Abstract
The discovery that the flavoprotein oxidase, Erv2p, provides oxidizing potential for disulfide bond formation in yeast, has led to investigations into the roles of the mammalian homologues of this protein. Mammalian homologues of Erv2p include QSOX (sulfhydryl oxidases) from human lung fibroblasts, guinea-pig endometrial cells and rat seminal vesicles. In the present study we show that, when expressed in mammalian cells, the longer version of human QSOX1 protein (hQSOX1a) is a transmembrane protein localized primarily to the Golgi apparatus. We also present the first evidence showing that hQSOX1a can act in vivo as an oxidase. Overexpression of hQSOX1a suppresses the lethality of a complete deletion of ERO1 (endoplasmic reticulum oxidase 1) in yeast and restores disulfide bond formation, as assayed by the folding of the secretory protein carboxypeptidase Y.
Keywords: disulfide bond, Ero1p, Erv2p, protein disulfide isomerase, sulfhydryl oxidase
Abbreviations: AMS, 4-acetamido-4′-maleimidylstilbene-2, 2′-disulfonic acid; CHO, Chinese hamster ovary; CPY, carboxypeptidase Y; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; Endo H, endoglycosidase H; ER, endoplasmic reticulum; Ero1, endoplasmic reticulum oxidase 1; Mal, maleimide; NEM, N-ethylmaleimide; PDI, protein disulfide-isomerase; PEG, poly(ethylene glycol); QSOX, sulfhydryl oxidase; SP, semi-permeabilized; tPA, tissue plasminogen activator; YNB, minimal medium containing 0.67% yeast nitrogen base; YP medium, 2% peptone, 1% yeast extract; YPD, YP medium containing 2% glucose; YPGal, YP medium containing 2% galactose
INTRODUCTION
Many secreted proteins contain disulfide bonds, which are required for their proper folding, function and stability. In eukaryotic cells disulfide bond formation usually occurs while the protein folds in the lumen of the ER (endoplasmic reticulum). While disulfide bonds can be formed spontaneously in vitro, an intermediary, such as a transition metal or flavin, is required to overcome this kinetically sluggish reaction. In vivo, the most common mechanism for the formation of protein disulfide bonds is a thiol-disulfide exchange reaction of free thiols with an already disulfide-bonded species. These reactions are catalysed by cellular enzymes know as thiol-disulfide oxidoreductases.
The Ero1 (endoplasmic reticulum oxidase 1)-PDI (protein disulfide-isomerase)-dependent pathway for oxidation of disulfide bonds in substrate proteins within the eukaryotic ER is now well-established [1]. PDI is the direct donor of disulfide bonds to reduced proteins and is in turn oxidized by Ero1. It is now widely accepted that Ero1 can use molecular oxygen as a terminal electron acceptor and functions with the aid of a bound FAD cofactor [2]. A second pathway for disulfide bond formation in the yeast ER was identified by screening for proteins that could rescue an Ero1p mutant when overexpressed [3]. Overexpression of a luminal ER protein, Erv2p, conferred increased resistance to DTT (dithiothreitol) and the restoration of CPY (carboxypeptidase Y) maturation in both ero1-1 and ero1Δ strains, leading to the realization that Erv2p forms the basis of a disulfide bond formation pathway that can function independently of Ero1p.
Erv2p is a FAD-binding QSOX (sulfhydryl oxidase) and belongs to the ERV (essential for respiration and vegetative growth)/ALR (augmenter of liver regeneration) family of proteins [4]. Proteins containing an Erv2p homologous domain, known as the QSOX, have been detected in all multicellular plants and animals for which complete genome sequences exist [5]. Two proteins belonging to the QSOX family have been discovered in humans. The first to be discovered was a gene that was up-regulated when human fibroblasts reached quiescence [6], hence called the Quiescin Q6 gene, now renamed as hQSOX1. Two splice variants of the human QSOX1 gene have been reported, one of 3314 bases, which encodes a peptide of 747 amino acids (QSOX1a), and another of 2588 bases, which encodes a peptide of 604 amino acids (QSOX1b). Both splice-variant mRNAs are expressed at approximately the same level in human fibroblasts whereas in other cell types the ratios vary [6]. The longer protein has a potential C-terminal transmembrane domain which has been spliced out to generate the short form of the enzyme [5]. A second gene was identified as sharing similarity with hQSOX1 and encoding another member of the QSOX family. The corresponding protein, SOXN or hQSOX2 has been studied in human neuroblastoma cells [7]. All members of the QSOX family have a thioredoxin domain fused to their C-terminal Erv2p homologous domain [5]. A second more weakly scoring thioredoxin domain lacking any redox-active disulfides adjacent to the first thioredoxin domain has been identified in human and avian QSOX sequences [8]. Thus, while the ER oxidation system requires interaction between Ero1/Erv2p and PDI, the QSOX enzymes have evolved by a fusion of these two proteins, providing them with the unique advantage of being able to introduce disulfide bonds into substrate proteins without the need to interact with additional proteins. Although it has been known for sometime that QSOX proteins are capable of directly introducing disulfide bonds into substrate proteins in vitro [9], the in vivo roles of these proteins still remain a mystery.
Expression and tissue distribution patterns of the rat and human QSOX enzymes indicate high levels of expression in tissues that are heavily involved in the secretion of disulfide-rich proteins [5,10,11], leading to the hypothesis that QSOX members could be involved in the oxidative folding of secreted proteins. The intracellular location of the human and rat QSOX1 has been investigated with antibodies that recognise both the soluble and membrane-associated forms. The results demonstrate that although some intracellular material can be seen a population of QSOX1 is secreted [6,12,13]. Intracellular QSOX1 proteins can be detected in the ER [5,14], Golgi [5,14,15] and secretory granules [14,15], consistent with a role for QSOX in the formation of disulfide bonds within proteins entering the secretory pathway. The widespread distribution of these enzymes in all multicellular organisms and the discovery that the yeast counterpart, Erv2p, is capable of introducing disulfide bonds into substrate proteins, independently of Ero1p, has created renewed interest into the in vivo role of these enzymes.
In order to investigate the role of mammalian QSOX in oxidative protein folding, we created a stable cell line expressing the longer form of the human QSOX1 (hQSOX1a) enzyme as well as a model protein, human tPA (tissue plasminogen activator). We have previously shown [16] that overexpression of Ero1, which provides oxidizing equivalents to the disulfide bond formation pathway, allows the formation of disulfide bonds in tPA to occur at concentrations of DTT which would normally result in synthesis of only the reduced form of tPA [16]. We hypothesized that if hQSOX1a was able to oxidize proteins then the overexpression of QSOX would have a similar effect on the folding of tPA. In the present study we describe the characterization of the hQSOX1a protein and present the first evidence to show that this protein is capable of disulfide bond formation in vivo.
EXPERIMENTAL
Cell culture and treatment conditions
CHO (Chinese hamster ovary) cells expressing tPA (CRL-9606; American Tissue Culture Collection) were cultured in HAM F-12 medium (tissue culture medium and supplements obtained from Invitrogen), while HT1080 cells (CCL-121; American Tissue Culture Collection) were cultured in DMEM 3 (Dulbecco's modified Eagle's medium 3). To disrupt the microtubule network, cells grown on coverslips were incubated at 4°C for 10 min. Nocodazole (5 μg/ml; Sigma) or DMSO (dimethyl sulfoxide; Sigma; solvent control) was added in medium to the cells followed by a further incubation for 2 h at 37°C.
Construction of a QSOX plasmid for mammalian expression and generation of stable cell lines
A pINCY plasmid containing the long form of the human QSOX1 (hQSOX1a; accession number AY358941) sequence was obtained from Genetech (a gift from Dr Hilary F. Clark, Genetech Inc., San Francisco, U.S.A.). The Invitrogen pcDNA3.1/V5-His-TOPO TA expression kit was used to construct the hQSOX1a expression plasmids. A PCR product was produced using primers that allowed amplification of all the coding sequence but removed the stop codon to allow in-frame addition of the V5-His tag. The PCR product was then cloned into the pcDNA3.1/V5-His-TOPO vector. The hQSOX1a-V5-KDEL construct was created by amplification of the coding sequence minus sequence C-terminal to the beginning of the transmembrane domain. This sequence was replaced with a V5-tag in addition to a KDEL sequence at the C-terminus and cloned into pcDNA 3.1. CHO-tPA cells overexpressing QSOX and QSOX-KDEL were generated in a similar manner to CHO-tPA cells overexpressing Ero1 as described in [16].
In vitro transcription and translations
The pcDNA3.1/V5-His-TOPO vector was linearized with PmeI (Roche Diagnostics) and transcribed with T7 RNA polymerase (Promega) as described in [17]. hQSOX1a mRNA transcripts were translated using a rabbit reticulocyte lysate (FlexiLysate; Promega) in the presence or absence of SP (semi-permeabilized) cells as described [18]. SP cells were prepared as described previously [19].
Endo H (endoglycosidase H) treatment and proteinase K digestion
After translations, isolated SP cells were solubilized in 0.5% SDS and 1% 2-mercaptoethanol for 5 min at 95°C. G5 buffer [1×50 mM sodium citrate buffer (pH 5.5)] was added and the sample was divided into two identical aliquots, which were incubated for 1 h at 37°C with either 500 units of Endo H or buffer alone. Cell lysates were treated with Endo H by first adding SDS and 2-mercaptoethanol to 0.5% and 1% respectively, followed by incubation for 5 min at 95°C and treatment with enzyme as described above. Proteinase K digestions were performed as described in [18]. Samples were analysed directly using SDS/PAGE.
Immunofluorescence and radiolabelling
Immunofluorescence and pulse–chase analysis of tPA was performed as described [16]. A mouse anti-V5 antibody (Upstate) was used for immunofluorescence to detect the V5-tagged hQSOX1a. Primary antibodies used for immunofluorescence included a rabbit anti-ERp57 antibody [20] as an ER marker or rabbit anti-GM130 antibody (raised against the first 73 amino acid residues at the N-terminus of GM130, a gift from Martin Lowe, Faculty of Life Sciences, The University of Manchester, U.K.), as a Golgi marker. Slides were viewed on an Olympus BX60 upright microscope equipped with a MicroMax cooled, slow-scan CCD (charge-coupled-device) camera driven by Metamorph software. For in vivo labelling of the V5-tagged hQSOX1a, 2×106 cells per 6 cm dish were washed twice with methionine- and cysteine-free DMEM, and then pre-incubated in the same medium for 20 min at 37°C. Each monolayer was pulse-labelled with 50 μCi of [35S]-EXPRESS label (Amersham Biosciences) for 45 min at 37°C and then chased with excess methionine and cysteine for 60 min. Cycloheximide (0.5 mM; Sigma) was included in the chase medium to block completion of labelled nascent chains. The cells were lysed in 1 ml of lysis buffer [50 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, 1% (v/v) Triton X-100, 0.1% (w/v) SDS, 1 mM PMSF and 20 mM NEM (N-ethylmaleimide)] for 10 min on ice. Lysates were centrifuged at 12000 g for 20 min to pellet nuclei and cell debris.
Immunoisolation of hQSOXa1 from translations and cell lysates
Each 25 μl translation mixture was made up to 1 ml in immunoisolation buffer [50 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl and 1% (v/v) Triton X-100]. Cell lysates and translations were pre-incubated with 50 μl Protein A-Sepharose [10% (w/v); Zymed laboratories] for 1 h at 4°C to pre-clear the samples of Protein A binding components. Pre-cleared samples were each incubated overnight at 4°C with 1 μl of mouse anti-V5 antibody and 50 μl of Protein A-Sepharose. The complexes were washed three times with fresh immunoisolation buffer and then prepared for analysis by SDS/PAGE.
Yeast strains, plasmids and growth conditions
Yeast strains were grown at the indicated temperature in YP medium (2% peptone, 1% yeast extract) containing either 2% glucose (termed YPD) or 2% galactose (termed YPGal) or in minimal medium (containing 0.67% yeast nitrogen base; termed YNB) with 2% glucose or galactose, plus appropriate supplements for selective growth. Solid media were supplemented with 2% Bacto-agar. The ERO1/ero1δ and PDI1/pdi1δ heterozygous diploid Saccharomyces cerevisiae strains were obtained from Steve Oliver (Faculty of Life Sciences, University of Manchester, U.K.). The parental wild-type yeast strain (BY4742) was obtained from Barrie Wilkinson (Faculty of Life Sciences, University of Manchester, U.K.). The ERO1-1 strain (CKY559) was from Chris Kaiser (Department of Biology, MIT, Cambridge, U.S.A.).
The coding sequence of yeast ERV2 was amplified from yeast genomic DNA and the hQSOX1a coding sequence was amplified from the pINCY plasmid and cloned into yeast expression vector, pYES2 (Invitrogen). The ero1δ and pdi1δ strain containing the PGAL1-ERV2 or PGAL1-hQSOX1a plasmid was constructed by transforming the heterozygous ERO1/ero1δ and PDI1/pdi1δ diploid strains with PGAL1-ERV2 or PGAL1-hQSOX1a. The transformants were sporulated and tetrads dissected on YPGal plates. As a control, the parental plasmid, pYES2, was transformed into the same strains in parallel.
CPY radiolabelling and immunoprecipitations
Yeast strains were grown in YNB medium containing 1 mM ammonium sulfate and the required supplements except methionine. For pulse labelling, 25 D600 equivalents of cells were pelleted, resuspended in 2.5 ml of culture supernatant, and 500 μCi of [35S]methionine added prior to a 10 min incubation at 30°C. DTT (5 mM) was added during the labelling where indicated. Samples were chased for appropriate times by adding an excess of cold methionine and cysteine to the culture medium. For labelling in the ERO1-1 strain, cells were grown logarithmically at 24°C and resuspended at 10 D600/ml media and pre-incubated at 38°C for 25 min. DTT was added to a final concentration of 62.5 μM for the last 10 min of this pre-incubation. Cells were labelled and chased as above but at 38°C. Labelling reactions were stopped by addition of an equal volume of ice-cold 20 mM sodium azide, and incubation on ice for 5 min. CPY was immunoisolated from cell lysates using a sheep anti-CPY antibody as described in [21].
CPY redox state
The in vivo redox state of CPY was assayed by modification with the thiol-reactive reagent AMS (4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid; Molecular Probes). Strains were grown and radiolabelled at 30°C for 10 min as described above. Where indicated, DTT (5 mM) was added during the labelling and samples were chased for 30 min by adding an excess of cold methionine and cysteine to the culture medium. Cells were harvested by centrifugation (5000 g for 5 min at 4°C) and resuspended in 100 μl of 20% (v/v) TCA (trichloroacetic acid). Cell membranes were disrupted by agitation with glass beads and proteins collected by centrifugation (20000 g for 10 min) at 4°C. Protein pellets were treated with 25 mM AMS as described [22]. At the end of the AMS treatment, 1 ml of immunoisolation buffer was added to each of the samples followed by immunoisolation of CPY.
PDI redox state
The in vivo redox state of PDI was assayed as described by Xiao et al. [23]. Yeast PDI was visualized by probing with a polyclonal anti-yeast PDI antibody (a gift from Jakob Winther, Institute of Molecular Biology and Physiology, Copenhagen, Denmark).
Electrophoresis
Samples for SDS/PAGE were resuspended in SDS/PAGE sample buffer [0.25 mM Tris/HCl (pH 6.8), 2% (w/v) SDS, 20% (v/v) glycerol and 0.004% (w/v) Bromophenol Blue]. DTT (50 mM) was added to reduce samples where indicated. Proteins resolved through 10% or 7.5% gels were either transferred on to nitrocellulose for Western blotting or fixed in 10% (v/v) acetic acid and 10% (v/v) methanol, and dried. Radiolabelled products were visualized by autoradiography using Kodak Biomax MR film (GRI).
RESULTS
hQSOX1a-V5 is a glycosylated, transmembrane protein localized to the Golgi apparatus
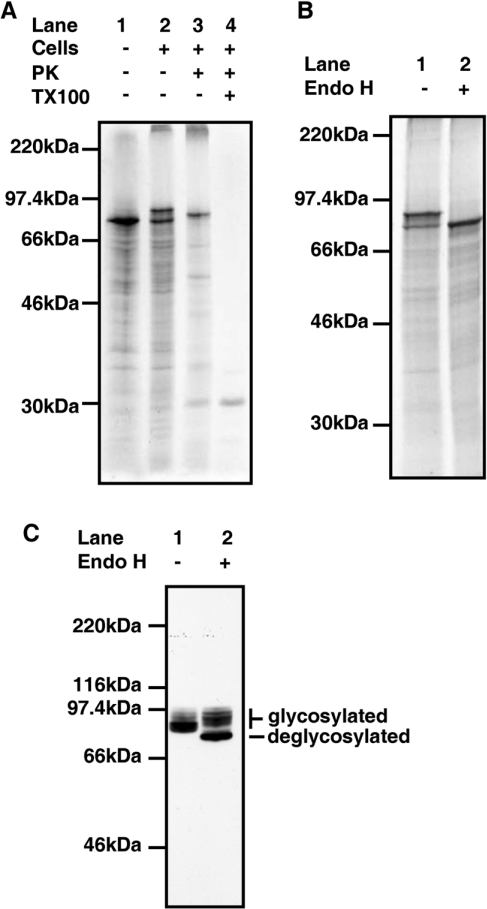
The predicted coding sequence for hQSOX1a contains a putative N-terminal signal peptide, a conserved N-glycosylation site and a C-terminal transmembrane domain [5]. To investigate whether the protein is indeed glycosylated and a transmembrane protein we performed in vitro translation of an RNA transcript coding for V5-tagged hQSOXa in the presence of SP HT1080 cells [19]. When mRNA coding for hQSOX1a-V5 was translated in the absence of SP cells, a single radiolabelled translation product of approximately 75 kDa was generated (Figure 1A, lane 1). An additional larger product was generated when translations were performed in the presence of cells (Figure 1A, lane 2). This larger product was sensitive to digestion with Endo H demonstrating that hQSOX1a is glycosylated (Figure 1B, lane 2).
Figure 1. In vitro translations and in vivo processing of hQSOX1a.
(A) hQSOX1a-V5 mRNA was translated in a rabbit reticulocyte lysate in the absence (lane 1) or presence of SP HT1080 cells (lanes 2–5) for 1 h at 30°C. Isolated cells were treated with Proteinase K (PK; lane 3) with the addition of 1% (w/v) Triton X-100 (TX100) as indicated (lane 4). (B) hQSOX1a-V5 mRNA was translated in a rabbit reticulocyte lysate in the presence of SP HT1080 cells for 1 h at 30°C. Isolated cells were incubated with (lane 2) or without (lane 1) Endo H at 37°C for 1 h. (C) Cells stably expressing hQSOX1a-V5 were lysed and proteins were either treated (lane 1) or not treated (lane 2) with Endo H. Proteins were detected by immunoblotting with V5-antibody.
To determine whether the N-terminal signal peptide of hQSOX1a resulted in the protein being translocated into the ER, translations were treated with proteinase K. When the hQSOX1a translation products synthesized in the presence of SP cells were treated with proteinase K (Figure 1A, lane 3), the lower molecular mass band was digested, indicating that this was untranslocated material. However, the higher molecular mass band was partially resistant to digestion indicating that this translation product was translocated and protected from digestion. When an identical digest was performed in the presence of Triton X-100 (Figure 1A, lane 4) no bands were detected, demonstrating that the translation product was not intrinsically resistant to proteolysis. An increase in electrophoretic mobility of the higher molecular mass band was observed on treatment with proteinase K (Figure 1A, compare lane 2 with lane 3). If the hQSOX1a gene product was anchored to the membrane by the predicted transmembrane domain then a short (21 amino acid) cytosolic domain would have been digested following proteinase K treatment potentially explaining the increased mobility of the translation product. Thus, consistent with protein prediction results for the hQSOX1a [5], translations in SP cells showed that the hQSOX1a gene product can be glycosylated and anchored to membranes via a C-terminal transmembrane domain.
In an attempt to determine the in vivo localization and role of hQSOX1a, we generated a stable cell line expressing V5-tagged hQSOX1a and a model disulfide-bonded protein, tPA. To determine whether hQSOX1a reached the Golgi apparatus we tested the sensitivity of the intracellular protein to Endo H. Cell lysates contained a mixture of glycosylated V5-tagged hQSOX1a species some of which were sensitive to digestion with Endo H but some of which were not (Figure 1C, lanes 1 and 2). The higher molecular mass species are characteristic of molecules that have had their oligosaccharide side chains modified in the Golgi apparatus. The material that remains sensitive to Endo H may also have reached the Golgi but had not been sufficiently modified to gain resistance to digestion. The result demonstrates that at steady state a proportion of the hQSOX1a synthesized is transported to the Golgi apparatus.
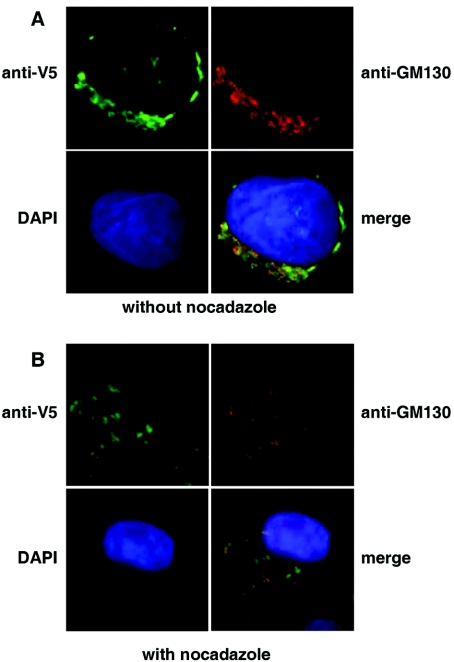
The Endo H experiments do not allow us to determine whether the protein was localized to post-ER compartments such as the Golgi apparatus or the cell-surface. To ascertain intracellular localization we carried out immunofluorescence studies on the CHO-tPA cells stably expressing the hQSOX1a-V5 protein (Figure 2A). We could not detect any hQSOX1a on the cell surface; however, a distinct perinuclear staining was observed (Figure 2A, upper left panel), which overlapped with that of the cis-Golgi marker protein GM130 (Figure 2A, upper and lower right panels). Treatment of cells with nocodazole, a compound which depolymerizes microtubules and hence causes disruption of the Golgi apparatus, led to a loss of the perinuclear staining pattern and scattered cytoplasmic distribution, with both the anti-V5 and the anti-GM130 antibody (Figure 2B), supporting the Golgi localization of the transfected hQSOX1a.
Figure 2. Intracellular localization of V5-tagged hQSOX1a.
(A) hQSOX1a-V5 expressing CHO-tPA cells were fixed and simultaneously stained with mouse anti-V5 antibody and rabbit anti-GM130 antibody. Secondary antibodies were conjugated to Alexa Fluor® 448 (green) or Alexa Fluor® 594 (red). Cell nuclei were stained with DAPI. (B) Golgi localization of the hQSOX1a-V5 was confirmed by incubating cells in the presence of 5 μg/ml nocadazole for 2 h. Cells were then fixed and stained as above.
Effects of overexpression of hQSOX1a on the folding of tPA
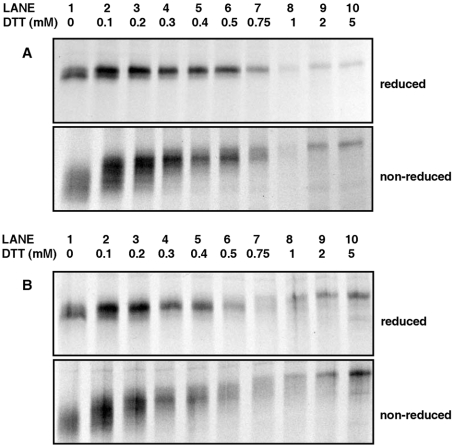
Whereas in vitro studies have shown that the avian QSOX and QSOX from the rat seminal vesicles [24] are capable of introducing disulfide bonds into substrate proteins, the ability of the SOXs to introduce disulfide bonds into proteins within cells has not been demonstrated. Using tPA as a model protein, we wished to determine whether the expression of hQSOX1a-V5 in theCHO-tPA cells would have any effect on the folding of tPA. We have previously shown that conditions preventing disulfide bond formation, such as addition of the reducing agent DTT to the culture medium of living CHO-tPA cells, leads to the complete glycosylation of a sequon within tPA that would otherwise undergo variable glycosylation [25]. However the overexpression of Ero1 conferred an increased resistance to the effects of reduction with DTT on tPA glycosylation and disulfide bond formation [16]. To determine whether the expression of hQSOX1a could function like Ero1 in vivo, hQSOX1a-V5 transfected stable CHO-tPA cells were radiolabelled for 10 min in the absence or presence of various concentrations of DTT. At the end of the labelling period, the cells were treated with the membrane permeable alkylating agent NEM to block free thiols, lysed and tPA was immunoisolated. The immunoisolated material was then subjected to both reducing and non-reducing SDS/PAGE (Figure 3).
Figure 3. Effect of hQSOX1a expression on the folding and glycosylation of tPA in the presence of DTT.
(A) Untransfected or (B) stable cell line expressing hQSOX1a-V5 was radiolabelled in the absence (lane 1) or presence (lanes 2–10) of various concentrations of DTT. Cells were lysed and newly synthesized tPA immunoisolated with an anti-tPA antibody. Immunoisolated proteins were separated by SDS/PAGE under reducing (upper panels) and non-reducing (lower panels) conditions.
In the untransfected CHO-tPA cells, the tPA synthesized during the 10 min labelling period in the absence of DTT migrated as a doublet band when reduced prior to electrophoresis (Figure 3A, upper panel, lane 1) indicating that variable glycosylation of tPA had occurred. When subjected to non-reducing SDS/PAGE (Figure 3A, lower panel, lane 1), tPA migrated with a greater mobility and with a more diffuse banding pattern than on reducing gels, indicating that in the absence of DTT, tPA forms intrachain disulfide bonds. As the concentration of DTT in the labelling medium was increased from 0–5 mM, the doublet pattern progressively disappeared until only a single band of the over-glycosylated (type 1) tPA was present at concentrations of DTT above 0.75 mM (Figure 3A, upper panel, lanes 2–10). This change in banding pattern was accompanied by a progressive reduction in the formation of disulfide bonds as indicated by the slower migrating sharper bands under non-reducing conditions (Figure 3A, lower panel, lanes 2–10). Thus at 1 mM DTT no disulfide bonds were formed as no faster migrating bands were seen when the sample was run under non-reducing conditions (Figure 3A, lane 8). A clear difference in mobility of type 1 tPA synthesized in the absence or presence of 5 mM DTT (Figure 3A, upper panel, lanes 1 and 10) and separated under reducing conditions was observed. This was because the tPA synthesized in the presence of 5 mM DTT is completely reduced, thus making its 34 cysteine residues available for alkylation by NEM, slowing the migration of the protein. The lower intensity of signal at higher concentrations of DTT is due to the antibody having a lower affinity during immunoisolation for the reduced and alkylated protein than the disulfide bonded form as characterized previously [16].
Similar results were obtained in the CHO-tPA cells expressing the V5-tagged hQSOX1a (Figure 3B). This is in contrast with results obtained with the overexpression of Ero1 [16]. In the cell line overexpressing Ero1-Lα, variable glycosylated tPA could be observed at DTT concentrations as high as 1 mM and formation of disulfide bonds even at 2 mM. Thus, unlike Ero1 overexpression, the overexpression of QSOX1a had little or no effect on the glycosylation or disulfide bond formation of tPA.
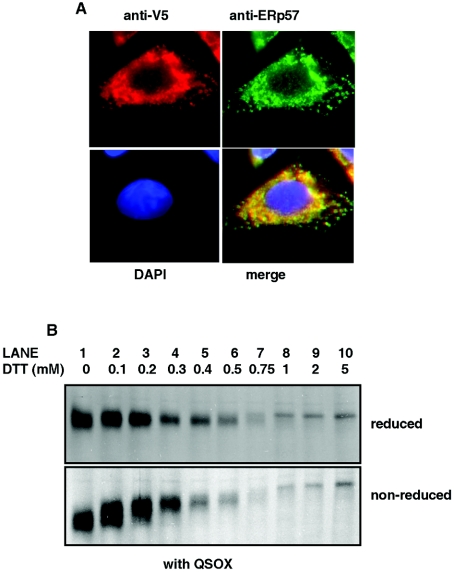
One explanation for the lack of effect of hQSOX1a overexpression on the disulfide bond formation within tPA may be that most of the exogenously expressed hQSOX1a-V5 was localized in the Golgi, while the folding and glycosylation of tPA occurs in the ER. To check whether the QSOX1a overexpression would provide resistance to DTT if the hQSOX1a-V5 was located in the ER, we created a CHO-tPA cell-line stably expressing hQSOX1-V5 with a C-terminal KDEL sequence for ER localization. Immunofluorescence studies on CHO-tPA cells stably expressing hQSOX1-V5-KDEL showed that the V5- tagged hQSOX1 lost its perinuclear staining and instead showed a characteristic ER-like distribution (Figure 4A). A similar staining pattern was seen when cells were stained with the ER marker ERp57 (Figure 4A). Thus replacement of the transmembrane and cytosolic tail sequence with a KDEL sequence caused redistribution of the hQSOX1-V5 from the Golgi into the ER, in turn trapping the newly synthesized tPA and the exogenously expressed hQSOX1a in the same cellular compartment.
Figure 4. Localization of hQSOX1a-V5-KDEL in CHO-tPA cells to the ER.
(A) CHO-tPA cells stably expressing hQSOX1-V5-KDEL were fixed and stained with a mixture of anti-V5 antibody (left-hand panels) and anti- ERp57 antibody (right-hand panels). Secondary antibodies were conjugated to Alexa Fluor® 448 (green) or Alexa Fluor® 594 (red). Cell nuclei were stained with DAPI (4,6-diamidino-2-phenylindole). (B) CHO-tPA cells expressing hQSOX-V5-KDEL were radiolabelled for 10 min in the absence (lane 1) or presence (lanes 2–10) of varying concentrations of DTT. Cells were lysed and tPA immunoisolated from lysates with anti-tPA antibody. Immunoisolated proteins were separated by SDS/PAGE under reducing (upper panel) and non-reducing conditions (lower panel).
In order to determine whether the trapping of the hQSOX1a-V5 in the ER has any effect on disulfide bond formation within tPA, cells stably expressing hQSOX1-V5-KDEL were assayed for tPA folding as described above. In the hQSOX1a-V5-KDEL expressing cells disulfide bond formation was as sensitive to the effects of DTT as in cells without hQSOX1a (compare Figure 4B with Figure 3A). Previous experiments looking at the in vitro activity of Ero1 and QSOX have shown that both enzymes are fully active at the DTT concentrations used in our experiments [26,27]. Based on our results we can conclude that, unlike Ero1, the hQSOX1a expressed in the CHO-tPA does not contribute to the formation of disulfide bonds during folding of tPA. Given the localization of hQSOX1a it is possible that the mammalian QSOX enzymes are not required for the initial steps of disulfide bond formation that occur in the ER, but rather in the formation of disulfide bonds within the Golgi apparatus.
Overexpression of hQSOX1a can rescue an ero1Δ strain
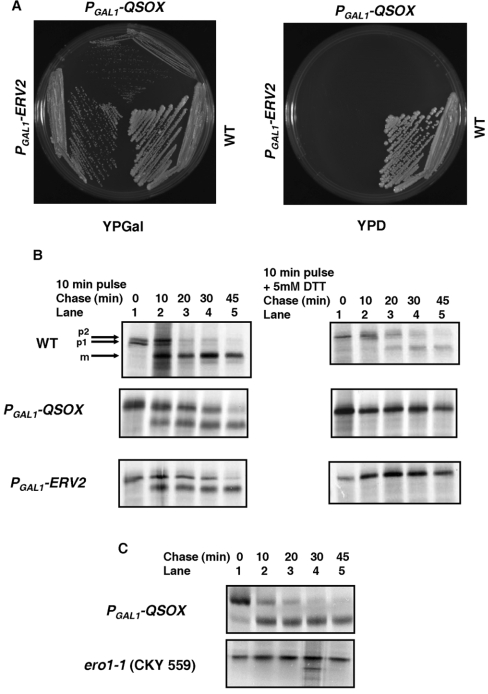
The inability to detect a role for QSOX in facilitating disulfide bond formation within mammalian cells led us to hypothesize that, similar to observations made in yeast, oxidation of proteins via Ero1 could be the major pathway for disulfide formation in the mammalian ER [28]. Thus the contribution of QSOX to the disulfide bond formation pathway may only be evident in cells with an impaired Ero1 function. To explore this possibility a heterozygous diploid ERO1/ero1δ strain was transformed with a yeast expression plasmid containing the hQSOX1a coding sequence under the control of the GAL1 promoter. The strain was also transformed with an empty plasmid to serve as a negative control and plasmid containing the Erv2p-coding sequence to serve as a positive control. The transformants were selected, allowed to sporulate and tetrads were dissected on to glucose and galactose media. At least 60 tetrads were dissected for each strain with identical results obtained for each tetrad. As reported previously [3], all the spores derived from the ERV2 transformants were able to grow on galactose media, while only two of the four spores from the strain transformed with the empty plasmid were alive (results not shown). When transformants containing the PGAL1-hQSOX1a plasmids were sporulated, all four spores were found to be viable on galactose medium (results not shown), although the ero1Δ PGAL1-hQSOX1a strain grew more slowly than the ero1Δ PGAL1-ERV2 strain (Figure 5A, left-hand panel). Growth of the ero1Δ PGAL1-hQSOX1a strain depended on the overexpression of hQSOX1a as this strain could not grow on glucose medium (Figure 5A, right-hand panel). These results demonstrate that hQSOX1a is capable of rescuing Ero1-deficient strains and is therefore capable of fulfilling the essential function of Ero1 within yeast cells.
Figure 5. hQSOX1a overexpression restores growth and formation of disulfide bonds in an Ero1p-deficient strain.
(A) Growth of wild-type (WT) or an ero1Δ strain containing PGAL1-hQSOX1a or PGAL1-ERV2I I on YPGal (left-hand panel). Growth is dependent on overexpression of Erv2p or hQSOX1a as the strains could not grow on glucose medium (right-hand panel). (B) The parental BY4742 (WT), ero1Δ PGAL1-hQSOX1a and ero1Δ PGAL1-ERV2I I strains were radiolabelled in the absence (left-hand panels) or presence (right-hand panels) of 5 mM DTT. Labelling was stopped, cells allowed to recover in the absence of DTT and samples taken at various time points as indicated. Cells were lysed and CPY folding analysed by separating the immunoisolated proteins by SDS/PAGE under reducing conditions. Positions of the ER (p1), Golgi (p2) and vacuolar (m) forms of CPY are indicated. (C) The ERO1–1 (CKY559) or the ero1Δ PGAL1-hQSOX1a strain growing logarithmically at 24°C were resuspended at 10 D600/ml and incubated at 38°C. After 15 min, DTT was added to a concentration of 65 μM and after a further 10 min cells were radiolabelled and then chased for the indicated times. Cells were lysed and CPY folding analysed as in (B).
To determine whether the overexpression of hQSOX1a was able to restore disulfide bond formation in the ero1δ PGAL1-hQSOX1a strain we evaluated the folding and transport of CPY. CPY contains five intrachain disulfide bonds whose formation is necessary for its folding and exit from the ER [29]. Three sub-cellular forms of CPY can be detected by reducing SDS/PAGE during its folding and maturation within the cell [30]: a 67 kDa ER form termed p1CPY, a 69 kDa Golgi form termed p2CPY and finally a 61 kDa mature (m) vacuolar form of the protein. When disulfide bond formation is compromised, only the p1 form of CPY is synthesized due to the absolute requirement for disulfide bonds to form prior to transport of CPY from the ER to the Golgi apparatus [29]. In the wild-type cells most of the CPY synthesized in the ER in the absence of DTT is efficiently converted into the p2 form and finally to the mature form within 20 min of the chase time (Figure 5B, lane 3), demonstrating correct folding, disulfide bond formation and transport of CPY in this strain. The mature form of CPY can also be detected in the ero1Δ PGAL1-ERV2 and the ero1Δ PGAL1-hQSOX1a strains, although it takes about 45 min for most of the CPY in these strains to be processed, possibly due to the slow growing nature of these strains (Figure 5B, lanes 2–5). CPY processing was also assessed in a temperature-sensitive strain of Ero1 grown at the non-permissive temperature (Figure 5C, lanes 2–5). As shown previously [31], no processing was seen in the Ero1-1 strain grown at the non-permissive temperature demonstrating that the lack of a functional Ero1 results in a lack of processing of CPY to the mature protein. In contrast, folding of CPY in the ero1Δ PGAL1-hQSOX1a strain occurred at 38°C with faster kinetics than at 30°C (compare Figure 5B with 5C). These results demonstrate that like Erv2p, hQSOX1a is capable of reversing the Ero1 defect and therefore supports disulfide bond formation in an Ero1p-independent pathway. Addition of 5 mM DTT to the medium during the pulse period resulted in accumulation of the p1 form of CPY in all three strains (Figure 5B, right-hand panel, lane 1). As observed previously [32], on removal of DTT in the wild-type yeast strains, the majority of the CPY is able to fold and reach its mature form within 45 min (Figure 5B, top right-hand panel, lanes 2–5). In contrast, the CPY in the ero1Δ PGAL1-ERV2 and the ero1Δ PGAL1-hQSOX1a strains were unable to resume folding on removal of the DTT and accumulated in the immature ER form (Figure 5B, middle and lower right-hand panels).
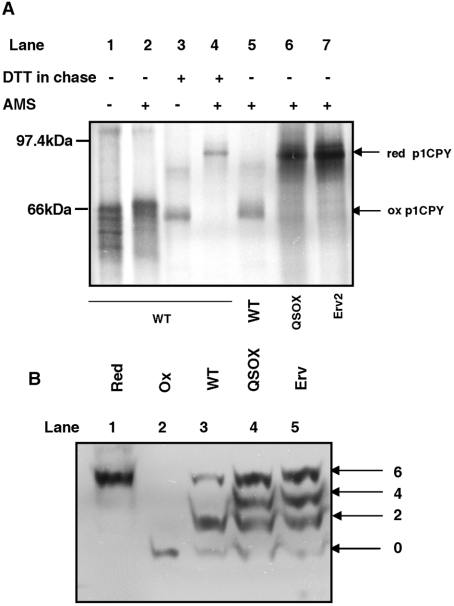
The accumulation of CPY in the ER form could either be due to the inability of the strains to recover from DTT treatment or due to the formation of non-native disulfide bonds in CPY on removal of the DTT. To address this point the oxidation state of CPY was assayed directly by modification of free thiols with the thiol-conjugating reagent AMS. Modification of a single cysteine residue by AMS increases the molecular mass of the protein by approximately 0.5 kDa [33]. When the wild-type yeast was radiolabelled for 10 min in the absence of DTT and chased for 30 min, only a slight shift in mobility was observed following AMS treatment (Figure 6A, lanes 1 and 2). This is consistent with modification of the single cysteine residue present in oxidized CPY [34] indicating that CPY was fully oxidized under these conditions. Similar results were obtained with the ero1Δ PGAL1-ERV2 and ero1Δ PGAL1-hQSOX1a strains (results not shown). In contrast, when 5 mM DTT was added to the pulse medium and the cells were allowed to recover from DTT for 30 min, only the CPY in wild-type cells was able to form disulfide bonds (Figure 6A, lane 5). The CPY from the ero1Δ PGAL1-ERV2 and ero1Δ PGAL1-hQSOX1a strains was modified with AMS to the same extent as the fully reduced CPY (Figure 6A, compare lanes 6 and 7 with lane 4) indicating that the removal of DTT in these strains does not result in rapid re-establishment of oxidizing conditions within the ER. Thus, although Erv2p and hQSOX1a overexpression can rescue Ero1p-deficient strains, challenging the ER in such strains with reducing agents seems to abolish their ability to carry out disulfide bond formation in substrate proteins, even after removal of the reducing agent.
Figure 6. Redox states of CPY and PDI.
(A) BY4742 wild-type (WT) cells were radiolabelled for 10 min in the absence (lanes 1 and 2) or presence of 10 mM DTT (lanes 3 and 4) to detect the fully oxidized and fully reduced p1CPY respectively. BY4742 (WT), ero1Δ PGAL1-hQSOX1a (QSOX) and ero1Δ PGAL1-ERV2 (Erv) were radiolabelled for 10 min in the presence of 5 mM DTT followed by a 30 min chase in the absence of DTT (lanes 5, 6 and 7). Cells were isolated at the end of the labelling and chase periods and proteins from cell pellets were TCA precipitated and solubilized in the presence (lanes 2 and 4–7) or absence (lanes 1 and 3) of 25 mM AMS prior to immunoisolation of CPY. Samples were resolved by non-reducing SDS/PAGE. The reduced (red) and oxidized (ox) forms of p1CPY are indicated. (B) TCA (20%) was added to logarithmically growing cultures to quench thiol/disulfide exchange and precipitate proteins. Free sulfhydryl groups were modified by resuspension in 5 mM Mal-PEG under denaturing conditions [3% SDS and 0.2 M Tris/HCl, (pH 8)]. Proteins were separated by SDS/PAGE and visualized by Western blotting using an anti-yeast PDI antibody. Apparent molecular mass shifts above the unmodified protein indicate the number of free sulfhydryl groups modified. The redox states of PDI in the parental BY4742 (WT), ero1Δ PGAL1-hQSOX1a (QSOX) and ero1Δ PGAL1-ERV2 (Erv) are compared with reduced (Red) and oxidized (Ox) controls that were treated with 10 mM DTT or 1 mM diamide before Mal-PEG modification.
Interaction of hQSOX1a with Pdi1p
The presence of a PDI-like thioredoxin domain in addition to a FAD-binding domain within the QSOX sequence has led to the suggestion that, unlike Ero1, the QSOX family of proteins have evolved to be able to interact directly with substrate proteins [5]. Indeed, it has been shown that in vitro QSOX1 is able to directly oxidize proteins in the absence of PDI [24]. To determine whether hQSOX1a can function in vivo in the absence of PDI, PDI1/pdi1δ heterozygous diploid yeast strains were transformed with PGAL1-ERV2 and PGAL1-hQSOX1a plasmids. Transformants were selected, sporulated and tetrads dissected on to glucose and galactose media. Only spores containing the chromosomal PDI1 were able to grow (results not shown), indicating that despite the ability of Erv2p and QSOX1 to carry out direct oxidation of substrate proteins, there is a requirement for PDI to be present for these cells to be viable. This result supports in vitro data obtained for the avian QSOX1a, where increased yields of RNase A were obtained by incorporation of reduced PDI into the reaction [24].
Both QSOX1 and Erv2p have been shown to oxidize PDI in vitro [3,24] and Ero1p has been shown to be necessary to maintain PDI in an oxidized state in vivo. To evaluate the possibility that hQSOX1a was acting through Pdi1p in the ero1Δ PGAL1-hQSOX1a strain we determined the redox state of Pdi1p within these cells. Mal-PEG [maleimide-poly(ethylene glycol)] was used to determine the in vivo redox state of yeast PDI as described by Xiao et al. [23]. Yeast PDI has six cysteine residues, four in the active sites and two which form a stable intrachain disulfide bond [23,35]. The predominant bands in the wild-type strain represent fully oxidized or reduced Pdi1p and a species shifted by two Mal-PEG modifications (Figure 6B, lane 3) representing partially reduced Pdi1p [23]. Pdi1p from the ero1Δ PGAL1-hQSOX1a and ero1Δ PGAL1-ERV2 was much more reduced, as bands corresponding to two, four and six cysteine modifications by Mal-PEG could be detected in these strains (Figure 6B, lanes 4 and 5). Hence both the active site and the structural disulfide are more reduced in the complemented strains compared with the wild-type. The presence of increased amounts of reduced Pdi1p in these strains would indicate that both Erv2p and hQSOX1a are not as efficient as Ero1p in oxidizing Pdi1p in vivo. The lack of formation of the structural disulfide within a proportion of the Pdi1p present in these strains would affect overall activity as this disulfide has been shown to destabilize the N-terminal active site [35]. Such destabilization improves both the oxidase and isomerase activities of Pdi1p and could provide a possible explanation for the observed slow growth of these two strains.
DISCUSSION
Previous studies on the QSOX family of enzymes have demonstrated their ability to introduce disulfide bonds into proteins in vitro [24]. However, a demonstration of enzymatic activity in vivo has been missing mainly due to a lack of knowledge of potential endogenous substrates. In the present study we show that hQSOX1a is able to complement the function of Ero1 at least in yeast. The ability to act as a replacement for Ero1 under these circumstances does not necessarily mean that the enzyme acts as an oxidase within the ER of mammalian cells. Indeed the lack of effect of overexpression of the protein on the sensitivity of the oxidative folding pathway of tPA to a reducing agent, coupled with a cellular localization to the Golgi apparatus, would argue against such a function within the ER.
Most disulfide-bonded proteins are normally folded and assembled in the ER and transported to other compartments of the secretory pathway in a fully disulfide-bonded form. The remarkable distribution of proteins of the QSOX family in many cellular compartments [5] indicate that new pathways for disulfide bond formation outside the eukaryotic ER remain to be investigated. The approach we have taken in the present study to investigate the localization of the hQSOXa form differs from previous approaches in that we specifically detect the transmembrane version rather than both the transmembrane and soluble proteins. Hence we see a clear localization in the Golgi apparatus suggesting that substrate proteins may exit the ER in a precursor form that requires further disulfide bond formation prior to, or during, secretion. Identification of the substrates for QSOX enzymes, will be crucial if we are to fully understand the role of these enzymes within the cell. Candidates proteins would be components of the extracellular matrix which are typically of a high molecular mass containing several disulfide bonds which mature at later stages of the secretory pathway, often forming higher order structures stabilized by disulfide bonds. One characteristic of the disulfides that would be formed by QSOX in these proteins is that they would be either between separate polypeptide chains or between already folded protein domains. Hence the chances of formation of the incorrect disulfide bonds would be minimal and therefore so would be the need for an isomerase.
While hQSOX1a may be involved in the formation of disulfide bonds within the cell or on the cell surface, the shorter QSOX1 form (hQSOX1b) may function as a secreted oxidase to counteract the effects of extracellular reducing agents. The overall homology between the QSOX2 and the QSOX1 proteins is ∼40% with 68% identity in functional regions such as the thioredoxin-like or the ERV-1 domain [7]. While initial studies indicate that hQSOX2 plays an important role in inducing apoptosis in neuroblastoma cells, it will be interesting to determine whether it can also act as an oxidase. While hQSOX1a is highly expressed in the placenta, stomach and lung [5], high levels of hQSOX2 were detected in the pancreas, brain, kidney and heart [7]. Thus it is possible that the two genes have different functions or take on redundant functions in different tissues.
Unlike Ero1 which acts through PDI [22], QSOX does not require any additional partners to introduce disulfides into proteins. However, the ability of QSOX to oxidize substrate proteins, rapidly and indiscriminately as demonstrated in vitro [24] would potentially make them unsuitable proteins to have in the ER. In addition to newly synthesized reduced proteins which have to be oxidized to their native forms, the ER is also the site at which misfolded proteins are reduced and targeted for degradation [36]. QSOX could potentially catalyse the oxidation of both of these kinds of substrates. The presence of a strong, indiscriminate oxidant will thus act as a load rather than a help in the ER, as the cell would need to continuously isomerize the non-native disulfides introduced into substrate proteins. As the product of the reaction of QSOX with substrate proteins is hydrogen peroxide [24], such futile formation and isomerization of disulfide bonds could lead to the oxidative stress, ultimately leading to cell-death. However, our results with tPA would suggest that QSOX has a minimal role in catalysis of disulfide bonds within the ER. The presence of a robust Ero1-dependent pathway for disulfide bond formation could prevent QSOX from playing a major role in disulfide bond formation within the ER under normal physiological conditions. Thus, while Ero1 and PDI are involved in oxidative folding of secretory proteins in the ER of eukaryotic cells, it is likely that QSOX acts at later stages of the secretory pathway, either to maintain disulfide bonds or to oxidize specific substrates.
Acknowledgments
We would like to thank Barrie Wilkinson and Jim Warwicker for helpful discussions. We would also like to thank Steve Oliver, Hilary Clark, Martin Lowe, Jakob Winther and Chris Kaiser for providing us with reagents. This work was supported by a grant from the Welcome Trust (#74081) and the BBSRC (Biotechnology and Biological Sciences Research Council; #D00764).
References
- 1.Sevier C. S., Kaiser C. A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 2.Tu B. P., Weissman J. S. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 3.Sevier C. S., Cuozzo J. W., Vala A., Aslund F., Kaiser C. A. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat. Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- 4.Gerber J., Muhlenhoff U., Hofhaus G., Lill R., Lisowsky T. Yeast ERV2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J. Biol. Chem. 2001;276:23486–23491. doi: 10.1074/jbc.M100134200. [DOI] [PubMed] [Google Scholar]
- 5.Thorpe C., Hoober K. L., Raje S., Glynn N. M., Burnside J., Turi G. K., Coppock D. L. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch. Biochem. Biophys. 2002;405:1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 6.Coppock D. L., Kopman C., Scandalis S., Gilleran S. Preferential gene expression in quiescent human lung fibroblasts. Cell Growth Differ. 1993;4:483–493. [PubMed] [Google Scholar]
- 7.Wittke I., Wiedemeyer R., Pillmann A., Savelyeva L., Westermann F., Schwab M. Neuroblastoma-derived sulfhydryl oxidase, a new member of the sulfhydryl oxidase/Quiescin6 family, regulates sensitization to interferon gamma-induced cell death in human neuroblastoma cells. Cancer Res. 2003;63:7742–7752. [PubMed] [Google Scholar]
- 8.Raje S., Thorpe C. Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: role of a thioredoxin domain in disulfide bond formation. Biochemistry. 2003;42:4560–4568. doi: 10.1021/bi030003z. [DOI] [PubMed] [Google Scholar]
- 9.Janolino V. G., Swaisgood H. E. Isolation and characterization of sulfhydryl oxidase from bovine milk. J. Biol. Chem. 1975;250:2532–2538. [PubMed] [Google Scholar]
- 10.Benayoun B., Esnard-Feve A., Castella S., Courty Y., Esnard F. Rat seminal vesicle FAD-dependent sulfhydryl oxidase. Biochemical characterization and molecular cloning of a member of the new sulfhydryl oxidase/quiescin Q6 gene family. J. Biol. Chem. 2001;276:13830–13837. doi: 10.1074/jbc.M010933200. [DOI] [PubMed] [Google Scholar]
- 11.Musard J. F., Sallot M., Dulieu P., Fraichard A., Ordener C., Remy-Martin J. P., Jouvenot M., Adami P. Identification and expression of a new sulfhydryl oxidase SOx-3 during the cell cycle and the estrus cycle in uterine cells. Biochem. Biophys. Res. Commun. 2001;287:83–91. doi: 10.1006/bbrc.2001.5440. [DOI] [PubMed] [Google Scholar]
- 12.Mairet-Coello G., Tury A., Fellmann D., Risold P. Y., Griffond B. Ontogenesis of the sulfhydryl oxidase QSOX expression in rat brain. J. Comp. Neurol. 2005;484:403–417. doi: 10.1002/cne.20411. [DOI] [PubMed] [Google Scholar]
- 13.Amiot C., Musard J. F., Hadjiyiassemis M., Jouvenot M., Fellmann D., Risold P. Y., Adami P. Expression of the secreted FAD-dependent sulfydryl oxidase (QSOX) in the guinea pig central nervous system. Brain Res. Mol. Brain Res. 2004;125:13–21. doi: 10.1016/j.molbrainres.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 14.Tury A., Mairet-Coello G., Poncet F., Jacquemard C., Risold P. Y., Fellmann D., Griffond B. QSOX sulfhydryl oxidase in rat adenohypophysis: localization and regulation by estrogens. J. Endocrinol. 2004;183:353–363. doi: 10.1677/joe.1.05842. [DOI] [PubMed] [Google Scholar]
- 15.Mairet-Coello G., Tury A., Esnard-Feve A., Fellmann D., Risold P. Y., Griffond B. FAD-linked sulfhydryl oxidase QSOX: topographic, cellular, and subcellular immunolocalization in adult rat central nervous system. J. Comp. Neurol. 2004;473:334–363. doi: 10.1002/cne.20126. [DOI] [PubMed] [Google Scholar]
- 16.Chakravarthi S., Bulleid N. J. Glutathione is required to regulate the formation of native disulfide bonds within proteins entering the secretory pathway. J. Biol. Chem. 2004;279:39872–39879. doi: 10.1074/jbc.M406912200. [DOI] [PubMed] [Google Scholar]
- 17.Gurevich V. V., Pokrovskaya I. D., Obukhova T. A., Zozulya S. A. Preparative in vitro mRNA synthesis using SP6 and T7 RNA polymerases. Anal. Biochem. 1991;195:207–213. doi: 10.1016/0003-2697(91)90318-n. [DOI] [PubMed] [Google Scholar]
- 18.Dalley J. A., Bulleid N. J. The endoplasmic reticulum (ER) translocon can differentiate between hydrophobic sequences allowing signals for glycosylphosphatidylinositol anchor addition to be fully translocated into the ER lumen. J. Biol. Chem. 2003;278:51749–51757. doi: 10.1074/jbc.M303978200. [DOI] [PubMed] [Google Scholar]
- 19.Wilson R., Allen A. J., Oliver J., Brookman J. L., High S., Bulleid N. J. The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem. J. 1995;307:679–687. doi: 10.1042/bj3070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jessop C. E., Bulleid N. J. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- 21.Tyson J. R., Stirling C. J. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frand A. R., Kaiser C. A. Ero1p oxidises protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 23.Xiao R., Wilkinson B., Solovyov A., Winther J. R., Holmgren A., Lundstrom-Ljung J., Gilbert H. F. The contributions of protein disulfide isomerase and its homologues to oxidative protein folding in the yeast endoplasmic reticulum. J. Biol. Chem. 2004;279:49780–49786. doi: 10.1074/jbc.M409210200. [DOI] [PubMed] [Google Scholar]
- 24.Hoober K. L., Sheasley S. L., Gilbert H. F., Thorpe C. Sulfhydryl oxidase from egg white. A facile catalyst for disulfide bond formation in proteins and peptides. J. Biol. Chem. 1999;274:22147–22150. doi: 10.1074/jbc.274.32.22147. [DOI] [PubMed] [Google Scholar]
- 25.Allen S., Naim H. Y., Bulleid N. J. Intracellular folding of tissue-type plasminogen activator. Effects of disulfide bond formation on N-linked glycosylation and secretion. J. Biol. Chem. 1995;270:4797–4804. doi: 10.1074/jbc.270.9.4797. [DOI] [PubMed] [Google Scholar]
- 26.Gross E., Sevier C. S., Heldman N., Vitu E., Bentzur M., Kaiser C. A., Thorpe C., Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. U.S.A. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoober K. L., Thorpe C. Egg white sulfhydryl oxidase: kinetic mechanism of the catalysis of disulfide bond formation. Biochemistry. 1999;38:3211–3217. doi: 10.1021/bi9820816. [DOI] [PubMed] [Google Scholar]
- 28.Frand A. R., Kaiser C. A. Two pairs of conserved cysteines are required for the oxidative activity of Ero1p in protein disulfide bond formation in the endoplasmic reticulum. Mol. Biol. Cell. 2000;11:2833–2843. doi: 10.1091/mbc.11.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamsa E., Simonen M., Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- 30.Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- 31.Frand A. R., Kaiser C. A. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 32.Simons J. F., Ebersold M., Helenius A. Cell wall 1,6-β-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 1998;17:396–405. doi: 10.1093/emboj/17.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joly J. C., Swartz J. R. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry. 1997;36:10067–10072. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 34.Endrizzi J. A., Breddam K., Remington S. J. 2.8-Å structure of yeast serine carboxypeptidase. Biochemistry. 1994;33:11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- 35.Wilkinson B., Xiao R., Gilbert H. F. A structural disulfide of yeast protein-disulfide isomerase destabilises the active site disulfide of the N-terminal thioredoxin domain. J. Biol. Chem. 2005;280:11483–11487. doi: 10.1074/jbc.M414203200. [DOI] [PubMed] [Google Scholar]
- 36.Tortorella D., Story C. M., Huppa J. B., Wiertz E. J., Jones T. R., Bacik I., Bennink J. R., Yewdell J. W., Ploegh H. L. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 1998;142:365–376. doi: 10.1083/jcb.142.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]