2,2′
The practitioner can review the images in each data set, select images from one or more data sets, and invoke one or more instances of the CAS and WOS to process the selected images.
-
Step 3
When a request is received by a CAS instance, the CAS instance retrieves the specified images.
-
Step 4
The CAS performs the requested analysis and submits the results to the RMS.
-
Step 3′
When a request is received by a WOS instance, the request is entered into a work-order queue. Requests stored in a WOS are retrieved and can be performed by a human reader. The human reader logs on to the system using his or her credentials and selects one or more of the outstanding requests stored in the WOS.
-
Step 4′
The images can then be displayed from the associated data set. The human reader then marks and edits the locations of nodules interactively on each image and submits the results to the RMS once the reading is complete.
-
Step 5
The practitioner can, at a later time, use the GUI to query the RMS to search for and retrieve the CAD and human mark-up results for a given data set. The corresponding images are retrieved and displayed and the associated mark-ups are overlaid on the monitor. Results from multiple readers, human or CAD, can be distinguished by the color and shape of mark-ups and annotations.
Additional use cases for GridIMAGE include the screening of large aggregates of image data sets, validation and comparison of algorithms, performance of parameter studies, and rapid prototyping of analysis algorithms. The components of the GridIMAGE system to support these use cases are described here in more detail. First, the data models used in GridIMAGE to represent input images, image data sets, and analysis results are presented, followed by descriptions of the individual GridIMAGE subsystems, their services, and the set of GUIs.
GridIMAGE Data Models
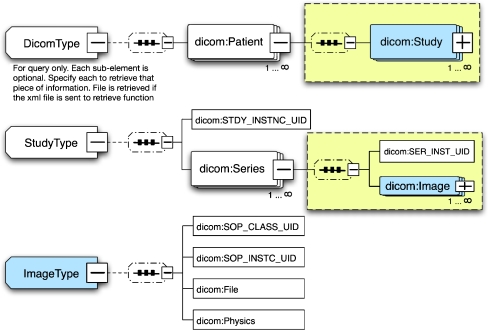
In the GridIMAGE system, DICOM images are exchanged in the environment using an XML representation that adheres to the schema shown in Figure 2. The schema mirrors the information hierarchy of the DICOM standard and includes elements to represent the patient, study, series, image, and acquisition physics. Acquisition physics entities such as peak kilovoltage, milliamperes per second, and spatial position come from the DICOM Standard PS3.3 C 8.2.1. Each element type also includes commonly used DICOM tags as attributes. Eighty-eight commonly used DICOM tags have been included in the current implementation. Unique image server identifiers are represented as elements to improve the performance of query and data access. In addition, the schema allows the original image to be attached to the XML document as a base64-encoded binary object.18 In practice, an XML document that is transported over the Grid typically includes only a subset of DICOM header tags, and the binary DICOM file is attached only for the result of a retrieve operation.
Fig 2.
The main structure of the schema for the DICOM data object (additional attributes are not shown).
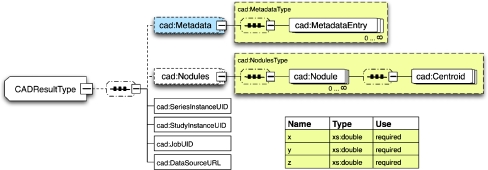
The human mark-up service of GridIMAGE necessitates a work-order data model for users of this service (eg, human readers). The schema for the work-order object is shown in Figure 3. The result of CAD analysis is represented by the schema in Figure 4. This schema enables description of the location of the nodule centroids in 3-dimensional space. It also specifies the unique identifier (UID) and location of the corresponding image data set. The XML schemas of the data models (Figs. 2, 3, and 4) used in the GridIMAGE implementation are registered and managed in the Mobius GME service.
Fig 3.
Work-order schema.
Fig 4.
Schema for nodule-detection results.
Data Management Subsystem
This subsystem provides support for managing image data sets and the results of the image analysis operations. It consists of two services: the DICOM Image Data Service, which provides methods to query for and access DICOM images, and the RMS, which provides Grid-enabled support for storing and retrieving image analysis results.
DICOM Image Servers Because the primary image data format for radiology is DICOM, it is essential that GridIMAGE be able to interact with DICOM servers. This also provides easier integration with existing clinical infrastructures. The image server uses a DICOM PACS as a backend image storage server and enables access by remote clients to images in the PACS through Grid service interfaces. The open-source DICOM library PixelMed is used as the backend PACS in the current implementation.19 PixelMed includes a set of Java classes for reading and writing DICOM objects, as well as communicating using DICOM messages. It also provides a simple DICOM PACS server. Communication between the image server and the PixelMed PACS is accomplished via standard DICOM messaging. The image server effectively provides a translation layer to convert between Grid service invocations and DICOM messages, which are then used to communicate with the PixelMed PACS.
The image server has been implemented as a strongly typed service using the Introduce toolkit (the image server was implemented using an earlier version of Introduce, which was available in caGrid version 0.5 and did not provide a data services template. Thus, its implementation is different from those of RMS and WOS, which were implemented using a later version of Introduce in caGrid 1.0. The image server will be ported to caGrid 1.0 using the new Introduce toolkit in the near future). It provides three primary service interfaces (or methods) for data access: query, retrieveSeriesImageMetadata, and retrieveImages. The query interface allows a remote client to search for data sets matching the specified attributes and returns records as data elements that adhere to the XML schema shown in Figure 2, without including binary images. The retrieveSeriesImageMetadata interface allows the retrieval of metadata associated with a DICOM series in an array of Image elements (Fig. 2), without including binary images. The retrieveImages interface allows for retrieval of a subset of images in a DICOM series ordered by their slice position. The images are returned as base64-encoded binary objects in an array of Image elements (Fig. 2).
Images are retrieved from the PACS and cached and sorted at the grid-enabled ImageServer before they are transmitted to the grid-enabled ImageClient GUI. The caching and sorting at the grid service side is required because several PACS implementations, including ours, do not have the capability to sort the images. As a result of sorting the images at the server side, the images could be retrieved on the client side by the GUI in an incremental manner. Incremental retrieval enables proper interactive visualization of the data set on the GUI side.
Results Management Service The purpose of the RMS is to provide storage and management of the CAD and human nodule detection results. The service utilizes a local Mobius Mako instance as the backend XML database system. The Mako database is instantiated by RMS if one does not exist already, and an XML collection is created using the lung nodule detection result XML schema shown in Figure 4. The RMS provides two Grid service interfaces: query and submit. The query interface is used by the user to query and retrieve nodule detection results for display. Queries are expressed in caBIG Query Language (CQL).20 CQL is an XML-based, recursively composed, object-oriented query language. It has been designed to handle a set of use cases within the caBIG community as opposed to a more general-purpose, commonly used query language such as Structured Query Language (SQL). While SQL allows a multitude of features and joins, CQL handles a simplified set of query features and it abstracts the user from the underlying data source. The submit interface is used by WOS and CAS for submission and storage of nodule detection results to the RMS.
CAD Subsystem
This subsystem consists of the CAS, which implements support for sharing CAD methods and facilitates access by clients to multiple CAD systems hosted at different locations.
CAD Application Service The purpose of the CAS is to expose CAD systems as Grid services. Most current CAD algorithms and applications are not Grid-aware, meaning they cannot be invoked remotely using Grid protocols. CAS implements the interface layer between Grid invocation and local algorithm execution. For the GridIMAGE system, a commercial CAD application from iCAD21 was used. Like image server, CAS has been developed using the Introduce toolkit as a strongly typed service. It provides only a single Grid service method: execute. The execute method (a) retrieves the images in the DICOM series from the specified data source service; (b) stores the images on the local file system and converts them to other formats if needed; (c) invokes the CAD algorithm as a system process; (d) retrieves the CAD result from the file system or from standard output; (e) converts the CAD result to the GridIMAGE output XML document, which conforms to the XML schema in Figure 4; and (f) submits the XML document to the specified RMS. When multiple CAD orders are received by a CAS, they are queued on the service for execution based on CAD order arrival time.
The Human Mark-up Subsystem
GridIMAGE allows review and annotation of images by human readers. The human mark-up subsystem represents the manual counterpart to the CAD subsystem. It implements support for practitioners to submit requests for mark-up of images by a human reader and for the human reader to retrieve such requests and mark-up images and submit the results for storage in the data management subsystem. The human reader chooses the time of his/her interpretation. With a sufficiently large volume of data, CAD may not be able to process all of its input. Hence, it is necessary to provide a queue for mark-up and CAD requests and to iterate through this queue for incomplete requests. The requests are referred to as work orders, for which the data model is shown in Figure 3. Work orders are stored in and accessed through the WOS. The WOS is implemented as a Grid data service using the caGrid Data Service template provided in the Introduce toolkit. It uses the Mobius Mako service as its backend database system. Work orders are stored in an XML collection and are created using the work order XML schema (Fig. 3) in Mobius Mako. Three service interfaces are provided in the WOS: query, submit, and update. The query interface is used to search for matching work orders. Queries are expressed in CQL. Results are returned in a result set that contains XML representations of the work order objects that satisfy the query. The submit interface is used to store new work orders, and the update interface is used to update an existing work order’s completion status.
Graphical User Interfaces
The suite of GUIs provides the mechanisms for the client user to interact with the GridIMAGE system. It consists of two main windows: the Mark-up GUI window and the Browse GUI window.
The Mark-up GUI window is implemented for human readers to mark-up the images. It allows the human reader to retrieve a list of work orders from the set of known work-order queues. Once a work order is selected, the specified images can be retrieved from the corresponding image server and displayed. The reader can mark-up nodules through simple mouse interactions and submit the nodule mark-up to RMS. The human mark-up user interface is shown in Figure 5. It should be noted that, to be consistent with the CAD markings, the human mark-up is limited to the nodule location information only and no nodule contour information is needed from the user.
Fig 5.
The user interface for human mark-up.
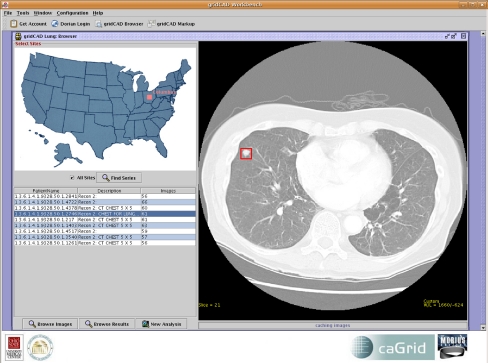
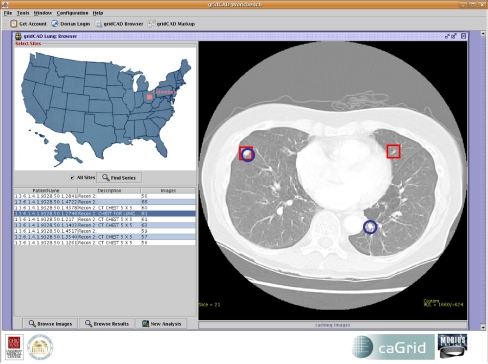
The Browse GUI window implementation (Fig. 6) provides functionality for the user to (a) log on using his or her Grid or institutional credentials, (b) select image server sites and query multiple image server instances for data sets, (c) display query results in a tabular view, (d) select a data set from the query results table, (e) review images in the data set by retrieving images from the image server or local cache, (f) review the detection results for the selected data set by retrieving images from the image server or local cache and the detection results from the RMS and display the nodule locations as overlays, and (g) invoke new analyses for the selected data set by specifying the algorithms to include in the study. If a CAD algorithm is selected, a request is submitted to the CAS with the data set identifier. On the other hand, if human mark-up is requested, a work order is constructed and submitted to the corresponding WOS. For image display, the GUI provides basic radiologic image review capabilities. A subset of the DICOM tags is overlaid on the image. The user can also modify the appearance of the images by changing the window and level of the images. Window and level presets are also selectable by pressing number keys.
Fig 6.
The browser user interface.
The Browse GUI retrieves one image at a time, beginning with the middle section (c) of the series, and incrementally retrieves the (c − n)th and (c + n)th images. It should be noted that the middleware has the capability to move images in any order and we arbitrarily chose to retrieve starting from the center slice in this implementation. In addition, a cache is used to store recently viewed image series. This avoids multiple retrievals of the same data set during a review session. The data sets are indexed in the cache using the DICOM study and series instance UID. The URLs of the image server, RMS, and WOS services are listed in a configuration file to allow easy customization.
Security
Because the GridIMAGE system is designed for Grid-based data sharing and analysis, security of the data and service access is important. In GridIMAGE, each service has its own credentials, consisting of a private key and certificate. The certificate is assigned by a Grid Certificate Authority (the certificate authority infrastructure is part of the caGrid system). Each user also is assigned an identity and a set of credentials. User credentials are managed and assigned by Dorian,7 the federated identity management service of caGrid. Services are trusted based on the service identity; thus, services have full access authorization to other caGrid-based secure services. User authorization is accomplished through the user’s identity.
DISCUSSION
In this paper, we described the design for grid services implemented by caGrid to enable geographically distributed human and CAD image review. CAD algorithms and PACS systems were utilized as Grid services, and caGrid was used to develop a service to interact with image review software used by human readers. This system is the foundation of the caBIG in vivo imaging testbed project. Our plans are to continue to extend the GridIMAGE infrastructure to support image-based cooperative research studies.
An early version of this system was demonstrated at the 2005 annual meeting of the Radiological Society of North America (RSNA) in Chicago, IL. The system described in this paper was also demonstrated at the NCI caBIG annual meeting, in March 2006 in Crystal City, VA. Improvements to the GridIMAGE system in the interim between these two demonstrations not only involved functionality enhancements but also transitioning the underlying infrastructure from caGrid version 0.5 to version 1.0 Beta. This transition enabled the use of caGrid Data Services for the implementation of RMS and WOS, thereby providing the standard CQL query capabilities. In addition, the introduction of human mark-up service and the requisite work-order queue have improved the architecture over the RSNA 2005 implementation of CAS, where the concept of the work queue did not exist. GridIMAGE, therefore, has undergone and will continue to undergo successive stages of technological development. This continued change effectively demonstrates the flexibility with which diverse Grid services of different design and implementation philosophies can be integrated together into a single application.
Demonstration installations typically include two to five Dicom Image data services, one or two CAD services, and one to three human readers. Image transfer rate is approximately 100 kBps over a 100-MB link with security and encryption options and including retrieval from PACS and sorting on the service side. The throughput includes time spent performing server-side tasks, during which no data are moved over the grid. Because these demonstrations typically included small numbers of nodes, the scalability in terms of number of nodes is not measured.
The future version of GridIMAGE will be layered on caGrid 1.0. caGrid 1.0 includes a security infrastructure that supports authentication and group-based authorization. The caGrid Dorian service provides a Grid service infrastructure for the management of Grid user accounts. Dorian provides an integration point between external security domains and the Grid security domain, enabling users to obtain Grid credentials using their locally provided authentication mechanism. The caGrid Grid Grouper provides a group-based authorization method where memberships in groups are defined and managed on the grid. Future versions of GridIMAGE will be able to leverage caGrid 1.0 security to provide secure access to patient data.
Further improvements to the GridIMAGE system are underway. One improvement will address the latency incurred in transmitting large DICOM images to the CAD algorithms. This latency governs how rapidly an end user can interact with different data sets. We are currently developing a caGrid-compatible high-performance data transfer mechanism using GridFTP, the de facto standard protocol for secure, reliable, high-performance data transfer in a Grid environment. Although high-performance data transfer lowers the latency associated with moving large amounts of data to an algorithm, an alternative solution could be the movement of the algorithm to the data source. Moving algorithms to data instead of moving data to algorithm servers reduces the amount of network transfer and may potentially increase the overall system performance. In addition, such a solution may avoid transmitting data with patient-identifying information from the data-source (eg, outside a hospital), which may be a more attractive solution considering Institutional Review Board and Health Insurance Portability and Accountability Act requirements.
CONCLUSION
GridIMAGE provides a useful prototype system for the continued development of Grid computing capabilities in medical imaging. Grid-based distributed compute servers can be used to support the execution of a variety of image processing applications, ranging from CAD, iterative reconstructions from CT, biomarker quantification, quantitative estimates of changes in tumor volume, and quality control algorithms to assess image quality. Grid service-based image processing applications could be supplied by the academic research community, by professional groups such as the RSNA, by the NCI, or by vendors. Use of the Grid service could make it practical to routinely run large-scale quality control studies using imagery acquired by large numbers of institutions. In addition to facilitating access to Grid-enabled resources, the GridIMAGE framework can be utilized to link resources within an institution through a unified, standards-based infrastructure. This would enable a flexible, scalable, and extensible enterprise-level image management and processing system, in which new image servers (eg, DICOM servers) and analytical resources can be added easily.
Grid-based authentication and authorization methods also will make it possible to offer image analysis services using grid-based application service provider models. For example, CAD manufacturers could make their algorithms available through the Grid for a per-use charge or by subscription. Finally, widespread adoption of grid technology will also make it increasingly convenient to carry out broad multi-institutional studies that use images, radiology text reports, and correlative medical data, such as current large-scale studies to evaluate the clinical significance of early detection of lung nodules.
Acknowledgments
This work was supported in part by funding from the National Institutes of Health NIBIB BISTI P20-EB000591, and The National Science Foundation (grants CNS-0426241, CNS-0615155 CNS-0509326, and ANI-0330612), Biomedical Research and Technological Transfer Commission (grant BRTT-02-0003), and Ohio Department of Development (grant ODOD AGMT TECH 04-049).
Footnotes
Although many grids are now operative and many more will be in the near future, the Grid (with a capital) is now routinely used to refer to the overall network of grid efforts.
References
- 1.Gurcan MN, Sahiner B, Petrick N, Chan HP, Kazerooni EA, Cascade PN, Hadjiiski L. Lung nodule detection on thoracic computed tomography images: preliminary evaluation of a computer-aided diagnosis system. Med Phys. 2002;29:2552–2558. doi: 10.1118/1.1515762. [DOI] [PubMed] [Google Scholar]
- 2.Saltz J, Oster S, Hastings S, Langella S, Kurc TM, Sanchez W, Kher M, Manisundaram A, Shanbhag K, Covitz P. caGrid: Design and implementation of the core architecture of the cancer Biomedical Informatics Grid. Bioinformatics. 2006;22(15):1910–1916. doi: 10.1093/bioinformatics/btl272. [DOI] [PubMed] [Google Scholar]
- 3.OSU Department of Biomedical Informatics: Biomedical Informatics Department Web Page. http://www.bmi.osu.edu. Accessed November 23, 2006
- 4.Hastings SL, Langella S, Oster S, Saltz JH: Distributed data management and integration framework: The Mobius Project. In: Proceedings of the Global Grid Forum 11 (GGF11) Semantic Grid Applications Workshop. Global Grid Forum, Lemont, 2004, p 20
- 5.Berman F, Hey AJ, Fox G. Grid Computing: Making the Global Infrastructure a Reality. New York: Wiley; 2003. [Google Scholar]
- 6.Foster I, Kesselman C. The Grid: Blueprint for a New Computing Infrastructure. San Francisco: Morgan Kaufmann; 1999. [Google Scholar]
- 7.Foster I, Kesselman S, Tuecke S. The anatomy of the grid: enabling scalable virtual organizations. Int J Supercomput Appl. 2001;15:200–222. doi: 10.1177/109434200101500302. [DOI] [Google Scholar]
- 8.Grethe JS, Baru C, Gupta A, James M, Ludaescher B, Martone ME, Papadopoulos PM, Peltier ST, Tajasekar A, Zaslavsky IN, Ellisman MH. Biomedical Informatics Research Network: building a national collaboratory to hasten the derivation of new understanding and treatment of disease. Stud Health Technol Inform. 2005;112:100–109. [PubMed] [Google Scholar]
- 9.Solomonides A, McClatchey R, Odeh M, Brady M, Mulet-Parada M, Schottlander D, Amendolia SR: MammoGrid and eDiamond: grids applications in mammogram analysis. In: Proceedings of the IADIS International Conference: e-Society. International Association for Development of Information Society, Lisbon, Portugal, 2003, pp 1032–1033
- 10.Amendolia SR, Brady M, McClatchey R, Mulet-Parada M, Odeh M, Solomonides T. MammoGrid: large-scale distributed mammogram analysis. Stud Health Technol Inform. 2003;95:194–199. [PubMed] [Google Scholar]
- 11.Espert IB, Garcia VH, Quilis DS. An OGSA middleware for managing medical images using ontologies. J Clin Monit Comput. 2005;19:295–305. doi: 10.1007/s10877-005-0675-0. [DOI] [PubMed] [Google Scholar]
- 12.Bottigli U, Cerello P, Delogu P, Fantacci ME, Fauci F, Forni G, Golosio B, Indovina PL, Lauria S, Torres EL, Magro R, Masala GL, Oliva P, Palmiero R, Raso G, Retico A, Stefanini A, Stumbo S, Tangaro S: A computer-aided detection system for mammographic images implemented on a GRID infrastructure. In: Proceedings of the 13th IEEE-NPSS Real Time Conference. IEEE, Piscataway, 2003
- 13.Langella S, Oster S, Hastings S, Siebenlist F, Kurc TM, Saltz JH: Dorian: grid service infrastructure for identity management and federation. In: IEEE International Symposium on Computer-Based Medical Systems (CBMS), Salt Lake City, 2006
- 14.Globus: Globus Web Page. http://www.globus.org. Accessed November 23, 2006
- 15.Covitz PA, Hartel F, Schaefer C, Coronado S, Fragoso G, Sahni H, Gustafson S, Buetow KH. caCORE: a common infrastructure for cancer informatics. Bioinformatics. 2003;19:2404–2412. doi: 10.1093/bioinformatics/btg335. [DOI] [PubMed] [Google Scholar]
- 16.Hastings SL, Oster S, Langella S, Kurc TM, Saltz JH: Introduce: an open source toolkit for rapid development of strongly-typed grid services. OSU Biomedical Informatics Department Technical Report, BMI-OSU-0001-0706, 2006
- 17.Langella S, Hastings S, Oster S, Kurc T, Catalyurek U, Saltz J: A distributed data management middleware for data-driven application systems. In: Proceedings of the 2004 IEEE International Conference on Cluster Computing. IEEE, Piscataway, 2004
- 18.Josefsson S: The Base16, Base32, and Base64 Data Encodings. http://tools.ietf.org/html/3548. Accessed November 23, 2006
- 19.Clunie DA: PixelMed Java Toolkit. http://idoimaging.com/cgi-bin/imaging/program.pl?ident=137com. Accessed November 23, 2006
- 20.CQL: caBIG Query Language. http://gforge.nci.nih.gov/plugins/scmcvs/cvsweb.php/cagrid-1-0/Documentation/docs/DataService/Technical/dataServiceTechnicalGuide.doc?cvsroot=cagrid-1-0. Accessed November 23, 2006
- 21.iCAD: iCAD Inc. Web Page. http://www.icadmed.com. Accessed November 23, 2006