Abstract
Recent genetic knock-in and pharmacological approaches have suggested that, of class IA PI3Ks (phosphatidylinositol 3-kinases), it is the p110α isoform (PIK3CA) that plays the predominant role in insulin signalling. We have used isoform-selective inhibitors of class IA PI3K to dissect further the roles of individual p110 isoforms in insulin signalling. These include a p110α-specific inhibitor (PIK-75), a p110α-selective inhibitor (PI-103), a p110β-specific inhibitor (TGX-221) and a p110δ-specific inhibitor (IC87114). Although we find that p110α is necessary for insulin-stimulated phosphorylation of PKB (protein kinase B) in several cell lines, we find that this is not the case in HepG2 hepatoma cells. Inhibition of p110β or p110δ alone was also not sufficient to block insulin signalling to PKB in these cells, but, when added in combination with p110α inhibitors, they are able to significantly attenuate insulin signalling. Surprisingly, in J774.2 macrophage cells, insulin signalling to PKB was inhibited to a similar extent by inhibitors of p110α, p110β or p110δ. These results provide evidence that p110β and p110δ can play a role in insulin signalling and also provide the first evidence that there can be functional redundancy between p110 isoforms. Further, our results indicate that the degree of functional redundancy is linked to the relative levels of expression of each isoform in the target cells.
Keywords: AS252424, insulin signalling, phosphatidylinositol 3-kinase (PI3K), PIK3CA, PIK3CB, protein kinase B/Akt
Abbreviations: CHO-IR cells, Chinese-hamster ovary cells expressing human insulin receptor; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; GPCR, G-protein-coupled receptor; NCS, newborn calf serum; PI, phosphatidylinositol; PI3K, phosphatidylinositol 3-kinase; PKB, protein kinase B; SH2, Src homology 2
INTRODUCTION
PI3Ks (phosphatidylinositol 3-kinases) catalyse the phosphorylation of the D-3 position of the inositol headgroup of PI (phosphatidylinositol) leading to the synthesis of second messengers PtdIns3P, PtdIns(3,4)P2, PtdIns(3,5)P2 and PtdIns(3,4,5)P3 [1,2]. A large part of our understanding of how PI3K participates in cell signalling is based on the use of two structurally distinct cell-permeable inhibitors of PI3K, LY294002 [3] and wortmannin [4]. In the case of insulin signalling, use of these inhibitors has provided strong evidence that PI3K activity is necessary for a wide range of insulin's effects on cells [1,5]. However, the PI3K lipid kinase family comprises eight enzymes, divided into three classes (I, II and III) based on sequence homology comparisons. These isoforms of PI3K have distinct substrate specificities, expression profiles and modes of regulation [6,7]. Class I PI3K is subdivided into two subclasses, class IA and class IB. There is only one class IB PI3K (p110γ) and this operates downstream of heterotrimeric GPCRs (G-protein-coupled receptors). Class IA PI3Ks are heterodimers consisting of a catalytic subunit (p110) and a regulatory subunit. The regulatory subunit has no catalytic activity, but has two SH2 (Src homology 2) domains which facilitate interactions with tyrosine phosphorylation and allows for activation by receptor tyrosine kinases. It is because of this that class IA PI3K has been thought of as the main form stimulated by insulin. In mammals, there are three different genes producing catalytic subunits of class IA PI3K: p110α, p110β and p110δ [6,7]. The p110α and p110β isoforms are ubiquitously expressed, whereas the p110δ isoform is predominantly expressed in leucocytes [8]. As p110α and p110β are the main forms expressed in insulin target tissues, they have long been thought of as the two forms of PI3K most likely to be involved in insulin signalling.
The recent observations that amplifications or activating mutations in p110α are found in many tumour types [9–15], that p110β is involved in thrombosis [16] and that p110δ and p110γ are involved in inflammatory processes [17,18] has fuelled interest in the development of strategies to target specific classes of PI3K. However, such strategies could have harmful side effects on normal cellular function. This has focused renewed effort in defining the roles of different isoforms of PI3K in cell signalling pathways. As both LY294002 and wortmannin are broad-spectrum inhibitors of PI3K, they have not been particularly useful in determining which isoforms are involved in insulin signalling. Gene targeting studies in mice were initially of little value in addressing these issues as both the p110α- and p110β-knockouts are lethal [19–21]. However, heterozygous mice are viable, and glucose metabolism and insulin action have been studied in these animals [22]. Neither p110α+/− nor p110β+/− mice are insulin-resistant, but combined heterozygous deletion of both of these isoforms results in mice that are slightly glucose-intolerant [22]. It could be argued that this shows functional redundancy between these two isoforms in insulin signalling in vivo. However, it is difficult to interpret these findings as levels of the p85 adapter subunit change dramatically in these animals and this could also be influencing the insulin signalling phenotype [23,24]. More recently, a transgenic gene knock-in approach has been used to generate mice in which the p110α gene is mutated at a single residue to produce an allele that results in a catalytically inactive p110α. Mice homozygous for this defect are embryonic lethal, but heterozygous mice have reduced p110α activity, and experiments in these mice suggest that p110α is the most important form present in insulin signalling complexes and is required for signalling to downstream events [21]. However, countering this there are several studies indicating that p110β plays the most important role in insulin signalling [25–27]. The situation is confused further by the finding that shRNA (short hairpin RNA) knockdown of either p110α or p110β in CHO-IR cells (Chinese-hamster ovary cells expressing human insulin receptor) had no effect on insulin-induced activation of PKB (protein kinase B, also known as Akt) [28]. One interpretation of these results might be that each of the isoforms might be able to substitute for the other. Nonetheless, the general conclusion of all these studies has been that each combination of p85α/p110 is engaged differently by growth factor signalling pathways to achieve distinct signalling outcomes.
The methods described above all have various limitations, and it is clear that the use of appropriate pharmacological approaches could provide important new insights. Recently, a number of compounds have been reported that have the ability to selectively inhibit different PI3K isoforms. These include inhibitors of p110α [29,30], p110β [16,30], p110δ [31] and p110γ [32]. Use of these inhibitors has provided evidence that p110α is necessary for insulin signalling pathways [21,30]. However, these studies have only focused on a limited range of cell types to date, and it remains to be seen whether this is a universal requirement.
We have used a range of isoform-selective PI3K inhibitors to investigate further the role of individual isoforms of PI3K in insulin signalling. Our results indicate that p110α is necessary for insulin stimulation of PKB in CHO-IR cells and 3T3-L1 cells. However, we find that, in HepG2 cells and J774.2 macrophages, other isoform-specific class IA PI3K inhibitors have attenuating effects on insulin signalling. This provides strong evidence that these isoforms can participate in insulin signalling and that, in some cases, there can be functional redundancy between class IA PI3K isoforms in insulin signalling. Further, our data indicate that the ability of an isoform to participate in signalling and the degree of redundancy is linked to the relative level of expression of the different class IA catalytic subunits.
MATERIALS AND METHODS
Materials
Unless stated otherwise, reagents were purchased from Sigma Chemicals. The antibodies directed to phospho-Ser473 PKB and phospho-Thr308 PKB were from Cell Signaling Technologies. Polyclonal antibodies to p110α, p110β and p110δ were kindly provided by Dr Bart Vanhaesebroeck, Ludwig Institute for Cancer Research, London, U.K. Polyclonal antibodies to p85α were as described previously [33]. Recombinant p85α/p110δ was purchased from Upstate Biotechnologies.
Production of recombinant PI3K
To produce other class IA PI3Ks, Sf21 insect cells were co-infected with baculovirus expressing N-terminal His-tagged human p85α and either wild-type murine p110α or wild-type human p110β. To produce class IB PI3K, Sf21 insect cells were infected with baculovirus expressing N-terminal His-tagged bovine p110γ. The PI3Ks were purified using an Ni-NTA (Ni2+-nitrilotriacetate) superflow (Qiagen) affinity column. The purity of the PI3K preparations was verified by Coomassie Blue staining of SDS/PAGE gels and the titres of baculovirus were adjusted such that the p85/p110 ratio was approx. 1:1 for the class IA PI3Ks. The functional authenticity of multiple preparations of the recombinant PI3Ks was verified by Western blotting and also by sensitivity to previously described isoform-selective PI3K inhibitors (Table 1).
Table 1. IC50 values for selected PI3K inhibitors against lipid kinase activity.
All IC50 values (in nM) were determined using the PI3K lipid kinase assays on multiple preparations of recombinant protein as described in the Materials and methods section. IC50 values for AS252424 have been reported previously [32]. Results are means±S.D. n≥3 for all determinations.
| Inhibitor | p110α | p110β | p110δ |
|---|---|---|---|
| PIK-75 | 7.8±1.7 | 343±23 | 907±32 |
| PI-103 | 3.7±0.5 | 18.2±1.8 | >500 |
| SN 30693 | 231±22 | 667±32 | 401±21 |
| TGX-221 | >1000 | 8.5±0.9 | 211±18 |
| IC87114 | >1000 | >1000 | 60.2±5.6 |
| AS252424 | 935 | >5000 | >5000 |
| Wortmannin | 0.57 | 2.33 | 0.40 |
| LY294002 | 500 | 973 | 570 |
Synthesis and biochemical characterization of PI3K inhibitors
PI3K inhibitors were synthesized following general procedures described as follows: PI-103 [34], PIK-75 [35], IC87114 [36], SN 30693 [37] and AS252424 [32]. TGX-221 was prepared as described in [38], with the single modification being the use of bis-2,4,6-trichlorophenyl malonate, instead of malonyl dichloride, in the first step. The bis-2,4,6-trichlorophenyl malonate was prepared and used according to the procedure published in [16].
IC50 values were measured using a standard lipid kinase activity with PI as a substrate, basically as described previously [39]. The differences were (i) that 100 μM cold ATP was used instead of 10 μM, (ii) the DMSO concentration was 1% rather than 2%, and (iii) [γ-33P]ATP (GE Healthcare) was used instead of [γ-32P]ATP. The TLC plates were quantified using a phosphorimager screen (StormImager, Amersham). The reported IC50 values were determined by non-linear regression analysis (GraphPad Prism software) on the basis of at least three independent experiments repeated across multiple preparations of recombinant protein.
Cell culture
HepG2 cells were grown in DMEM (Dulbecco's modified Eagle's medium) (Invitrogen) supplemented with 10% (v/v) NCS (newborn calf serum), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). CHO-IR cells were grown in Ham's F12 (Invitrogen) supplemented with 10% (v/v) NCS, 100 units/ml penicillin and 100 μg/ml streptomycin. 3T3-L1 fibroblasts were grown in DMEM supplemented with 10% (v/v) FBS (fetal bovine serum) (Invitrogen), 100 units/ml penicillin and 100 μg/ml streptomycin. J774.2 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) NCS, 100 units/ml penicillin and 100 μg/ml streptomycin.
Differentiation of 3T3-L1 cells
3T3-L1 fibroblasts were obtained from the American Type Culture Collection (A.T.C.C., Manassas, VA, U.S.A.). Cells were allowed to reach confluence and were induced to differentiate as described previously [40]. Briefly, at 2 days post-confluence (day 0), differentiation was initiated by the addition of 100 nM insulin, 1 μM dexamethasone and 0.25 mM IBMX (isobutylmethylxanthine) in DMEM with 10% (v/v) FBS. After 3 days (day 3), the induction medium was replaced by DMEM supplemented with 10% (v/v) FBS and 100 nM insulin only, and was replaced every 2 days by DMEM with 10% (v/v) FBS. Cells were seeded in six-well plates for Western blotting and glucose-uptake experiments and were starved in serum-free medium with 0.2% (w/v) BSA (ICP Bio), 100 units/ml penicillin and 100 μg/ml streptomycin, 16 h before stimulation.
Protein isolation, immunoprecipitation and immunoblotting
Cells were washed twice with ice-cold PBS (140 mM NaCl, 3 mM KCl, 6 mM Na2HPO4, 1 mM KH2PO4, pH 7.4) and were solubilized with lysis buffer {20 mM Tris/HCl, 138 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5% (v/v) glycerol, 1% (v/v) Nonidet P40, 5 mM EDTA, 20 μM leupeptin, 18 μM pepstatin, 1 mM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride], 4 μg/ml aprotinin, 2 mM Na3VO4, 20 mM NaF and 1 mM DTT (dithiothreitol), pH7.4}. Lysates were kept on ice for 20 min, and insoluble material was removed by centrifugation at 14000 g for 10 min. Protein concentration was determined by colorimetric assay [BCA (bicinchoninic acid); Pierce].
Proteins were separated by SDS/PAGE and transferred on to PVDF membranes (Pall Corporation). The membranes were incubated for 1 h in blocking buffer [20 mM Tris/HCl, pH 7.4, 137 mM NaCl and 0.5% (v/v) Tween 20] containing 3% (w/v) BSA or non-fat dried milk powder and were then incubated overnight in blocking buffer containing antibodies. Immunoreactive proteins were detected using horseradish-peroxidase-linked secondary antibodies (Dako) and ECL® (enhanced chemiluminescence) according to the manufacturer's instructions (GE Healthcare). Signals were analysed and quantified using a Fuji FLA-3000 phosphorimager and Fuji Image Gauge software.
For immunoprecipitation, lysates were submitted to pre-clearing by incubation at 4 °C for 30 min with Protein A–Sepharose. Polyclonal antibodies to the N-SH2 domain of p85α were pre-incubated with Protein A–Sepharose before the addition of cleared lysates and incubation overnight at 4 °C. Immune complexes were washed twice with lysis buffer and then solubilized in 1× Laemmli sample buffer.
Statistical analysis
Results are presented as means±S.E.M. with the number of experiments indicated in the legend. Statistical significance was assessed using one-way ANOVA and Dunnett's multiple comparison test.
RESULTS
Characterization of isoform-specific PI3K inhibitors
Class IA isoform-specific inhibitors (Figure 1) were synthesized as described in the Materials and methods section, and their activity against the different isoforms was measured in an in vitro PI3K assay using multiple preparations of recombinant p85/p110 (Table 1). This is the first report of the selectivity of the PIramed compound SN 30693 and we found that it is a broad-spectrum PI3K inhibitor, but it has some selectivity for p110α. Our results are broadly in agreement with previous studies that found that PIK-75 and PI-103 are selective inhibitors of p110α [30], that TGX-221 is selective for p110β [16] and that IC87114 is selective for p110δ [30,31]. However, it is worth noting that our results diverge slightly from those of Knight et al. [30] in terms of absolute IC50 values for PI-103 and PIK-75, particularly in the relative sensitivities of p110β and p110δ. The reason for this is not clear, but could relate to slight differences in assay methodologies or in the source of enzyme. For example, we used 100 μM ATP, whereas the study of Knight et al. [30] used 10 μM ATP.
Figure 1. Structures of the selected PI3K inhibitors.
p110α is the major PI3K isoform responsible for insulin signalling in CHO-IR and 3T3-L1 cells
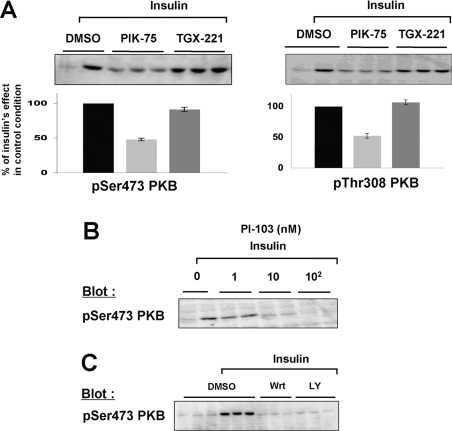
CHO-IR cells have been shown to possess 105 insulin receptors per cell [41,42] and are consequently extremely sensitive to insulin stimulation. In our hands, 1 nM insulin induces 50% of the maximal PKB phosphorylation on both sites (results not shown). Using this limiting dose of insulin (1 nM), we found that the p110α-specific inhibitor PIK-75 blocked the phosphorylation of PKB induced by insulin on both Ser473 and Thr308 in CHO-IR cells (Figure 2A) in a dose-dependent manner (Figure 2B), with an IC50 of 78 nM (Figure 2C). The phosphorylation of PKB Ser473 was also blocked using a second, structurally unrelated, inhibitor selective for p110α (PI-103) (Figure 2D). As a control, wortmannin (100 nM) and LY294002 (5 μM) were also shown to block insulin-induced phosphorylation of PKB Ser473 in CHO-IR cells (Figure 2E). In contrast, the inhibitor of p110β (TGX-221) was not able to inhibit PKB phosphorylation, even when used at high concentrations (Figures 2A and 2B). Similar results were obtained using 0.1, 10 or 100 nM insulin (results not shown).
Figure 2. Effect of isoform-specific inhibitors on insulin-induced phosphorylation of PKB in CHO-IR cells.
Overnight-starved CHO-IR cells were incubated for 5 min with the indicated PI3K inhibitors or DMSO and stimulated or not with insulin (1 nM, 10 min). Whole-cell lysates were then analysed by Western blotting using specific antibodies. (A) Effect of p110α-specific inhibitor (PIK-75, 100 nM) and p110β-specific inhibitor (TGX-221, 100 nM) on the insulin-induced stimulation of PKB phosphorylation on Ser473 (left) and Thr308 (right). A representative Western blot is shown in each of the upper panels and quantification is presented below. The relative stimulation of insulin over the basal level was taken as 100% and all values are calculated as a percentage of this. The results are means±S.E.M. for three independent experiments each performed in triplicate. (B) Dose–response effect of p110α-specific inhibitor (PIK-75) and p110β-specific inhibitor (TGX-221) on the insulin-induced stimulation of PKB (1 nM, 10 min). (C) Sigmoidal concentration–response curve of PKB phosphorylation obtained in the presence of increasing concentrations of PIK-75. (D) Dose–response effect of another structurally unrelated p110α-selective inhibitor (PI-103) on the insulin-induced stimulation of PKB phosphorylation (100 nM insulin, 10 min). (E) Effect of wortmannin (Wrt; 100 nM) and LY294002 (LY; 5 μM) on the insulin-stimulated phosphorylation of PKB (100 nM insulin, 10 min). These broadly active PI3K inhibitors were used as a control.
Knight et al. [30] have shown previously that p110α is required for insulin signalling to PKB in 3T3-L1 adipocytes, a cell model that possesses a number of insulin receptors intermediate between CHO and CHO-IR [43]. We sought to extend these findings and to determine whether this function is acquired during the differentiation process. We found that, in 3T3-L1 pre-adipocytes, inhibition of p110α by PIK-75 decreases the insulin-induced phosphorylation of PKB on both sites (Ser473 and Thr308) (Figure 3A). PI-103 had a very similar effect (Figure 3B). On the other hand, the p110β inhibitor TGX-221 had no effect on PKB phosphorylation status (Figure 3A), while wortmannin and LY294002 completely inhibited the phosphorylation of PKB (Figure 3C). In fully differentiated 3T3-L1 adipocytes, a similar pattern was observed with inhibitors of p110α, but not of p110β, decreasing the insulin-induced phosphorylation of PKB on both Ser473 and Thr308 (Figure 4).
Figure 3. Effect of isoform-specific inhibitors on insulin signalling in 3T3-L1 fibroblasts.
Overnight-starved 3T3-L1 fibroblasts were incubated for 5 min with the indicated PI3K inhibitors or DMSO and stimulated or not with insulin (100 nM, 10 min). Whole-cell lysates were then analysed by Western blotting using specific antibodies. (A) Effect of p110α-specific inhibitor (PIK-75, 100 nM) and p110β-specific inhibitor (TGX-221, 100 nM) on the insulin-induced stimulation of PKB phosphorylation. A representative Western blot is shown in each of the upper panels and quantification is presented below. The relative stimulation of insulin over the basal level was taken as 100% and all values are calculated as a percentage of this. The results are means±S.E.M. for three independent experiments each performed in triplicate. (B) Dose–response effect of another structurally unrelated p110α-selective inhibitor (PI-103) on the insulin-induced stimulation of PKB. (C) Effect of wortmannin (Wrt; 100 nM) and LY294002 (LY; 5 μM) on the phosphorylation of PKB. These broadly active PI3K inhibitors were used as a control.
Figure 4. Effect of isoform-specific inhibitors on insulin signalling in 3T3-L1 adipocytes.
Overnight-starved 3T3-L1 adipocytes were incubated for 5 min with the indicated PI3K inhibitors or DMSO and stimulated or not with insulin (100 nM, 10 min). Whole-cell lysates were then analysed by Western blotting using specific antibodies. (A) Effect of p110α-specific inhibitor (PIK-75, 100 nM) and p110β-specific inhibitor (TGX-221, 100 nM) on the insulin-induced stimulation of PKB phosphorylation. A representative Western blot is shown in each of the upper panels and quantification is presented below. The relative stimulation of insulin over the basal level was taken as 100% and all values are calculated as a percentage of this. The results are means±S.E.M. for three independent experiments each performed in triplicate. (B) Dose–response effect of two other structurally unrelated p110α-selective inhibitors (PI-103 and SN 30693) on the insulin-induced stimulation of PKB phosphorylation.
In HepG2 cells, p110α is required but is not sufficient to mediate insulin signalling
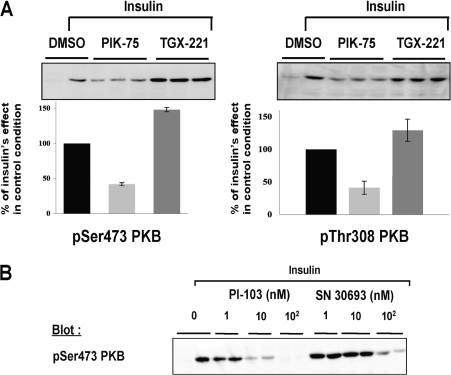
In contrast with what was seen in 3T3-L1 and CHO-IR cells, in the human hepatoma HepG2 cell line, none of the p110α selective inhibitors was able to diminish the insulin-induced phosphorylation of PKB (Figures 5A and 5B). The same was true even when the exposure time to inhibitors before insulin stimulation was increased to 30 min (results not shown). As a control, 100 nM wortmannin and 5 μM LY294002 completely inhibited the phosphorylation of PKB (Figure 5C), demonstrating that simultaneous inhibition of all PI3K isoforms does block insulin signalling in these cells. Therefore we investigated the effect of inhibitors specific for p110β, p110δ and p110γ on insulin-induced phosphorylation of PKB. However, these inhibitors alone were also not able to diminish the effects of insulin-induced phosphorylation on PKB (Figures 5A and 5C).
Figure 5. Effect of isoform-specific inhibitors on insulin signalling in HepG2 cells.
Overnight-starved human heptoma cells (HepG2) were incubated for 5 min with the indicated PI3K inhibitors or DMSO and stimulated or not with insulin (100 nM, 10 min). Whole-cell lysates were then analysed by Western blotting using specific antibodies. (A) Dose–response effect of p110α-specific inhibitor (PIK-75) and p110β-specific inhibitor (TGX-221) on the insulin-induced stimulation of PKB phosphorylation. (B) Dose–response effect of two other structurally unrelated p110α-selective inhibitors (PI-103 and PI-SN 30693) on the insulin-induced stimulation of PKB phosphorylation. (C) Dose–response effect of p110δ-specific inhibitor (IC87114) and p110γ-specific inhibitor (AS 252424) on the insulin-induced stimulation of PKB phosphorylation. Wortmannin (Wrt) and LY294002 (Ly) are shown as controls. (D) Effect of inhibitors in combination on the insulin-induced stimulation of PKB phosphorylation. A representative Western blot is shown in the upper panel and quantification is presented below. The relative stimulation of insulin over the basal level was taken as 100% and all values are calculated as a percentage of this. The results are means±S.E.M. for three independent experiments each performed in triplicate. ***P<0.001; n.s, not significant.
Given the effect of wortmannin and LY294002, we next investigated whether there might be some degree of redundancy in the participation of PI3K isoforms in insulin signalling to PKB in these cells. To perform these experiments, we used combinations of inhibitors at concentrations at which they were specifically inhibiting their target isoform (Figure 5D). Inhibition of either p110α/p110β or p110α/p110δ reduced the phosphorylation of PKB to near basal, whereas inhibition of p110β/p110δ did not (Figure 5D).
In J774.2 cells, all class IA PI3K isoforms can mediate insulin signalling
In J774.2 macrophage cells, insulin strongly increases the phosphorylation of PKB and this is completely abolished by 100 nM wortmannin, indicating that PI3K is required for this effect (Figure 6). In these cells, PIK-75, TGX-221 and IC87114 all partially attenuate insulin-induced phosphorylation of Ser473 of PKB, indicating that p110α, p110β and p110δ all contribute to insulin signalling in these cells and all three are required for insulin to be fully effective.
Figure 6. Effect of isoform-specific inhibitors on insulin signalling in J774.2 cells.
Overnight-starved J774.2 cells were incubated for 5 min with wortmannin (wrt; 100 nM), PIK-75 (50 nM), TGX-221 (50 nM), IC87114 (100 nM) or DMSO and stimulated or not with insulin (100 nM, 10 min). Whole-cell lysates were then analysed by Western blotting using antibodies recognizing phospho-Ser473 (pS473) of PKB. A representative Western blot is shown in the upper panel and quantification is presented below. The relative stimulation of insulin over the basal level was taken as 100% and all values are calculated as a percentage of this. Results are means±S.E.M. for three independent experiments each performed in duplicate. ***P<0.001.
Isoform-dependence is related to the relative level of expression and activity of each isoform
The variability in dependence on different class IA isoforms could be due to differences in levels of expression within different cell types. We used two methods to investigate this. The first was Western blotting which allows a comparison of the relative expression levels of a given isoform between different cell types (Figure 7A). This revealed some interesting findings. The first was that J774.2 cells express high levels of p110β and p110δ. HepG2 cells also express readily detectable levels of p110δ. This probably explains why p110δ inhibitors only attenuate insulin signalling in HepG2 and J774.2 cells. Another observation from these studies was that the cell types in which p110α-selective inhibitors completely blocked insulin signalling were the cell types which expressed relatively high levels of p110α and relatively lower levels of p110β and p110δ (Figure 7A, lanes 2–4).
Figure 7. Comparison of the relative levels of expression and activity of different class IA PI3K isoforms.
(A) Western blotting using isoform-specific antibodies was used to compare the levels of expression of class IA p110 isoforms in different cell types: lane 1, J774.2; lane 2, 3T3-L1 fibroblasts; lane 3, 3T3-L1 adipocytes; lane 4, CHO-IR; lane 5, HepG2. For p110α detection, total class IA activity was immunoprecipitated from 200 μg of lysate using an antibody recognizing the p85α adapter subunit, and recombinant protein was loaded as a control (0.2 μg, lane marked +). For p110β and p110δ detection, 20 μg of total cell lysate was loaded in each case. (B–E) Total class IA activity was immunoprecipitated from 20 μg of lysate from the indicated cell type using an antibody recognizing the p85α adapter subunit. PI3K activity was then assayed in the presence of 100 nM of the indicated inhibitor. Cell types were J774.2 (B), 3T3-L1 fibroblasts (C), 3T3-L1 adipocytes (D), CHO-IR (E) and HepG2 (F). Results are expressed relative to the activity in the presence of DMSO and are means±S.E.M. for three independent determinations. WORT, wortmannin.
The different antibody reactivities mean that Western blotting does not allow a direct comparison of the relative levels of each isoform within a given cell type. Therefore we used a second method in which we immunoprecipitated all class IA PI3K activity using a p85α antibody. We then used isoform-selective PI3K inhibitors to assess the relative amount of the total activity that was attributable to any particular isoform (Figures 7B–7E). The other p110α-selective inhibitor, PI-103, gave very similar results to those obtained using PIK-75 (results not shown). One finding from these studies was that there was no significant p110β activity detectable in 3T3-L1 fibroblasts (Figure 7B), but p110β became a small component of overall class IA PI3K activity after differentiation into adipocytes (Figure 7C). This provides functional evidence to support the previous Western blotting studies of Asano et al. [27], who demonstrated an increase in p110β expression during differentiation. However, the overall amount of p110β activity remains much less than that of p110α. Following from this, p110α activity accounted for a majority of class IA activity in all the cell types where p110α-selective inhibitors preferentially block insulin signalling (Figures 7C–7E). Similarly, the cell types in which p110β and p110δ inhibitors are effective possess the largest amount of activity of these isoforms. Together with the Western blotting data, these findings suggest that the total amount of a given isoform present in a cell type is an important determinant of whether that isoform plays a role in insulin signalling.
DISCUSSION
Previous studies using the broad-specificity PI3K inhibitors wortmannin and LY294002 provided strong evidence that PI3K activity is essential for almost all of the effects of insulin [1,5]. However, the role of the different isoforms of PI3K has not been investigated extensively, and the results have been somewhat conflicting. Initial studies in 3T3-L1 adipocytes suggested that p110β was more important than p110α in insulin signalling [27]. These conclusions were based on three major lines of evidence: (i) p110β levels greatly increased during differentiation of 3T3-L1 cells into insulin-sensitive adipocytes, whereas p110α activity levels remained unchanged; (ii) p110β was increased by insulin stimulation, whereas p110α activity was not; and (iii) microinjection of neutralizing antibodies targeting p110β blocked insulin-stimulated GLUT4 translocation, whereas p110α antibodies did not. The latter finding was taken as direct evidence that p110β played a major role in insulin signalling. However, these findings have been challenged by two different studies which indicate that p110α is necessary for insulin signalling, whereas p110β is not. One of these studies used knock-in mice, which were heterozygous for a kinase-dead form of p110α [21]. These mice had defects in glucose metabolism and insulin signalling, implying an important role for p110α in insulin action. The second study utilized isoform-selective inhibitors of PI3K [30]. In that study, isoform-selective pharmacological inhibitors of p110α blocked a range of insulin's actions in vitro and in vivo, whereas p110β inhibitors were without effect.
The present study uses a range of structurally distinct isoform-specific inhibitors of class IA PI3Ks to extend the investigations of the role of different PI3K isoforms in insulin signalling in a range of cell types. Our studies using PI-103 and PIK-75 extend the range of cell types in which the role of p110α in insulin signalling is studied. These have both previously been shown to be highly potent p110α-selective inhibitors and their potential off-target activities have also been investigated extensively [30]. These studies show that they have very different patterns of off-target activity. This means that using these in combination gives a high degree of confidence that the effects being seen are due to p110α. Also, our studies extend the previous work [30] by adding biological data on a novel PI3K inhibitor, SN 30693, which we show has some selectivity for p110α.
Further, the studies of Knight et al. [30] used two compounds that they described as p110β/p110δ inhibitors (TGX-115 and TGX-286), but that had some selectivity for p110β. They found that these compounds did not have a significant effect on insulin action in the cell types they tested and concluded that p110β was not important for insulin signalling. To test more extensively the involvement of p110β in insulin signalling, we have used an alternative compound, TGX-221 [16], as this is a more selective and potent inhibitor of p110β. Using this compound, we have provided further lines of evidence that p110β activity is not in fact necessary for insulin signalling in CHO-IR and 3T3-L1 cells.
The major finding of our studies is that p110α is not necessary for insulin signalling in all cell types and that p110β and p110δ can participate in some cell types. Importantly, we have demonstrated that none of the p110α inhibitors blocks insulin signalling to PKB in HepG2 hepatoma cells. To our knowledge, this is the first example of p110α inhibitors having no effect on growth factor signalling. The findings in the hepatoma cells may have functional correlates in hepatocytes as there is some evidence that insulin does not rely on p110α activity as much in liver as in other tissues. This comes from heterozygous p110α knock-in mice, where insulin signalling to PKB is relatively normal in liver but is severely impaired in muscle and fat [21].
Our initial hypothesis was that one of the other class IA isoforms would take the place of p110α, but this was not the case in HepG2 cells, as inhibitors of p110β and p110δ also had no effect on insulin's stimulation of PKB. However, p110α inhibitors in combination with either p110β or p110δ inhibitors were able to block this signalling to PKB, which provides evidence that functional redundancy between p110 isoforms exists in these cells and that when one isoform is suppressed another can at least in part compensate. However, the results imply that p110α is a necessary component in this mixture, as a combination of p110β and p110δ inhibitors had no effect. J774.2 cells are another cell type where p110α is not the only class IA isoform involved in insulin signalling, but in these cells, all three isoforms can play a role.
How do these different patterns of dependence on particular PI3K isoforms arise? One explanation could lie in the fact that the isoform-dependence correlates with the levels of expression and activity of p110α, p110β and p110δ in particular cells. For example, cells where p110α inhibitors largely attenuate insulin signalling (i.e. CHO-IR, 3T3-L1 fibroblasts and adipocytes) all have a very high relative level of p110α compared with other isoforms. This contrasts with HepG2 cells and J774.2 cells, which express significant levels of p110β and p110δ. Therefore the most likely explanation for our results is that the ability to participate in insulin signalling is linked to the level of activity of a given isoform in the cell.
In support of this, it was observed that, in tissues of knock-in mice where p110α was found to be the predominant isoform in insulin signalling, the activity attributed to p110α was very high and that attributed to the other isoforms was very low [21]. This concept is also supported by previous studies which show that overexpression of either p110β or p110δ can change the response of cells to serum, indicating that the level of the enzyme present in the cell is a critical factor in the ability to participate in growth factor signalling pathways [44]. It is, however, important to note that the degree of expression of a given isoform will be tempered by the kinetic properties of that isoform [39]. For example, the specific activity of p110α towards its physiological substrate PtdIns(4,5)P2 is more than five times higher than those of p110β or p110δ [45]. Therefore it is likely that p110β levels would need to increase significantly more than p110α to achieve the same effect. This may explain why the small observed increase in p110β levels as 3T3-L1 cells differentiate into adipocytes does not result in p110β playing an important functional role in insulin signalling in these cells.
This then raises the question of why cells might need to express different levels of the different class IA PI3K isoforms. We speculate that the purpose of this is not so much to alter the direct growth-factor-mediated engagement and activation of PI3K, as to allow different linkages to occur with other signalling pathways, most likely via differences in the p110 isoforms themselves. For example, there is strong evidence that p110β (but not p110α or p110δ) can also be activated by GPCRs [28,46]. Thus the presence of significant levels of p110β in a cell may allow synergies between growth factor and GPCR signalling. It has also been observed that p110β has different requirements for Ras activation than either p110α or p110δ [44], indicating that the presence of p110β might allow different connections between growth factor and Ras signalling pathways. Further, it has been proposed that differences in catalytic properties between the isoforms could result in activation of different effector pathways [39].
In summary, our results demonstrate that all three class IA PI3K isoforms can participate in insulin-induced activation of PKB and suggest that the levels of expression and activity are key factors in determining the extent of a particular isoform's involvement. These findings suggest that specificity in class IA PI3K signalling does not arise from differences in the ability of different p85/p110 combinations to couple to upstream tyrosine phosphorylated proteins. We suggest that specificity instead could arise as a result of the ability of different p110 subunits to interact with different signalling pathways.
Acknowledgments
Funding was provided by the Auckland Medical Research Foundation, the Maurice Wilkins Centre for Molecular Biodiscovery, the Auckland Cancer Society Research Centre and the Health Research Council of New Zealand (Project Grant 06/062A). We thank Dr Phill Hawkins (The Babraham Institute, Cambridge, U.K.) and Dr Osamu Hazeki (Graduate School of Pharmaceutical Sciences, The University of Tokyo, Tokyo, Japan) for providing reagents. We thank Dr Cenk Suphioglu (Department of Allergy and Clinical Immunology, Monash University) for assistance in preparing recombinant PI3Ks.
References
- 1.Shepherd P. R., Withers D. J., Siddle K. Phosphoinositide 3-kinase: the key switch mechanism in insulin signalling. Biochem. J. 1998;333:471–490. doi: 10.1042/bj3330471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B., Leevers S., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Vlahos C. J., Matter W. F., Hui K. Y., Brown R. F. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morphinyl)-8-phenyl-4H-1-benzopyranol-one (LY294002) J. Biol. Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 4.Arcaro A., Wymann M. P. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidyl 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 1993;296:297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepherd P. R. Mechanisms regulating phosphoinositide 3-kinase signalling in insulin sensitive tissues. Acta Physiol. Scand. 2005;183:3–12. doi: 10.1111/j.1365-201X.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- 6.Vanhaesebroeck B., Waterfield M. D. Signaling by distinct classes of phosphoinositide 3-kinases. Exp. Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 7.Wyman M. P., Pirola L. Structure and function of phosphoinositide 3-kinase. Biochem. Biophys. Acta. 1998;1436:127–150. doi: 10.1016/s0005-2760(98)00139-8. [DOI] [PubMed] [Google Scholar]
- 8.Vanhaesebroeck B., Welham M. J., Kotani K., Stein R., Warne P. H., Zvelebil M. J., Higashi K., Volinia S., Downward J., Waterfield M. D. p110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shayesteh L., Lu Y., Kuo W., Baldocchi R., Godfrey T., Collins C., Pinkel D., Powell B., Mills G. B., Gray J. W. PI3KCA is implicated as an oncogene in ovarian cancer. Nat. Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 10.Levine D. A., Bogomolniy F., Yee C. J., Lash A., Barakat R. R., Borgen P. I., Boyd J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin. Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 11.Velho S., Oliveira C., Ferreira A., Ferreira A. C., Suriano G., Schwartz S., Duval A., Carneiro F., Machado J. C., Hamelin R., Seruca R. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur. J. Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 12.Campbell I. G., Russell S. E., Choong D. Y. H., Montgomery K. G., Ciavarella M. L., Hooi C. S. F., Cristiano B. E., Pearson R. B., Phillips W. A. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 13.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S. M., Riggins G. J., et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 14.Bachman K. E., Argani P., Samuels Y., Silliman N., Ptak J., Szabo S., Konishi H., Karakas B., Blair B. G., Lin C., et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Can. Biol. Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 15.Wu G., Mambo E., Guo Z., Hu S., Huang X., Gollin S. M., Trink B., Ladenson P. W., Sidransky D., Xing M. Uncommon mutation but common amplification of the PIK3A gene in thyroid tumours. J. Clin. Endocr. Metab. 2005;90:4688–4693. doi: 10.1210/jc.2004-2281. [DOI] [PubMed] [Google Scholar]
- 16.Jackson S. P., Schoenwaelder S. M., Goncalves I., Nesbitt W. S., Yap C. L., Wright C. E., Kenche V., Anderson K. E., Dopheide S. M., Yuan Y., et al. PI 3-kinase p110β: a new target for antithrombotic therapy. Nat. Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 17.Ruckle T., Schwarz M. K., Rommel C. PI3Kγ inhibition: towards an ‘aspirin of the 21st century’? Nat. Rev. Drug Discov. 2006;5:903–918. doi: 10.1038/nrd2145. [DOI] [PubMed] [Google Scholar]
- 18.Okkenhaug K., Bilancio A., Farjot G., Priddle H., Sancho S., Peskett E., Pearce W., Meek S. E., Salpekar A., Waterfield M. D., et al. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 19.Bi L., Okabe I., Bernard D. J., Nussbaum R. L. Early embryonic lethality in mice deficient in the p110β catalytic subunit of PI 3-kinase. Mamm. Genome. 2002;13:169–172. doi: 10.1007/BF02684023. [DOI] [PubMed] [Google Scholar]
- 20.Bi L., Okabi I., Bernard D. J., Wynshaw-Boris A., Nussbaum R. L. Proliferative defects and embryonic lethality in mice homozygous for a deletion in the p110α subunit of phosphoinositide 3-kinase. J. Biol. Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- 21.Foukas L. C., Claret M., Pearce W., Okkenhaug K., Meek S., Peskitt E., Sancho S., Smith A. J. H., Withers D. J., Vanhaesebroeck B. Critical role for the p110α phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 22.Brachman S. M., Ueki K., Engelman J. A., Kahn C. R., Cantley L. C. Phosphoinositide 3-kinase catalytic subunit deletion and regulatory subunit have opposite effects on insulin sensitivity in mice. Mol. Cell. Biol. 2005;25:1596–1607. doi: 10.1128/MCB.25.5.1596-1607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueki K., Fruman D. A., Yballe C. M., Fasshauer M., Klein J., Asano T., Cantley L. C., Kahn C. R. Positive and negative roles of p85α and p85β regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J. Biol. Chem. 2003;278:48453–48466. doi: 10.1074/jbc.M305602200. [DOI] [PubMed] [Google Scholar]
- 24.Terauchi Y., Tsuji Y., Satoh S., Minoura H., Murakami K., Okuno A., Inukai K., Asano T., Kaburagi Y., Ueki K., et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85α subunit of phosphoinositide 3-kinase. Nat. Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 25.Roche S., Downward J., Raynal P., Courtneidge S. A. A function for phosphatidylinositol 3-kinase β (p85α-p110β) in fibroblasts during mitogenesis: requirement for insulin- and lysophosphatidic acid-mediated signal transduction. Mol. Cell. Biol. 1998;18:7119–7129. doi: 10.1128/mcb.18.12.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooshmand-Rad R., Hajkova L., Klint P., Karlsson R., Vanhaesebroeck B., Claesson-Welsh L., Heldin C. H. The PI 3-kinase isoforms p110α and p110β have differential roles in PDGF- and insulin-mediated signaling. J. Cell Sci. 2000;113:207–214. doi: 10.1242/jcs.113.2.207. [DOI] [PubMed] [Google Scholar]
- 27.Asano T., Kanda A., Katagiri H., Nawano M., Ogihara T., Inukai K., Anai M., Fukushima Y., Yazaki Y., Kikuchi M., et al. p110β is up-regulated during differentiation of 3T3-L1 cells and contributes to the highly insulin-responsive glucose transport activity. J. Biol. Chem. 2000;275:17671–17676. doi: 10.1074/jbc.M910391199. [DOI] [PubMed] [Google Scholar]
- 28.Kubo H., Hazeki K., Takasuga S., Hazeki O. Specific role for p85/p110β in GTP-binding-protein-mediated activation of Akt. Biochem. J. 2005;392:607–614. doi: 10.1042/BJ20050671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayakawa M., Kaizawa H., Moritomo H., Koizumi T., Ohishi T., Okada M., Ohta M., Tsukamoto S., Parket P., Workman P., Waterfield M. D. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3-kinase p110α inhibitors. Bioorgan. Med. Chem. 2006;14:6847–6858. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 30.Knight Z. A., Gonzalez B., Feldman M. E., Zunder E. R., Goldenberg D. D., Williams O., Loewith R., Stokoe D., Balla A., Toth B., et al. A pharmacological map of the PI3-K family defines a role for p110α in insulin signalling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadhu C., Masinovsky B., Dick K., Sowell C. G., Staunton D. E. Essential role of phosphoinositide 3-kinase δ in neutrophil directional movement. J. Immunol. 2003;170:2647–2654. doi: 10.4049/jimmunol.170.5.2647. [DOI] [PubMed] [Google Scholar]
- 32.Pomel V., Klicic J., Covini D., Church D. D., Shaw J. P., Roulin K., Burgat-Charvillon F., Valognes D., Camps M., Chabert C., et al. Furan-2-ylmethylene thiazolidinediones as novel, potent, and selective inhibitors of phosphoinositide 3-kinase γ. J. Med. Chem. 2006;49:3857–3871. doi: 10.1021/jm0601598. [DOI] [PubMed] [Google Scholar]
- 33.Shepherd P. R., Navé B. T., Rincon J., Nolte L. A., Bevan A. P., Siddle K., Zierath J. R., Wallberg-Henriksson H. Differential regulation of phosphoinositide 3-kinase adapter subunit variants by insulin in human skeletal muscle. J. Biol. Chem. 1997;272:19000–19007. doi: 10.1074/jbc.272.30.19000. [DOI] [PubMed] [Google Scholar]
- 34.Hayakawa M., Kaizawa H., Moritomo H., Kawaguchi K.-i., Koizumi T., Yamano M., Matsuda K., Okada M., Ohta M. Condensed heteroaryl derivatives. Int. Pat. WO/2001/083456. 2001 [Google Scholar]
- 35.Hayakawa M., Kaizawa H., Kawaguchi K.-i., Matsuda K., Ishikawa N., Koizumi T., Yamano M., Okada M., Ohta M. Imidazopyridine derivatives. Int. Pat. WO/2001/083481. 2001 [Google Scholar]
- 36.Sadhu C., Dick K., Treiberg J., Sowell C. G., Kesicki E. A., Oliver A. Inhibitors of human phosphatidyl-inositol 3-kinase δ. Int. Pat. WO/2001/081346. 2001 [Google Scholar]
- 37.Shuttleworth S. J., Folkes A. J., Chuckowree I. S., Wan N. C., Hancox T. C., Baker S. J., Sohal S., Latif M. A. Pharmaceutical compounds. Int. Pat. WO/2006/046040. 2006 [Google Scholar]
- 38.Jackson S. P., Robertson A. D., Kenche V., Thompson P., Prabaharan H., Anderson K., Abbott B., Goncalves I., Nesbitt W., Schoenwaelder S., Saylik D. Inhibition of phosphoinositide 3-kinase β. Int. Pat. WO/2004/016607. 2004 [Google Scholar]
- 39.Beeton C. A., Chance E. M., Foukas L. C., Shepherd P. R. Comparison of the kinetic properties of the lipid and protein kinase activities of the p110α and p110β catalytic subunits of class Ia phosphoinositide 3-kinases. Biochem. J. 2000;350:353–359. [PMC free article] [PubMed] [Google Scholar]
- 40.Navé B. T., Haigh R. J., Hayward A. C., Siddle K., Shepherd P. R. Compartment-specific regulation of phosphoinositide 3-kinase by platelet-derived growth factor and insulin in 3T3-L1 adipocytes. Biochem. J. 1996;318:55–60. doi: 10.1042/bj3180055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konstantopoulos N., Clark S. Insulin and insulin-like growth factor-1 stimulate dephosphorylation of paxillin in parallel with focal adhesion kinase. Biochem. J. 1996;314:387–390. doi: 10.1042/bj3140387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawley D. M., Maddux B. A., Patel R. G., Wong K. Y., Mamula P. W., Firestone G. L., Brunetti A., Verspohl E., Goldfine I. D. Insulin receptor monoclonal antibodies that mimic insulin action without activating tyrosine kinase. J. Biol. Chem. 1989;264:2438–2444. [PubMed] [Google Scholar]
- 43.Wang Q., Bilan P. J., Klip A. Opposite effects of insulin on focal adhesion proteins in 3T3-L1 adipocytes and in cells overexpressing the insulin receptor. Mol. Biol. Cell. 1998;9:3057–3069. doi: 10.1091/mbc.9.11.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang S., Denley A., Vanhaesebroeck B., Vogt P. K. Oncogenic transformation induced by the p110β, -γ, and -δ isoforms of class I phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meier T. I., Cook J. A., Thomas J. E., Radding J. A., Horn C., Lingaraj T., Smith M. C. Cloning, expression, purification and characterization of the human class-IA phosphoinositide 3-kinase isoforms. Protein Expression Purif. 2004;35:218–224. doi: 10.1016/j.pep.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Kurosu H., Maehama T., Okada T., Yamamoto T., Hoshino S., Fukui Y., Ui M., Hazeki O., Katada T. Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110β is synergistically activated by the βγ subunits of G-proteins and phosphotyrosyl peptide. J. Biol. Chem. 1997;272:24252–24256. doi: 10.1074/jbc.272.39.24252. [DOI] [PubMed] [Google Scholar]