Abstract
The development of nematicides targeting parasitic nematodes of animals and plants requires the identification of biochemical targets not found in host organisms. Recent studies suggest that Caenorhabditis elegans synthesizes phosphocholine through the action of PEAMT (S-adenosyl-L-methionine:phosphoethanolamine N-methyltransferases) that convert phosphoethanolamine into phosphocholine. Here, we examine the function of a PEAMT from C. elegans (gene: pmt-1; protein: PMT-1). Our analysis shows that PMT-1 only catalyses the conversion of phosphoethanolamine into phospho-monomethylethanolamine, which is the first step in the PEAMT pathway. This is in contrast with the multifunctional PEAMT from plants and Plasmodium that perform multiple methylations in the pathway using a single enzyme. Initial velocity and product inhibition studies indicate that PMT-1 uses a random sequential kinetic mechanism and is feedback inhibited by phosphocholine. To examine the effect of abrogating PMT-1 activity in C. elegans, RNAi (RNA interference) experiments demonstrate that pmt-1 is required for worm growth and development and validate PMT-1 as a potential target for inhibition. Moreover, providing pathway metabolites downstream of PMT-1 reverses the RNAi phenotype of pmt-1. Because PMT-1 is not found in mammals, is only distantly related to the plant PEAMT and is conserved in multiple parasitic nematodes of humans, animals and crop plants, inhibitors targeting it may prove valuable in human and veterinary medicine and agriculture.
Keywords: Caenorhabditis elegans, kinetic mechanism, methyltransferase, parasitic nematode, phosphocholine biosynthesis, product identification
Abbreviations: dsRNA, double-stranded RNA; ESI-Q-TOF, electrospray ionization–quadrupole–time-of-flight; GFP, green fluorescent protein; IPTG, isopropyl β-D-thiogalactoside; NGM, nematode growth media; P-DME, phospho-dimethylethanolamine; PEAMT, S-adenosyl-L-methionine:phosphoethanolamine N-methyltransferase(s) (EC 2.1.1.103); P-MME, phospho-monomethylethanolamine; PMT-1, Caenorhabditis elegans PEAMT; RNAi, RNA interference; SAH, S-adenosylhomocysteine; SAM, S-adenosyl-L-methionine
INTRODUCTION
Parasitic nematodes cause a range of human, animal and plant diseases [1–4]. In human and animal medicine, the reported resistance to certain anthelmintics, side effects, or limited efficacy resulting from life cycle differences underscores the need for the continued development of nematicidal compounds [1–4]. In agriculture, few nematode control options are available. Although breeding strategies can be effective, sources of broad-spectrum resistance are unavailable for many crops [5]. In addition, most agricultural nematicides present serious safety and environmental drawbacks [3].
Currently, a lack of metabolic information on parasitic nematodes limits efforts to develop new nematicides. Identifying biochemical targets that differ between the parasite and host species is a promising approach for finding effective and specific new compounds. For this purpose, the free-living nematode Caenorhabditis elegans serves as a useful system for studying nematode biology and for analysing the biochemistry of enzymes in potential target pathways [6–8]. Comparative genomic approaches have identified multiple genes, many of uncharacterized function, that differ between nematodes and their host organisms [9–11]. Recent studies suggest that C. elegans may use a route different from that used by mammals for the synthesis of phosphatidylcholine, a major component in membranes and a precursor in the production of glycoconjugates secreted by parasitic nematodes to avoid host immune responses [9,12–14].
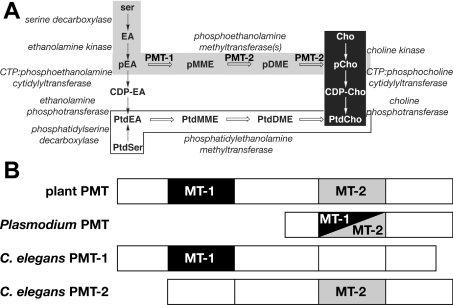
Phosphatidylcholine synthesis occurs through three metabolic routes (Figure 1A). In mammals, fungi and some bacteria, the de novo choline or Kennedy pathway converts choline into phosphatidylcholine [15–18]. Yeast and mammalian liver cells use the Bremer–Greenberg pathway, which involves the methylation of phosphatidylethanolamine to phosphatidylcholine [19,20]. In plants, the multiple methylation of phosphoethanolamine to phosphocholine by PEAMT (S-adenosyl-L-methionine:phosphoethanolamine N-methyltransferase; EC 2.1.1.103) accounts for nearly all of the metabolic flux into the Kennedy pathway, thus circumventing the need for choline phosphorylation [21–26]. This pathway is also how Plasmodium falciparum (the protozoan parasite that causes malaria) synthesizes phosphocholine [27,28]. Although the PEAMT from plants and P. falciparum catalyse a common chemical reaction, their functional organization differs (Figure 1B). The PEAMT from plants contain two tandem methyltransferase domains, with the N-terminal domain methylating phosphoethanolamine to P-MME (phospho-monomethylethanolamine) and the C-terminal domain converting P-MME into P-DME (phospho-dimethylethanolamine) and P-DME to phosphocholine [25,26]. The Plasmodium PEAMT differs from the plant enzymes because it contains a single methyltransferase domain that accepts all three substrates of the pathway [27,28].
Figure 1. Overview of phosphatidylcholine biosynthesis.
(A) Phosphatidylcholine biosynthesis in different organisms. The de novo choline or Kennedy pathway (white on black), the Bremer–Greenberg pathway (black on white) and the phosphobase methylation pathway (black on grey) are shown. Metabolite names consist of a prefix (p, phospho; CDP-, cytidine 5′-diphosphate; or Ptd, phosphatidyl) and a core name (EA, ethanolamine; MME, monomethylethanolamine; DME, dimethylethanolamine; Cho, choline). (B) Overview of the domain organization and reactions catalysed by the PEAMT from plants, P. falciparum and C. elegans. Methyltransferase domains are indicated by shaded boxes. Methylation of phosphoethanolamine to P-MME is catalysed by the first methyltransferase domain (MT-1) and the next two methylation reactions that convert P-MME into phosphocholine are catalysed by the second methyltransferase domain (MT-2). The Plasmodium PEAMT uses a single domain (black/grey) to perform all three methylation reactions.
Earlier reports describe the presence of two genes in C. elegans with low sequence identity (<30%) with the PEAMT from plants and protozoa [25–27], suggesting that nematodes may use a plantlike methylation pathway for phosphocholine synthesis. Interestingly, the two C. elegans PEAMT-related proteins are unrelated (12% sequence identity) to each other. Biochemical and kinetic analyses of the protein encoded by one of the C. elegans PEAMT-like genes (gene: pmt-2; protein: PMT-2; accession number AAB04824.1; Wormbase locus F54D11.1) showed that the purified protein did not accept phosphoethanolamine as a substrate, but catalysed the methylation of P-MME to P-DME and P-DME to phosphocholine (Figure 1A) [13]. Moreover, RNAi (RNA interference) experiments with the pmt-2 gene showed that the activity of PMT-2 was required for normal growth and development of C. elegans and that only supplementation with choline, not ethanolamine-derived precursors, rescued the RNAi phenotype [13]. Therefore, unlike the PEAMT in plants and Plas-modium, PMT-2 alone was incapable of constituting the entire phosphobase pathway.
To determine if the other PEAMT-related protein (gene: pmt-1; protein: PMT-1; accession number AAA81102.1; Wormbase locus ZK622.3a/b) catalyses the missing first step of this metabolic pathway in C. elegans, we examined the biochemical and kinetic properties of PMT-1. Analysis of purified recombinant PMT-1 demonstrates that it catalyses only the initial methylation step in the pathway (Figure 1A), thus providing the missing activity for a new route to phosphocholine in C. elegans. RNAi of the pmt-1 gene shows that the activity of PMT-1 is required for normal development of the worm, which suggests that the phosphobase pathway in C. elegans is physiologically important. This also validates PMT-1 as a potential target for nematicide development. Both PMT-1 and PMT-2, which are not found in mammals and are only distantly related to the plant PEAMT, are conserved in parasitic nematodes of humans, animals and crop plants. Therefore inhibitors targeting PEAMT activity in parasitic nematodes may be effective nematicides for human and veterinary medicine and agriculture.
EXPERIMENTAL
Materials
Escherichia coli Rosetta II (DE3) pLysS cells were purchased from Novagen. Benzamidine–Sepharose resin, the HiTrap Chelating HP FPLC column and the Superdex-200 16/60 size-exclusion FPLC column were from Amersham Biosciences. Radiolabelled [methyl-14C]SAM (S-adenosyl-L-methionine) (∼60 mCi·mmol−1) was bought from either American Radiochemicals or Amersham Biosciences. P-MME and P-DME were synthesized by Gateway Chemical. Kanamycin and IPTG (isopropyl β-D-thiogalactoside) were purchased from Research Products International. Phosphoethanolamine and all other reagents were from Sigma–Aldrich.
Cloning of pmt-1 from C. elegans and generation of bacterial expression vector
The coding region of pmt-1 (accession number AAA81102.1, Wormbase locus ZK622.3a/b) was amplified by PCR from C. elegans cDNA using 5′-dGAGGAATTCCATATGTCGACCGACCAACAATC-3′ as the forward primer (NdeI site is underlined; coding region start site is in boldface) and 5′-dGACCGCTCG-AGCTAATGAGTCAACTCAAGAAG-3′ as the reverse primer (XhoI site is underlined; coding region stop site is in boldface). The 1.4 kb PCR product was gel-extracted (QIAquick Spin Gel Extraction kit; Qiagen) and cloned into pCRII-TOPO vector (Invitrogen). Automated nucleotide sequencing confirmed the fidelity of the PCR product. Digesting the pCRII-TOPO-CePMT-1 vector with XhoI and NdeI, then ligating the 1.4 kb DNA fragment into XhoI/NdeI-digested pET28a yielded the pET28a-pmt-1 expression vector. According to the guidelines for gene and protein nomenclature for C. elegans (http://www.cbs.umn.edu/CGC/Nomenclature/nomenguid.htm), the isolated gene is designated pmt-1 and the encoded protein named PMT-1.
Expression in E. coli and protein purification
The pET28a-pmt-1 expression construct was transformed into E. coli Rosetta II (DE3) pLysS cells. Transformed E. coli were grown at 37 °C in Terrific broth containing 50 μg·ml−1 kanamycin and 35 μg·ml−1 chloramphenicol until a D600 of ∼0.8–1.2 was reached. Following induction with 1 mM IPTG, the cultures were grown at 30 °C for 4–6 h. Cells were harvested by centrifugation (10000 g for 10 min) and resuspended in lysis buffer [50 mM Tris, pH 8.0, 500 mM NaCl, 25 mM imidazole, 10% (v/v) glycerol and 1% (v/v) Tween 20]. After sonication and centrifugation (50000 g for 1 h), the supernatant was loaded on to a 5 ml HiTrap Chelating HP FPLC column, which was previously charged with 0.1 M nickel sulfate and equilibrated with lysis buffer, using an Akta Explorer FPLC system. The column was washed with 10 column volumes of lysis buffer and 10 column volumes of wash buffer (lysis buffer minus Tween 20). Bound protein was eluted using an imidazole gradient (25–300 mM) in the wash buffer. To remove the hexahistidine tag, His-tagged PMT-1 was incubated with thrombin during dialysis for 24 h at 4 °C against wash buffer. Dialysed protein was reapplied to the nickel-charged HiTrap Chelating FPLC column, which was equilibrated in wash buffer, to remove undigested protein. The eluent was next loaded on to a 0.2 ml benzamidine–Sepharose column that was equilibrated in wash buffer to deplete the sample of thrombin. The flow-through was passed through a Superdex-200 16/60 size-exclusion FPLC column equilibrated in 25 mM Hepes (pH 7.5) and 500 mM NaCl. Fractions containing PMT-1 were pooled, dialysed into 30% glycerol, 50 mM Hepes (pH 7.5), 500 mM NaCl and 2 mM Na2EDTA and stored at −80 °C. Protein concentration was determined by the Bradford protein assay method (Bio-Rad) with BSA as the standard.
Enzyme assays
A radiochemical assay was used to measure enzymatic activity [13]. Standard assay conditions were 0.1 M Hepes·KOH (pH 8), 2 mM Na2EDTA, 10% glycerol, 1.5 mM SAM (100 nCi of [methyl-14C]SAM) and 100 μM phosphoethanolamine in 100 μl. Assays contained 1 μg of protein and were incubated for 2.5, 5 or 7 min at 30 °C. Protein amount and time of reaction provided a linear rate of product formation. Reactions were terminated by addition of 1 ml of ice-cold water. The phosphorylated product was purified by loading the quenched reaction mixture on to a 1 ml Dowex (50WX8–100)-resin column, which was washed with 2 ml of ice-cold water and then eluted with 10 ml of 0.1 M HCl. For scintillation counting, 2 ml of the eluent was mixed with 3 ml of Ecolume scintillation fluid (ICN Biomedicals). Steady-state kinetic parameters for SAM (15–1500 μM) were determined under standard assay conditions at 200 μM phosphoethanolamine. The kcat and Km values of PMT-1 for phosphoethanolamine (2–200 μM) were determined at 1.5 mM SAM. All data were fitted to the Michaelis–Menten equation, v=(kcat[S])/(Km+[S]), using Kaleidagraph (Synergy Software).
Analysis of the kinetic mechanism of PMT-1 used global fitting analysis [13]. Reaction rates were measured under standard assay conditions with a matrix of substrate concentrations (15–1500 μM SAM and 5–200 μM phosphoethanolamine). Global curve fitting in SigmaPlot (Systat Software) was used to model the kinetic data to rapid equilibrium rate equations describing Ping Pong, v=(Vmax[A][B])/(KB[A]+KA[B]+[A][B]), ordered sequential, v=(Vmax[A][B])/(KAKB+KB[A]+[A][B]), and random sequential, v=(Vmax[A][B])/(αKAKB+KB[A]+KA[B]+[A][B]), kinetic mechanisms, where v is the initial velocity, Vmax is the maximum velocity, KA and KB are the Km values for substrates A and B respectively, and α is the interaction factor if the binding of one substrate changes the dissociation constant for the other substrate [29].
Reaction rates of PMT-1 in the presence of inhibitors, i.e. SAH (S-adenosylhomocysteine), P-MME and phosphocholine, were determined under standard assay conditions. Enzymatic activity was measured in reaction mixtures containing various substrate concentrations, as above, containing SAH (0–20 μM), P-MME (0–25 mM) or phosphocholine (0–30 mM) as inhibitors. Data from these experiments were fitted to the equations for competitive, v=Vmax[S]/{Km(1+[I]/Kis)+[S]}, or non-competitive, v=Vmax[S]/{Km(1+[I]/Kis)+[S](1+[I]/Kii)}, inhibition by global fitting analysis in SigmaPlot.
Characterization of reaction products
TLC and ESI-Q-TOF (electrospray ionization–quadrupole–time-of-flight) MS were used to confirm the chemical identity of the PMT-1 reaction product. To screen the substrate specificity of PMT-1, the enzyme was assayed under standard reaction conditions containing 400 nCi of [methyl-14C]SAM and 10 mM of phosphoethanolamine, P-MME or P-DME. Reaction products were purified as described above, collected and evaporated to dryness. The dry product was resuspended in methanol and applied to a Whatman LK6D silica TLC plate, which was then developed with n-butanol/methanol/concentrated HCl/water (7.5:7.5:1:1, by vol.). Phosphoimaging was used to visualize the radiolabelled products on the TLC plate. Migration of reaction products was compared with authentic standards.
To confirm the chemical identity of the PMT-1 reaction product by ESI-Q-TOF MS, a scaled-up assay (1 ml) was performed under standard reaction conditions without the use of radiolabelled SAM. The reaction product was purified as described above, collected and evaporated to dryness. The sample was resuspended in 100 μl of 10% (v/v) acetonitrile and analysed by an ABI QSTAR XL (Applied Biosystems/MDS Sciex) hybrid QTOF MS/MS (tandem MS) mass spectrometer equipped with a nanoelectrospray source (Protana XYZ manipulator). Positive mode nanoelectrospray was generated from borosilicate nanoelectrospray needles at 1.5 kV. TOF MS spectra were obtained using the Analyst QS software with an m/z range of 30–500. The observed m/z values of the parent ion and fragmentation pattern, i.e. neutral loss of HPO3 (80 Da) and H3PO4 (98 Da), were determined: P-MME, [M+H]+ 156.05, [(M−HPO3)+H]+ 76.08, [(M−H3PO4)+H]+ 58.07.
RNAi of pmt-1 by feeding and chemical rescue in C. elegans
For RNAi in C. elegans, a 960-bp nucleotide fragment was amplified from the pmt-1 gene using oligonucleotide primers containing the sequences 5′-dATGGTGAACGTTCGTCGTGC-3′ and 5′-dCATACGTATTTCTCATCATC-3′ respectively. The pmt-1 DNA fragment was cloned into the L4440 vector [30] between opposing T7 polymerase promoters. The resulting vector was transformed into E. coli strain HT115, which contains a gene encoding T7 RNA polymerase. The dsRNA (double-stranded RNA) molecule corresponding to pmt-1 was delivered by feeding C. elegans E. coli engineered to produce the dsRNA mole-cule [31,32]. For control experiments, worms were fed E. coli transformed with the L4440 vector encoding GFP (green fluorescent protein). Feeding RNAi was initiated from C. elegans larva at 20 °C on NGM (nematode growth medium)-agar plates containing 2.5 mg·ml−1 IPTG and E. coli expressing either pmt-1 or control GFP dsRNA.
To test if providing PEAMT pathway metabolites reversed the pmt-1 RNAi-generated phenotype, ethanolamine, monomethylethanolamine, dimethylethanolamine or choline was incorporated into the NGM-agar. Plates were seeded with E. coli expressing dsRNA homologous with pmt-1. In one set of experiments, either a stage-one (L1) or a dauer larva was placed on each plate and the P0 and F1 progeny examined for 5 days. The dauer larva were daf-7(e1372) mutants, which are constitutive dauers at 25 °C but are wild-type at 20 °C [33]. Upon placing the dauers on the RNAi-feeding plates, the incubation temperature was shifted from 25 to 20 °C to induce the worms to exit dauer and start feeding. In another set of experiments, a single stage-four (L4) C. elegans hermaphrodite was placed on each plate and allowed to lay eggs for 24 h. The phenotype of the P0 and F1 progeny was scored 48 and 72 h after the initial 24 h egg-laying period.
RESULTS
Analysis of the pmt-1 cDNA from C. elegans
A 1425 base-pair cDNA encoding a 475-amino-acid polypeptide with a calculated molecular mass of 55.1 kDa and pI 5.2 was isolated from mixed stage C. elegans cDNA. The transcript encoded by Wormbase locus ZK622.3 is now designated pmt-1. The predicted amino acid sequence of PMT-1 from C. elegans shares 23.2, 27.8 and 25.5% identity with the PEAMT from Arabidopsis, wheat and spinach respectively and less than 10% identity with the Plasmodium enzyme. Comparison of PMT-1 and C. elegans PMT-2 [13] showed only 12.4% amino acid sequence identity. Database searching identified PMT-1 ESTs (expressed sequence tags) in a range of parasitic nematodes, including Ancylostoma caninum (dog hookworm) (GenBank®: 15766091), Ascaris suum (large roundworm of pigs) (GenBank®: 17993264), Strongyloides stercoralis (human threadworm) (GenBank®: 12714760), Haemonchus contortus (sheep barber-pole worm) (GenBank®: 27590930) and Meloidogyne incognita (plant root-knot nematode) (GenBank®: 21652426). No PEAMT sequences were identified in humans or other animals, and searches with PMT-1 did not isolate any PMT-2 sequences.
Amino acid sequence comparisons reveal differences in the alignment and homology of the methyltransferase domains of PMT-1, PMT-2 and the PEAMT from plants and Plasmodium (Figure 2). A set of consensus sequence motifs (identified as I, post-I, II and III) defines the SAM binding site of various methyltransferases [34,35]. Unlike the bipartite plant enzymes, C. elegans PMT-1 appears to only contain the N-terminal methyltransferase domain. Alignment of the consensus motifs shows that the C-terminal half of PMT-1 lacks the sequence regions indicative of a methyltransferase domain. By analogy with the plant PEAMT [25,26], these sequence alignments suggest that C. elegans PMT-1 only catalyses the methylation of phosphoethanolamine, and reveals a third type of structural organization for the enzymes of the phosphobase pathway.
Figure 2. Amino acid sequence comparison of PEAMT.
Multiple sequence comparison of C. elegans PMT-1 (CePMT1; AAA81102.1), C. elegans PMT-2 (CePMT2; AAB04824.1), Spinacia oleracea (spinach) PEAMT (SoPMT; AAF61950.1), Triticum aestivum (wheat) PEAMT (TaPMT; AAL40895.1), Arabidopsis thaliana (thale cress) PEAMT (AtPMT; AAG41121.1) and P. falciparum PEAMT (PfPMT; AAR08195.1). Invariant amino acids are highlighted. Sequence motifs defining the methyltransferase domains are labelled I, post-I, II and III. These motifs in the first and second methyltransferase domains are indicated with dark grey and light grey boxes respectively. Highly conserved positions within the methyltransferase sequence motifs are shown in white text. Sequence alignment was performed using Multalin (http://prodes.toulouse.inra.fr/multalin/multalin.html).
Expression, purification and substrate preference of C. elegans PMT-1
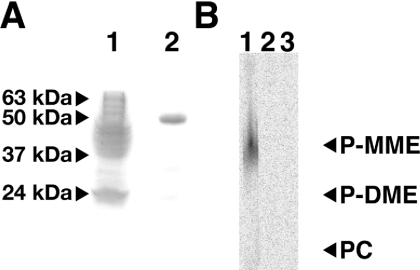
For biochemical and kinetic analyses of PMT-1, recombinant protein was expressed in E. coli and purified using nickel-affinity and size-exclusion chromatography. The hexahistidine tag was removed from PMT-1 by digestion with thrombin. SDS/PAGE analysis of the purified recombinant protein showed that it migrated with a molecular mass of approx. 52 kDa, which approximately corresponds to the predicted mass of the expressed protein (Figure 3A). Gel-filtration chromatography of PMT-1 showed that the protein is a monomer in solution (results not shown), similar to C. elegans PMT-2 and the PEAMT from spinach [13,25].
Figure 3. Protein expression and substrate specificity of PMT-1.
(A) SDS/PAGE analysis. Samples were stained for total protein using Coomassie Blue. Arrows correspond to the indicated molecular mass markers. Lane 1 shows the sonicated material (50 μg of protein) and lane 2 shows the final size-exclusion purified protein (2 μg of protein). (B) TLC analysis of the PMT-1 reaction products. PMT-1 (5 μg) was incubated for 30 min with phosphoethanolamine (lane 1), P-MME (lane 2) or P-DME (lane 3) and the products were isolated as described in the Experimental section. Positions of authentic standards for P-MME, P-DME and phosphocholine (PC) are indicated on the right.
To establish the substrate specificity of PMT-1, phosphoethanolamine, P-MME and P-DME were tested as potential substrates (Figure 3B). PMT-1 catalyses the SAM-dependent methylation of phosphoethanolamine, but does not accept either P-MME or P-DME as a substrate. No product was observed in the absence of SAM (results not shown). The reaction product generated by PMT-1 from phosphoethanolamine was confirmed as P-MME by ESI-Q-TOF MS, as described in the Experimental section. The specific activity of PMT-1 using phosphoethanolamine and SAM as substrates was 27±3 nmol product·min−1·mg of protein−1.
For comparison with other PEAMT enzymes, the kinetic parameters (kcat and Km) of PMT-1 for SAM and phosphoethanolamine were measured using a radiometric assay (Table 1). The Km value of PMT-1 for SAM is comparable with the range reported for the spinach, wheat and Plasmodium PEAMT (56–153 μM) [24–27]. PMT-1 displayed 6–10-fold lower Km values for phosphoethanolamine compared with the other PEAMT [24–27]. In addition, the turnover rate of PMT-1 was 25-fold faster than that reported for the Plasmodium enzyme [27]. Neither specific activities nor turnover rates have been described for any purified plant PEAMT. Compared with C. elegans PMT-2, the Km for SAM of PMT-1 is 2-fold lower and the turnover rate of PMT-1 is 100-fold slower [13].
Table 1. Steady-state kinetic parameters of PMT-1.
Assays (n=3) were performed as described in the Experimental section.
| Substrate | kcat (min−1) | Km (μM) | kcat/Km (M−1·s−1) |
|---|---|---|---|
| SAM | 3.5±0.3 | 145±18 | 402 |
| Phosphoethanolamine | 3.1±0.2 | 9.9±0.8 | 5220 |
Kinetic mechanism of PMT-1
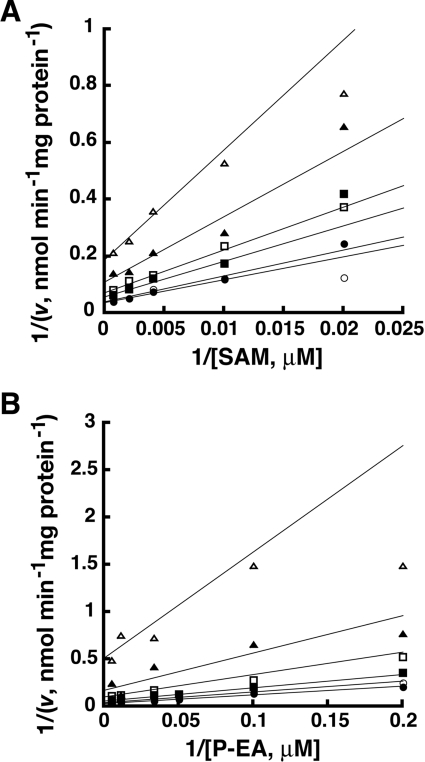
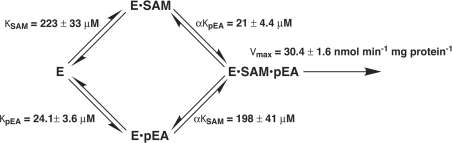
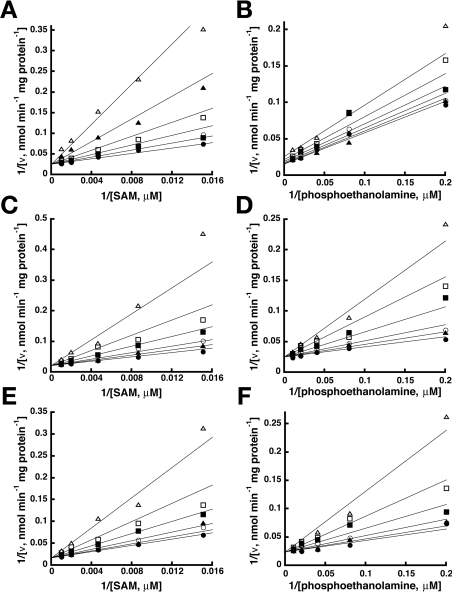
To determine if the different domain architecture affects the kinetic mechanism of PMT-1, a matrix of SAM and phosphoethanolamine concentrations were used to test possible kinetic models including Ping Pong, ordered sequential and random sequential for a two-substrate to two-product (Bi Bi) reaction [29]. The observed data (Figure 4) were simultaneously fitted to the equations for each Bi Bi kinetic mechanism. The quality of the fit was determined by examining Lineweaver–Burk plots, residual errors, standard error of the fitted parameters and the fit correlation coefficient. The best fit of the data was to a random sequential mechanism (r2=0.992) (Figure 5). The Ping Pong model was eliminated because of the intersecting pattern observed in the Lineweaver–Burk plots. Likewise, the lower quality of the fit to an ordered sequential mechanism (r2=0.912) removed that model from consideration. The fitted parameters for a random sequential mechanism suggest that the binding of either phosphoethanolamine or SAM only modestly enhances the binding of the other substrate (α=0.89). The model-derived values of αKSAM and αKphosphoethanolamine were similar to the Km values determined experimentally (Table 1).
Figure 4. Initial velocity variation of PMT-1 substrates.
Experimental data are indicated by the symbols in the double reciprocal plots of the substrate variation experiments. The lines shown represent the global fit of all data to the equation for a random sequential Bi Bi mechanism. Assays were performed as described in the Experimental section. (A) Double reciprocal plot of 1/v versus 1/[SAM] at 5, 10, 20, 30, 90 and 200 μM phosphoethanolamine (top to bottom). (B) Double reciprocal plot of 1/v versus 1/[phosphoethanolamine (P-EA)] at 15, 50, 100, 250, 500 and 2500 μM SAM (top to bottom).
Figure 5. Kinetic mechanism of PMT-1.
The fitted kinetic parameters are indicated on the Figure. KSAM and KpEA are the equilibrium dissociation constants for the binding of either SAM or phosphoethanolamine (pEA). α is the interaction factor between the substrates.
To provide further evidence for a random sequential kinetic mechanism, the effect of reaction products (SAH and P-MME) and phosphocholine, which is the final product of the PEAMT pathway, on the activity of PMT-1 was examined. Product inhibition by SAH versus SAM and phosphoethanolamine showed competitive and non-competitive inhibition patterns respectively (Figures 6A and 6B), yielding the Ki values summarized in Table 2. The non-competitive inhibition pattern observed with SAH versus phosphoethanolamine indicates that a dead-end complex, i.e. E·SAH·phosphoethanolamine, forms. With P-MME, competitive inhibition was observed with both substrates (Figures 6C and 6D; Table 2). In general, the Ki values for SAH were approx. 5-fold lower than the IC50 values determined for the Plasmodium PEAMT for this reaction product [27]. The observed product inhibition patterns are indicative of a random sequential Bi Bi kinetic mechanism [29].
Figure 6. Inhibition studies of PMT-1.
Inhibition assays were performed as described in the Experimental section. The lines show the fit to the data. (A) Double reciprocal plot of 1/v versus 1/[SAM] at 0, 1, 2.5, 5, 10 and 20 μM SAH (top to bottom). (B) Double reciprocal plot of 1/v versus 1/[phosphoethanolamine] at 0, 1, 2.5, 5, 10 and 20 μM SAH (top to bottom). (C) Double reciprocal plot of 1/v versus 1/[SAM] at 0, 1, 2.5, 6.25, 12.5 and 20 mM P-MME (top to bottom). (D) Double reciprocal plot of 1/v versus 1/[phosphoethanolamine] at 0, 1, 2.5, 6.25, 12.5 and 20 mM P-MME (top to bottom). (E) Double reciprocal plot of 1/v versus 1/[SAM] at 0, 1, 3, 7.5, 15 and 30 mM phosphocholine (top to bottom). (F) Double reciprocal plot of 1/v versus 1/[phosphoethanolamine] at 0, 1, 3, 7.5, 15 and 30 mM phosphocholine (top to bottom).
Table 2. Inhibition patterns of PMT-1.
All assays (n=3) were performed as described in the Experimental section. The inhibition patterns are as follows: C, competitive; NC, non-competitive. P-EA, phosphoethanolamine; PC, phosphocholine.
| Varied substrate | Inhibitor | Fixed substrate | Inhibition | Kis (μM) | Kii (μM) |
|---|---|---|---|---|---|
| SAM | SAH | P-EA (50 μM) | C | 9.1±1.2 | |
| SAM | P-MME | P-EA (50 μM) | C | 5110±407 | |
| SAM | PC | P-EA (50 μM) | C | 7810±773 | |
| P-EA | SAH | SAM (500 μM) | NC | 4.8±1.1 | 16.2±1.5 |
| P-EA | P-MME | SAM (500 μM) | C | 4230±751 | |
| P-EA | PC | SAM (500 μM) | C | 6940±615 |
Assays in the presence of phosphocholine showed competitive inhibition compared with SAM and phosphoethanolamine (Figures 6E and 6F) with inhibition constants slightly higher than those observed with P-MME (Table 2). The Ki values of PMT-1 for phosphocholine were roughly 10-fold higher than the IC50 value (0.5 mM) described for the spinach enzyme [25]. These experiments suggest that PMT-1 is subject to feedback inhibition by the final product of the PEAMT pathway.
RNAi of pmt-1 by feeding and chemical rescue in C. elegans
As a model organism, C. elegans is a valuable tool for the functional characterization of novel drug targets identified by genomics approaches [36]. Importantly, systematic comparison of RNAi experiments in C. elegans suggest that off-target effects are minor and that specific RNAi to generate a knockdown phenotype for a targeted gene can be used to validate if inhibition of the encoded activity is of potential therapeutic value [36].
To test the potential effect of disrupting the physiological function of PMT-1 in C. elegans, RNAi was used. C. elegans were raised on E. coli producing the dsRNA corresponding to either pmt-1 or GFP, as a control. Normal life cycle development was observed in all the control experiments. With bacteria expressing the dsRNA corresponding to pmt-1, when the parental worm (P0) exposure began either as an L1 or a daf-7 dauer larva, the phenotype was complete or highly penetrant (>95%) P0 sterility. When exposure began as a P0 L4 larva, the observed phenotype was arrested development and lethality in the progeny (F1) at the L1/L2 or the L3 larva stage. Since the pmt-1 RNAi experiments show that C. elegans of different larval stages were all developmentally impaired, this gene and the corresponding enzyme activity are necessary at multiple stages in the worm's life cycle.
To determine whether providing metabolites of the PEAMT pathway could reverse the RNAi-generated phenotype, C. elegans grown on NGM media or NGM media supplemented with ethanolamine, monomethylethanolamine, dimethylethanolamine or choline were fed E. coli expressing dsRNA homologous with either pmt-1 or GFP (control) (Figure 7; Table 3). To facilitate uptake into the worms, the free base forms of the metabolites were used, with conversion into the corresponding phosphobases presumably catalysed by endogenous ethanolamine and choline kinases in the worm [37]. In control experiments, nematodes (L1, dauer and L4) developed normally on either standard or supplemented media. Addition of ethanolamine to the media did not reverse the pmt-1 RNAi-generated phenotype for the L1, dauer or L4 worms. Supplementing the growth media with monomethylethanolamine (5–10 mM), dimethylethanolamine (5–10 mM) or choline (30 mM) rescued the P0 sterility and F1 larval arrest observed with the pmt-1 RNAi-generated phenotype. These results suggest that PMT-1 functions at a step before monomethylethanolamine enters the plant-like phosphobase methylation pathway.
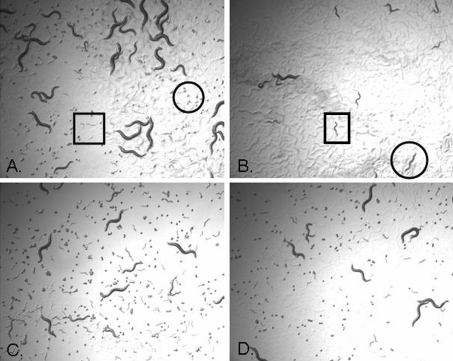
Figure 7. Chemical rescue of the pmt-1 RNAi phenotype with pathway intermediates.
C. elegans L4 hermaphrodites were placed on NGM-agar plates containing bacteria producing GFP dsRNA (A), pmt-1 dsRNA (B), pmt-1 dsRNA plus 10 mM DME (C) or pmt-1 dsRNA plus 30 mM choline (D). Photographs show the phenotype of the F1 progeny four days after introduction of the original L4 to the plate. (A) F1 progeny have developed to the adult stage and many F2 eggs (circle) and some F2 L1 (box) larvae are present. (B) Some F1 progeny have arrested at the L1/L2 stage of development (box) and some have arrested at the L3 stage (circle). F2 eggs and larva are absent. (C, D) F1 progeny have developed to the adult stage, and as in (A), many F2 eggs and some F2 L1 larvae are present.
Table 3. Summary of pmt-1 feeding-RNAi and chemical rescue phenotypes in C. elegans.
C. elegans at the L1, dauer or L4 stage were placed on NGM-agar plates containing the indicated compound (EA, ethanolamine; MME, monomethylethanolamine; DME, dimethylethanolamine), as described in the Experimental section. The observed phenotypes are indicated.
| Starting worm (P0) | Added compound | Phenotype |
|---|---|---|
| L1 | None | P0 sterility |
| 5, 10 or 30 mM EA | P0 sterility | |
| 5 or 10 mM MME | Fertile adults | |
| 5 or 10 mM DME | Fertile adults | |
| 30 mM Choline | Fertile adults | |
| Dauer | None | P0 sterility |
| 5, 10 or 30 mM EA | P0 sterility | |
| 5 or 10 mM MME | Fertile adults | |
| 5 or 10 mM DME | Fertile adults | |
| 30 mM Choline | Fertile adults | |
| L4 | None | F1 L1/L2 or L3 arrest; lethality |
| 5, 10 or 30 mM EA | F1 L1/L2 or L3 arrest; lethality | |
| 5 or 10 mM MME | Wild-type F1 | |
| 5 or 10 mM DME | Wild-type F1 | |
| 30 mM Choline | Wild-type F1 |
DISCUSSION
In animals and plants, the metabolic routes leading to phospholipid synthesis are well studied [21–23]; however, only limited information on these pathways in C. elegans and other nematodes is available. Surveys of lipid content in C. elegans indicate that one-third of total phospholipid content derives from phosphocholine and that this organism contains phosphorylcholine oligosaccharides and glycosphingolipids [38–40]. As in other eukaryotes, the de novo choline or Kennedy pathway functions in this nematode [14,37,41,42], but genomic analysis of C. elegans implies the presence of an alternative metabolic route to phosphatidylcholine. Genome-wide RNAi experiments in C. elegans showed that when the choline kinase or phosphatidylethanolamine methyltransferase genes were targeted by RNAi, there was no effect on worm growth and development [43], which raises the possibility of another pathway for phosphocholine synthesis and entry into the Kennedy pathway.
The presence of two transcribed loci in C. elegans that share sequence similarity with the plant PEAMT suggested the presence of a plant-like route to phosphocholine [25–27]. Previous biochemical studies of the protein encoded by one of these genes (pmt-2) showed that PMT-2 catalysed the last two steps in the pathway (Figure 1A) [13], which raised the possibility that the other
PEAMT-related protein catalysed the first reaction. Our results demonstrate that PMT-1 performs the missing first chemical step in the pathway, as shown in vitro using purified recombinant pro-tein and in vivo by chemical rescue of the pmt-1 RNAi phenotype. Importantly, although the two PEAMT from C. elegans are both methyltransferases and share a common nomenclature, they are distinct enzymes (not isoforms of the same activity) that are unrelated to each other in sequence (12% identity), use different substrates and yield different products.
Unlike the multifunctional PEAMT from plants and Plasmodium [24–28], C. elegans PMT-1 uses phosphoethanolamine as a substrate, not P-MME or P-DME. This difference in activity is reflected in the organization of the methyltransferase domains of the various PEAMT. PMT-1 is of similar amino acid length to the plant PEAMT, but only the N-terminal methyltransferase domain is found in this protein. Previous experiments with the plant PEAMT showed that removal of the C-terminal domain results in a functional truncated enzyme that catalyses the methylation of phosphoethanolamine to P-MME, not the conversion of P-MME or P-DME [25,26]. By analogy with the plant enzymes, sequence alignments suggested that PMT-1 from C. elegans catalyses only the initial reaction in the pathway, as demonstrated here using purified recombinant protein.
To date, three types of organization for PEAMT in the phosphobase pathway have been reported (Figure 1B) [13,24–28]. The plant PEAMT (type I) contain tandem methyltransferase domains in a single polypeptide. The Plasmodium enzyme (type II) performs all three methylation reactions at one active site. Analysis of PMT-1 and PMT-2 suggests that the type III PEAMT evolved distinct substrate specificities for two proteins to convert phosphoethanolamine into phosphocholine. The low sequence homology between PMT-1 and PMT-2 implies structural differences in their active sites, which may have consequences for the biochemical regulation and inhibition of the PEAMT pathway in nematodes.
The kinetic mechanism of PMT-1 was examined using initial velocity studies and analysis of product inhibition patterns. The results of these experiments indicate that PMT-1 employs a random sequential Bi Bi kinetic mechanism, as observed with PMT-2 [16]. Of the three possible Bi Bi kinetic mechanisms, the Ping Pong mechanism was eliminated based on the pattern of lines observed in the initial velocity experiments (Figure 4). Comparison of the global fitting analysis of the data with equations for either ordered or random sequential mechanisms revealed that a random mechanism model best fits the kinetic data (Figure 5). Moreover, product inhibition experiments provide additional evidence for a random sequential kinetic mechanism. Competitive inhibition by both P-MME and phosphocholine against either SAM or phosphoethanolamine as the varied substrate indicates that these inhibitors compete with both substrates for the same form of PMT-1, either free enzyme or the enzyme in complex with the saturating co-substrate [29,44]. Because P-MME and phosphocholine are larger than phosphoethanolamine, i.e. methylated, these molecules would occupy the phosphobase binding site and part of the SAM binding site. Likewise, competitive inhibition by SAH versus SAM can also be explained in this way. The noncompetitive inhibition by SAH versus phosphoethanolamine indicates formation of a dead-end complex between these two molecules [44]. Since the PEAMT from different organisms catalyse a common chemical reaction and are related by sequence similarity, it is likely that the enzymes from plants and Plasmodium also use a random sequential kinetic mechanism.
Product and/or feedback inhibition may biochemically regulate PEAMT activity in C. elegans, as was suggested for the spinach and Plasmodium enzymes [25,27]. Both PMT-1 and PMT-2 [13] are sensitive to SAH inhibition. Similarly, SAH may regulate the activity of the Plasmodium enzyme, in which the estimated IC50 value for SAH was roughly 3–5-fold lower than the Km value for SAM [27]. In addition, phosphocholine inhibits both PMT-1 (Ki=7 mM) and PMT-2 (Ki=1.5 mM) [13]. In plants, cytosolic phosphocholine concentrations reach up to 10 mM [25], which would inhibit PEAMT activity; however, it is unclear if C. elegans accumulates phosphocholine at similar levels. Nonetheless, biochemical and kinetic information on PMT-1 (and PMT-2) will be valuable for evaluating the effect of inhibitors of either enzyme.
The in vivo function of PMT-1 was examined by disruption of the endogenous pmt-1 gene by feeding RNAi. Since the two C. elegans PEAMT (PMT-1 and PMT-2) are related by less than 15% sequence identity and extensive database searching using the pmt-1 gene failed to detect the pmt-2 gene or any other related genes in C. elegans, the likelihood of cross-reactivity of the dsRNA is minimal [7,8]. In these experiments, C. elegans L1 and dauer larva grown in the presence of E. coli expressing dsRNA from the pmt-1 gene yielded P0 sterility. Likewise, feeding RNAi to L4 larva resulted in arrested development and lethality in the F1 progeny at the L1/L2 stage or in some progeny at the L3 stage (Figure 7 and Table 3). In previous genome-wide RNAi experiments with C. elegans, the pmt-1 gene, which had an unknown function at the time, was targeted [43]. Disruption of the gene yielded a non-viable RNAi phenotype. The RNAi-generated phenotypes observed with pmt-1 are similar to those reported in experiments targeting enzymes that function after choline kinase in the de novo choline pathway in C. elegans that caused drastic reductions in fertility, which are consistent with the role of phosphatidylcholine in embryo development [12].
The RNAi experiment validates PMT-1 as a potential nematicide target because knockdown of the pmt-1 gene results in a severe developmental phenotype in C. elegans, which indicates that efforts aimed at obtaining compounds inhibiting PMT-1 may be a viable strategy for nematicide development. The described experiments are a first step towards this goal; similar work in parasitic nematodes will need to be performed.
Chemical rescue experiments of the RNAi-mediated pmt-1 phenotype also support a role for PMT-1 in phosphocholine synthesis. Growing C. elegans in media supplemented with ethanolamine did not restore normal growth, whereas providing monomethylethanolamine, dimethylethanolamine or choline reversed the RNAi-generated phenotype. The RNAi-generated phenotypes of PMT-1 and PMT-2 are identical [13], demonstrating that both enzymes (and the PEAMT pathway) are needed for normal growth and development of C. elegans. Moreover, the results of chemical rescue experiments parallel the in vitro substrate specificity of each protein, i.e. PMT-1 accepts phosphoethanolamine and PMT-2 uses both P-MME and P-DME [13]. The arrested development and lethality effects of knocking out either PMT-1 or PMT-2 activity in C. elegans are in contrast with the results of RNAi targeting choline kinase [39], suggesting that the PEAMT pathway may be the primary provider of phosphocholine to the de novo choline pathway, as occurs in plants.
As demonstrated in the chemical rescue experiments, the RNAi phenotype is only reversed by addition of high concentrations (30 mM) of choline, which is much higher than levels found physiologically in blood and tissue of host organisms [45–49]. For example, human plasma choline levels vary in response to diet and range from 7 to 20 μM [45,46]. In livestock, such as sheep, adding the highest estimates of all choline forms together yields approx. 1 μM choline [47,48]. Ruminants have particularly low levels of choline in their intestinal lumen (almost none) due to microbial breakdown [49]; they synthesize choline in the liver via a phosphatidylcholine methyltransferase, a completely different pathway from the PEAMT-based route. Ultimately, the dose required to rescue the PMT RNAi phenotype in C. elegans is more than a million-fold higher than the amount of choline physiologically available in potential host organisms of parasitic nematodes.
If the physiologically essential role of PMT-1 in C. elegans is conserved in parasitic nematodes, such as An. caninum, As. suum, S. stercoralis, H. contortus and M. incognita where cDNAs predicted to encode PMT-1 homologues have been identified, then PMT-1 offers a biochemical target for the development of nematicidal molecules. The identification of new protein targets will complement anthelmintic and nematicide discovery and development programmes. Ultimately, the next generation of molecules targeting parasitic nematodes will improve human, animal and plant health worldwide.
Acknowledgments
This work was supported by a grant from the Environmental Protection Agency (X-83228201) to J. P. M., D. J. W., M. C. H. and J. M. J and a grant from the U.S. Department of Agriculture (2005-33610-16465) to M. C. H.
References
- 1.Chan M. S. The global burden of intestinal nematode infections – fifty years on. Parasitol. Today. 1997;13:438–443. doi: 10.1016/s0169-4758(97)01144-7. [DOI] [PubMed] [Google Scholar]
- 2.Jasmer D. P., Goverse A., Smant G. Parasitic nematode interactions with mammals and plants. Annu. Rev. Phytopathol. 2003;41:245–270. doi: 10.1146/annurev.phyto.41.052102.104023. [DOI] [PubMed] [Google Scholar]
- 3.Geerts S., Coles G. C., Gryseels B. Anthelmintic resistance in human helminths: learning from the problems with worm control in livestock. Parasitol. Today. 1997;13:149–151. doi: 10.1016/s0169-4758(97)01024-7. [DOI] [PubMed] [Google Scholar]
- 4.Prichard R. Anthelmintic resistance. Vet. Parasitol. 1994;54:259–268. doi: 10.1016/0304-4017(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 5.Chitwood D. J. Phytochemical based strategies for nematode control. Annu. Rev. Phytopathol. 2002;40:221–249. doi: 10.1146/annurev.phyto.40.032602.130045. [DOI] [PubMed] [Google Scholar]
- 6.Bakhetia M., Charlton W. L., Urwin P. E., McPherson M. J., Atkinson H. J. RNA interference and plant parasitic nematodes. Trends Plant Sci. 2005;10:363–367. doi: 10.1016/j.tplants.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Jones A. K., Buckingham S. D., Sattelle D. B. Chemistry-to-gene screens in Caenorhabditis elegans. Nat. Rev. Drug Discov. 2005;4:321–330. doi: 10.1038/nrd1692. [DOI] [PubMed] [Google Scholar]
- 8.Burglin T. R., Lobos E., Blaxter M. L. Caenorhabditis elegans as a model for parasitic nematodes. Int. J. Parasitol. 1998;28:395–411. doi: 10.1016/s0020-7519(97)00208-7. [DOI] [PubMed] [Google Scholar]
- 9.McCarter J. P. Genomic filtering: an approach to discovering novel antiparasitics. Trends Parasitol. 2004;20:462–468. doi: 10.1016/j.pt.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Behm C. A., Bendig M. M., McCarter J. P., Sluder A. E. RNAi-based discovery and validation of new drug targets in filarial nematodes. Trends Parasitol. 2005;21:97–100. doi: 10.1016/j.pt.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Mitreva M., Appleton J., McCarter J. P., Jasmer D. P. Expressed sequence tags from life cycle stages of Trichinella spiralis: application to biology and parasite control. Vet. Parasitol. 2005;132:13–17. doi: 10.1016/j.vetpar.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 12.Lochnit G., Bongaarts R., Geyer R. Searching new targets for anthelminthic strategies: interference with glycosphingolipid biosynthesis and phosphorylcholine metabolism affects development of Caenorhabditis elegans. Int. J. Parasitol. 2005;35:911–923. doi: 10.1016/j.ijpara.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Palavalli L. H., Brendza K. M., Haakenson W., Cahoon R. E., McLaird M., Hicks L. M., McCarter J. P., Williams D. J., Hresko M. C., Jez J. M. Defining the role of phosphomethylethanolamine N-methyltransferase from Caenorhabditis elegans in phosphocholine biosynthesis by biochemical and kinetic analysis. Biochemistry. 2006;45:6056–6065. doi: 10.1021/bi060199d. [DOI] [PubMed] [Google Scholar]
- 14.Jez J. M. Phosphatidylcholine biosynthesis as a potential target for inhibition of metabolism in parasitic nematodes. Curr. Enzyme Inhib. 2007 in the press. [Google Scholar]
- 15.Kent C. Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochim. Biophys. Acta. 2005;1733:53–66. doi: 10.1016/j.bbalip.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Kent C. Eukaryotic phospholipid biosynthesis. Annu. Rev. Biochem. 1995;64:314–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- 17.Carman G. M., Henry S. A. Phospholipid biosynthesis in yeast. Annu. Rev. Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 18.Sohlenkamp C., Lopez-Lara I. M., Geiger O. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 2003;42:115–162. doi: 10.1016/s0163-7827(02)00050-4. [DOI] [PubMed] [Google Scholar]
- 19.Kanipes M. I., Henry S. A. The phospholipid methyltransferases in yeast. Biochim. Biophys. Acta. 1997;1348:138–141. doi: 10.1016/s0005-2760(97)00121-5. [DOI] [PubMed] [Google Scholar]
- 20.Vance D. E., Walkey C. J., Cui Z. Phosphatidylethanolamine N-methyltransferase from liver. Biochim. Biophys. Acta. 1997;1348:142–150. doi: 10.1016/s0005-2760(97)00108-2. [DOI] [PubMed] [Google Scholar]
- 21.Mudd S. H., Datko A. H. Phosphoethanolamine bases as intermediates in phosphatidylcholine synthesis by Lemna. Plant Physiol. 1986;82:126–135. doi: 10.1104/pp.82.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datko A. H., Mudd S. H. Phosphatidylcholine synthesis: differing patterns in soybean and carrot. Plant Physiol. 1988;88:854–861. doi: 10.1104/pp.88.3.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datko A. H., Mudd S. H. Enzymes of phosphatidylcholine synthesis in Lemna, soybean, and carrot. Plant Physiol. 1988;88:1338–1348. doi: 10.1104/pp.88.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolognese C. P., McGraw P. The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phospho-ethanolamine N-methyltransferase. Plant Physiol. 2000;124:1800–1813. doi: 10.1104/pp.124.4.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuccio M. L., Ziemak M. J., Henry S. A., Weretilnyk E. A., Hanson A. D. cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J. Biol. Chem. 2000;275:14095–14101. doi: 10.1074/jbc.275.19.14095. [DOI] [PubMed] [Google Scholar]
- 26.Charron J. B., Breton G., Danyluk J., Muzac I., Ibrahim R. K., Sarhan F. Molecular and biochemical characterization of a cold-regulated phosphoethanolamine N-methyltransferase from wheat. Plant Physiol. 2002;129:363–373. doi: 10.1104/pp.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pessi G., Kociubinski G., Mamoun C. B. A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6206–6211. doi: 10.1073/pnas.0307742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pessi G., Choi J. Y., Reynolds J. M., Voelker D. R., Mamoun C. B. In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J. Biol. Chem. 2005;280:12461–12464. doi: 10.1074/jbc.M414626200. [DOI] [PubMed] [Google Scholar]
- 29.Segal I. H. New York: John Wiley & Sons; 1975. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems. [Google Scholar]
- 30.Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 31.Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:research0002.1–research0002.10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabara H., Grishok A., Mello C. C. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 33.Golden J. W., Riddle D. L. A pheromone-induced developmental switch in Caenorhabditis elegans: temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc. Natl. Acad. Sci. U.S.A. 1984;81:819–823. doi: 10.1073/pnas.81.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagan R. M., Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- 35.Shields D. J., Altarejos J. Y., Wang X., Agellon L. B., Vance D. E. Molecular dissection of the S-adenosylmethionine-binding site of phosphatidylethanolamine N-methyltransferase. J. Biol. Chem. 2003;278:35826–35836. doi: 10.1074/jbc.M306308200. [DOI] [PubMed] [Google Scholar]
- 36.Kaletta T., Hengartner M. O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 37.Gee P., Kent C. Multiple isoforms of choline kinase from Caenorhabditis elegans: cloning, expression, purification, and characterization. Biochim. Biophys. Acta. 2003;1648:33–42. doi: 10.1016/s1570-9639(03)00106-7. [DOI] [PubMed] [Google Scholar]
- 38.Satouchi K., Hirano K., Sakaguchi M., Takehara H., Matsuura F. Phospholipids from the free-living nematode Caenorhabditis elegans. Lipids. 1993;28:837–840. doi: 10.1007/BF02536239. [DOI] [PubMed] [Google Scholar]
- 39.Gerdt S., Dennis R. D., Borgonie G., Schnabel R., Geyer R. Isolation, characterization and immunolocalization of phosphorylcholine-substituted glycolipids in developmental stages of Caenorhabditis elegans. Eur. J. Biochem. 1999;266:952–963. doi: 10.1046/j.1432-1327.1999.00937.x. [DOI] [PubMed] [Google Scholar]
- 40.Cipollo J. F., Awad A., Costello C. E., Robbins P. W., Hirschberg C. B. Biosynthesis in vitro of Caenorhabditis elegans phosphorylcholine oligosaccharides. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3404–3408. doi: 10.1073/pnas.0400384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lochnit G., Geyer R. Evidence for the presence of the Kennedy and Bremer–Greenberg pathways in Caenorhabditis elegans. Acta Biochim. Pol. 2003;50:1239–1243. [PubMed] [Google Scholar]
- 42.Friesen J. A., Liu M. F., Kent C. Cloning and characterization of a lipid-activated CTP:phosphocholine cytidylyltransferase from Caenorhabditis elegans: identification of a 21-residue segment critical for lipid activation. Biochim. Biophys. Acta. 2001;1533:86–98. doi: 10.1016/s1388-1981(01)00145-7. [DOI] [PubMed] [Google Scholar]
- 43.Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 44.Rudolph F. B. Product inhibition and abortive complex formation. Methods Enzymol. 1979;63:411–436. doi: 10.1016/0076-6879(79)63018-5. [DOI] [PubMed] [Google Scholar]
- 45.Zeisel S. H., Growdon J. H., Wurtman R. J., Magil S. G., Logue M. Normal plasma choline responses to ingested lecithin. Neurology. 1980;30:1226–1229. doi: 10.1212/wnl.30.11.1226. [DOI] [PubMed] [Google Scholar]
- 46.Zeisel S. H. Choline: needed for normal development of memory. J. Am. Coll. Nutr. 2000;19:528S–531S. doi: 10.1080/07315724.2000.10718976. [DOI] [PubMed] [Google Scholar]
- 47.Dawson R. M., Grime D. W., Lindsay D. B. On the insensitivity of sheep to the almost complete microbial destruction of dietary choline before alimentary-tract absorption. Biochem. J. 1981;196:499–504. doi: 10.1042/bj1960499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson B. S., Snoswell A. M., Runciman W. B., Upton R. N. Uptake and output of various forms of choline by organs of the conscious chronically catheterized sheep. Biochem. J. 1984;217:399–408. doi: 10.1042/bj2170399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson B. S., Snoswell A. M., Runciman W. B., Kuchel T. R. Choline biosynthesis in sheep: evidence for extrahepatic synthesis. Biochem. J. 1987;244:367–373. doi: 10.1042/bj2440367. [DOI] [PMC free article] [PubMed] [Google Scholar]