Abstract
We here report on a human myopathy associated with a mutation in a fast myosin heavy chain (MyHC) gene, and also the genetic defect in a hereditary inclusion body myopathy. The disorder has previously been described in a family with an “autosomal dominant myopathy, with joint contractures, ophthalmoplegia, and rimmed vacuoles.” Linkage analysis and radiation hybrid mapping showed that the gene locus (Human Genome Map locus name: IBM3) is situated in a 2-Mb region of chromosome 17p13, where also a cluster of MyHC genes is located. These include the genes encoding embryonic, IIa, IIx/d, IIb, perinatal, and extraocular MyHCs. Morphological analysis of muscle biopsies from patients from the family indicated to us that the type 2A fibers frequently were abnormal, whereas other fiber types appeared normal. This observation prompted us to investigate the MyHC-IIa gene, since MyHC-IIa is the major isoform in type 2A fibers. The complete genomic sequence for this gene was deduced by using an “in silico” strategy. The gene, found to consist of 38 exons, was subjected to a complete mutation scan in patients and controls. We identified a missense mutation, Glu-706 → Lys, which is located in a highly conserved region of the motor domain, the so-called SH1 helix region. By conformational changes this region communicates activity at the nucleotide-binding site to the neck region, resulting in the lever arm swing. The mutation in this region is likely to result in a dysfunctional myosin, compatible with the disorder in the family.
We have recently identified a Swedish family with an autosomal dominant myopathy, which is manifested at birth by multiple joint contractures (1). In childhood and adolescence the myopathy appeared to be mild in most cases. In several affected family members the disease showed a progressive course from 30 to 50 years of age with increasing muscle weakness and elevated levels of creatine kinase in serum. The muscle weakness was predominantly proximal, and external ophthalmoplegia was a consistent finding. While young individuals showed minor histopathological changes, such as variability in fiber size, central nuclei, and focal disorganization of myofilaments, the oldest patients showed marked pathological alterations (1). These included dystrophic changes and the occurrence of rimmed vacuoles, which were very frequent in one case. In these cases cytoplasmic and nuclear inclusions of 15- to 20-nm tubulofilaments were present. These similarities with sporadic inclusion body myositis (sIBM), suggested that this disorder should be considered as a variant of hereditary inclusion body myopathy (hIBM) (2).
Myosin is a molecular motor protein that transduces chemical energy of ATP hydrolysis into mechanical force. Several genes encode myosin heavy chains (MyHCs) in striated muscle of mammals. In humans, α and slow/β MyHCs are encoded by genes located on chromosome 14, whereas the genes encoding embryonic, IIa, IIx/d, IIb, perinatal, and extraocular MyHCs are located in a cluster on chromosome 17 (3). Numerous point mutations in the gene encoding slow/β MyHC have been reported in association with familial hypertrophic cardiomyopathy (FHC; ref. 4). Such mutations may also affect slow, type 1, muscle fiber structure and function (5, 6), but myopathies associated with mutations in any of the fast MyHC genes located on chromosome 17 have so far not been reported (7).
By linkage analysis and radiation hybrid mapping we localized the disease locus (Human Genome Map locus IBM3) to a 2-Mb chromosomal region in 17p13.1 and we also showed that the MyHC gene cluster colocalized to the same region (8). The region covering the MyHC gene cluster has recently been sequenced, and this made it possible for us to deduce the genomic structure of the different MyHC genes in the cluster. Thus, more specific primers, situated in the introns, could be constructed and used in the analysis. This enabled us to explore the genes for mutations in greater detail.
In the present study further analysis of muscle biopsies from patients from the family indicated to us that the type 2A fibers frequently were abnormal, whereas other fiber types appeared normal. Because myosin type IIa is the main myosin isoform in type 2A fibers (9) we investigated the MyHC-IIa gene by using a complete mutation scan of all 38 exons. Subsequently, we identified a missense mutation in the MyHC-IIa gene located in a highly conserved region of the motor domain, the so-called SH1 helix region.
We here present a human myopathy associated with a mutation in a fast MyHC, and also we have identified a genetic defect in hereditary inclusion body myopathy (10). Furthermore, we present the complete genomic structure of the MyHC-IIa gene.
Materials and Methods
Patient Material.
The family has been described in detail (1, 8). Genomic DNA was extracted from EDTA-anticoagulated blood samples of all investigated family members by using phenol. Muscle tissue was obtained by open biopsy, and the tissue specimens were immediately frozen in liquid nitrogen. The numbering of the cases is in agreement with that reported earlier (1, 8).
Enzyme Histochemical Staining and Electron Microscopy.
Sections of fresh-frozen muscle tissue were stained with hematoxylin/eosin and modified Gomori trichrome. Sections were also incubated for NADH-tetrazolium reductase and myofibrillar ATPase at pH 10.4, 4.3, and 4.6 to identify structural changes in different muscle fiber types (11). Electron microscopy was performed as previously described (1).
“In Silico” Determination of MyHC-IIa Genomic Sequence.
The complete 5,956-bp cDNA for the MyHC-IIa gene was derived from GenBank (accession no. AF111784). The complete raw genomic sequence of the chromosome 17 MyHC gene cluster region has recently been made available on the Internet through the large-scale sequencing efforts of the Whitehead Institute/MIT Center for Genome Research. A bacterial artificial chromosome (BAC) clone containing the complete genomic sequence for this gene was derived by analysis using blast through URL http://www.ncbi.nlm.nih.gov. The BAC clone, 799N11 (GenBank AC005323), and its neighboring BAC clones 279N7 (AC002347) and 401O9 (AC005291) has been connected to a 533,641-bp contig (NT_000757). Six MyHC genes could all be found on this contig. The detailed analysis of the exon–intron boundaries of 38 exons could be deduced by comparison of the cDNA sequence and the genomic DNA sequence of clone 799N11 and by applying the GT-AG rule (Table 1, which is published as supplementary material on the PNAS web site, www.pnas.org).
PCR Conditions, DNA Sequencing, and Mutation Screening.
PCR primer pairs flanking all 38 exons were constructed by using the primerselect software (Lasergene, DNAStar, Madison, WI). Some exons situated close to each other were amplified together (Table 2, which is published as supplementary material on the PNAS web site). The PCR amplifications were carried out in total volumes of 20 μl including 1× PCR buffer (Pharmacia–Amersham) with 1.5 mM MgCl2, 100 ng of genomic DNA, 0.2 mM dNTPs, Taq DNA polymerase (0.3 units), and 13 pmol of each primer (Table 2). The annealing temperature was 57°C for all primer pairs used. The PCR fragments were purified by using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). The sequencing reactions were performed by using Big Dye Terminator chemistry (PE Biosystems, Foster City, CA) with the same primers as for the PCRs, forward and reverse primers in separate experiments. The sequencing reactions were analyzed on an ABI 310 Sequencer (PE Biosystems). All exons were sequenced from both directions. Screening for the 2116G → A (Glu-706 → Lys) mutation in DNA from all available family members (1, 8) and in 129 healthy control individuals was performed with single-strand conformation polymorphism (SSCP) analysis using the Phast system (Pharmacia) as described earlier (12).
Results
Enzyme Histochemical Staining and Electron Microscopy.
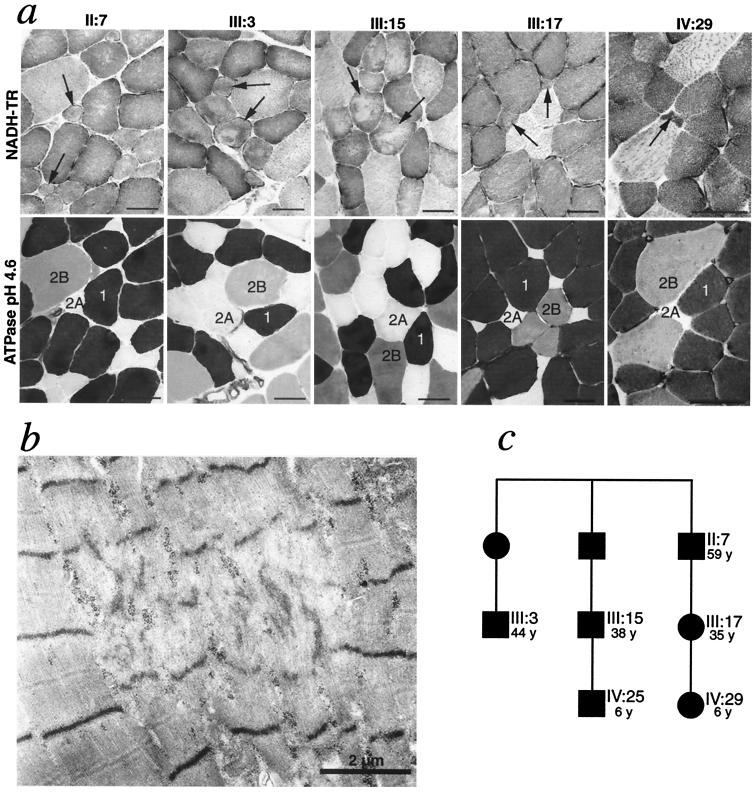
The involvement of different fiber types was analyzed by enzyme histochemical staining of myofibrillar ATPase and NADH-tetrazolium reductase on serial sections in muscle biopsies from six affected patients. In two patients (cases IV:29 and III:17; Fig. 1c) the type 2A fibers were small, infrequent (<1% and 8% of all fibers, respectively), and showed structural alterations (Fig. 1a). The other fiber types appeared structurally normal. In less affected muscles and muscle fascicles of the older patients (cases II:7, III:3, and III:15) morphological changes were mainly restricted to type 2A fibers, which varied in size and showed structural changes (Fig. 1a). Ultrastructural investigation in these five patients revealed as the main alteration focal disorganization of contractile filaments (Fig. 1b). In one additional case (IV:25) no type 2A fibers were identified but otherwise there were no light microscopical changes. There was no apparent loss of type 2B fibers in any case. In more severely affected muscles and muscle fascicles of the three oldest individuals all fiber types showed morphological changes, including rimmed vacuoles and inclusion consisting of 15- to 20-nm tubulofilaments (Fig. 2) as previously described (1). Because MyHC-IIa is the main MyHC isoform in type 2A fibers (9), and MyHC-IIa is a major constituent of extraocular muscles (13), which were involved in all affected family members, the MyHC-IIa was regarded a very strong candidate gene for the disease.
Figure 1.
Analysis of involvement of different muscle fiber types. (a) Analysis by enzyme histochemical staining of myofibrillar ATPase (Lower) and NADH-tetrazolium reductase (NADH-TR; Upper). The type 2A fibers are small and show structural alterations. In case III:17 and IV:29 they were reduced in number. In case IV:25 no type 2A fibers were identified in the biopsy. (Bars represent 40 μm.) (b) Ultrastructural investigation of case III:17 revealed scattered small muscle fibers with multiple foci of disorganization of myofilaments. (c) Part of the pedigree, showing the age and relation of the family members, all with myopathy, illustrated in a. Numbering of the patients refer to the original description of this family (1). Squares and circles correspond to males and females, respectively.
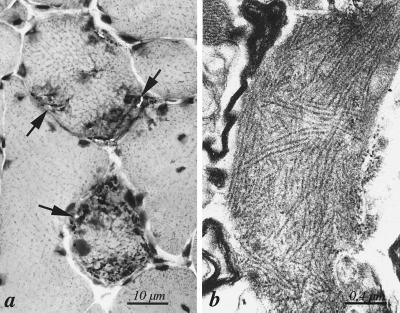
Figure 2.
Morphological features of hereditary inclusion body myopathy in one patient. (a) Section of quadriceps femoris muscle of patient III:15 showing two muscle fibers with multiple rimmed vacuoles (arrows). Hematoxylin–eosin. (b) Collections of cytoplasmic 15- to 20-nm tubulofilaments in association with rimmed vacuoles in patient III:15
“In Silico” Determination of the Genomic Organization of the MyHC-IIa Gene.
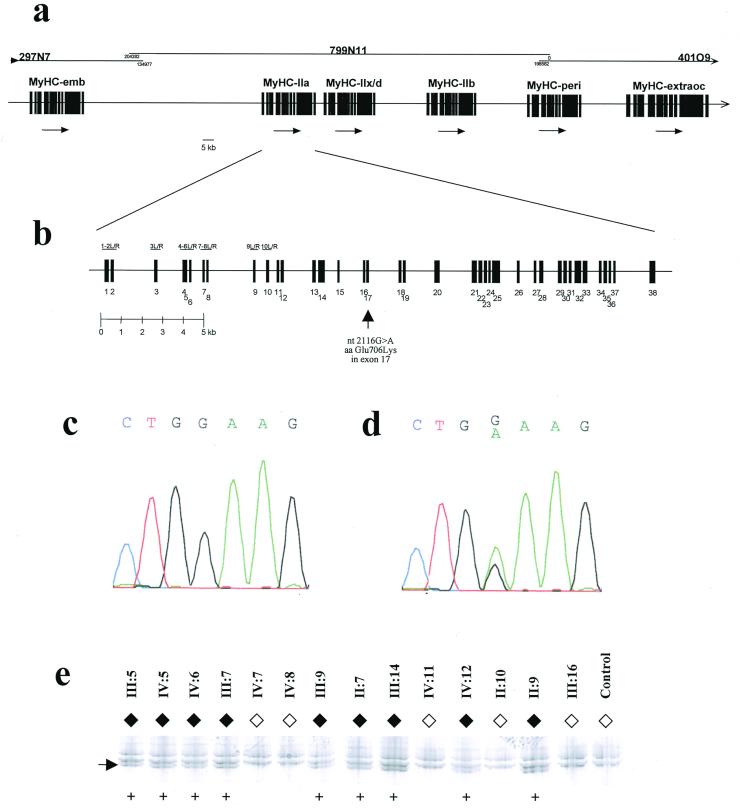
The cDNA sequence for the MyHC-IIa gene was scanned against GenBank by using blast, and an almost perfect hit came up from a genomic region on a recently sequenced 204,282-bp BAC (799N11). Several hits from the same BAC and the two flanking BACs (297N7 and 401O9) also came up. However, these did not have 100% match to the MyHC-IIa gene but instead to one each of five other members of the MyHC gene family. By using this “in silico” strategy it was possible to deduce the genomic structure of six members of the MyHC cluster on the three BAC clones (Fig. 3a). The three completely sequenced BAC clones are shown in Fig. 3a, together with an overview of the genomic organization of six members of the MyHC gene cluster. The detailed organization of the MyHC-IIa gene is shown in Fig. 3b. MyHC-IIa was shown to contain 38 exons (Fig. 3b and Table 1), and PCR primers flanking these exons were constructed (Table 2).
Figure 3.
Genomic analysis of the MyHC-IIa gene in patients and control. (a) Genomic organization of the six MyHC genes in 17p13.1. The three completely sequenced BAC clones are shown at the top, and the detailed organization of the MyHC-IIa gene is shown in b. Also in b, the location of the mutation 2116G → A in exon 17 is depicted with an arrow. PCR fragments used are indicated for exons 1–10 only. (c) DNA sequencing of the region of the 2116G → A mutation from a healthy control individual. (d) DNA sequencing from the same region in patient IV:15. (e) SSCP detection of the 2116G → A mutation in several other members of the studied family. The numbering refers to the family pedigree presented earlier (8). The aberrant band indicated by an arrow was present only in affected family members (♦) and never in unaffected (⋄). Presence of aberrant band is indicated (+). None of 129 control individuals (258 chromosomes) carried this mutation (data not shown).
Screen for MyHC-IIa Mutations in DNA from Patients with Hereditary Myopathy.
Subsequently all exons were sequenced in patients and healthy control individuals. One mutation was identified in patients (Fig. 3d) and never detected in controls (Fig. 3c)—i.e., a missense mutation in exon 17 nucleotide 2116, G → A, giving rise to an amino acid shift aa Glu-706 → Lys (E706K). Glu-706 is located in the SH1 helix region, which is located in the core of the myosin head (14, 15) (Fig. 4a).
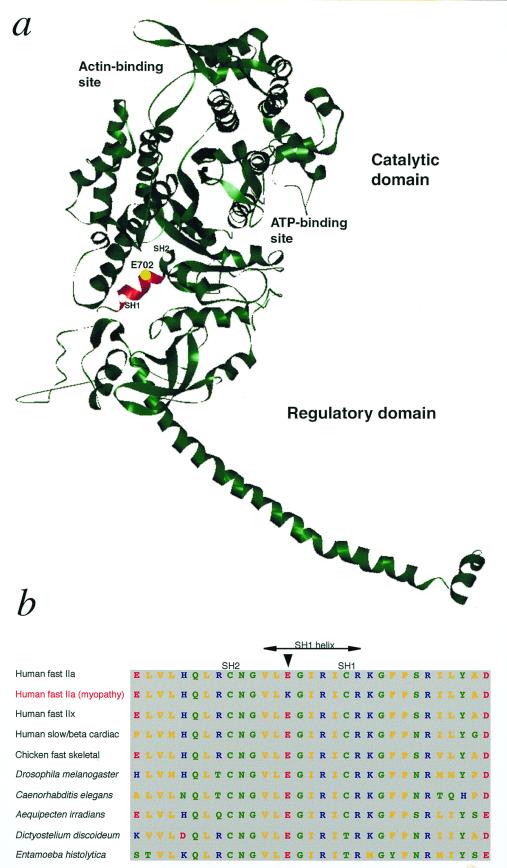
Figure 4.
MyHC with the position of the mutation. (a) Ribbon model of MyHC subfragment 1 (S1) of chicken skeletal muscle (15). The ATP- and actin-binding sites are indicated. The highly conserved SH1 helix (Val-700 through Arg-708) is shown in red, and a yellow sphere indicates the position of E702 (corresponding to E706 in human MyHC-IIa), which was mutated (Glu-706 → Lys) in the family members with myopathy. SH1 and SH2 refer to two reactive sulfhydryl groups (Cys-707 and Cys-697 of chicken skeletal muscle MyHC), which may be crosslinked in the presence of nucleotide. The structure of the MyHC molecule was obtained at http://www.mrc-lmb.cam.ac.uk/and the ribbon diagram was prepared by WebLab viewerlite 3.2 software (http://www.msi.com). (b) Comparison of amino acid sequences in the conserved SH1 helix region of myosin class II (conventional myosin) in various species and organisms (data derived from The Myosin Home Page, http://www.mrc-lmb.cam.ac.uk/myosin/trees/color.html; T. Hodge and J. Cope). An arrow indicates the residue E706 in human myosin, which was mutated in MyHC-IIa in family members with myopathy. Color codes for the amino acids: yellow, nonpolar; green, uncharged polar and glycine; red, acidic; and blue, basic.
The 2116G → A mutation was also readily detected as an aberrant band with the SSCP technique, and several of the members of the studied family were analyzed (Fig. 3e). The aberrant band was present only in affected family members and never in unaffected family members. None of 129 control individuals (258 chromosomes) carried this mutation (data not shown).
Discussion
This study presents a myopathy that is associated with a mutation in a gene encoding a fast MyHC. By screening of the entire MyHC-IIa gene we identified a mutation, 2116G → A (Glu-706 → Lys), in affected family members. There are several lines of evidence that this mutation in the MyHC-IIa gene is the cause of the congenital myopathy in this family. (i) The mutation segregates with the disease in the family and was not identified in controls. (ii) Glu-706 is located in the SH1 helix, in the core of the myosin head, which is of major importance for the function of myosin (Fig. 4a). Conformational changes upon binding and hydrolysis of ATP in the catalytic region are transmitted via the SH1 helix region, which constitutes a joint or fulcrum, to the regulatory domain, which acts like a lever arm (3, 15, 16). According to recent data the SH1 helix is unwound at one state of the contractile cycle, which may correspond to a prehydrolysis state (17). (iii) Glu-706 is 100% conserved in conventional class II myosins and 100% conserved in all known myosin classes (Fig. 4b) (14). (iv) The mutation causes a change from a negatively charged amino acid to a positively charged residue. (v) Morphological changes were in some patients completely restricted to type 2A fibers, and predominantly affected type 2A fibers in the other examined patients. This would be expected if there is a pathogenic mutation in the MyHC-IIa gene, because type IIa MyHC is the main constituent of type 2A fibers (9). Accordingly, mutations in β cardiac/slow MyHC have been reported to affect slow, type 1, muscle fiber structure and function (5, 6). (vi) External ophthalmoplegia is compatible with a pathogenic mutation affecting IIa MyHC because this is a major isoform of myosin in extraocular muscle (13). (vii) Targeted disruption of fast MyHC genes in mice causes myopathy with muscle weakness and disorganization of myofilaments (18), and point mutations in MyHC genes cause muscle dysfunction in Drosophila, Caenorhabditis elegans, and Dictyostelium (16).
MyHC gene missense mutations may cause dominant negative effects on muscle function by various mechanisms, including interference with filament assembly and sarcomeric structure, functional defects in ATPase activity, impaired myosin–actin interaction, and perturbation of conformational changes during ATP hydrolysis (16, 19, 20). In familial hypertrophic cardiomyopathy associated with β MyHC mutations it has been considered that myocardial hypertrophy and disarray of myocytes are secondary changes caused by defective sarcomeric function (20). The mechanism by which the MyHC-IIa Glu-706 → Lys mutation causes myopathy remains to be elucidated. Because the mutation is located in a region that is important for the conformational changes that takes place during ATP hydrolysis, it may cause a perturbation of motility.
There are several clinical and morphological observations in this family that raise questions on the relation between genotype and phenotype. The early involvement with congenital joint contractures, which resolve more or less spontaneously during childhood, may be due to developmental factors involved in the regulation of myosin isoform expression (21). In several family members the myopathy showed a progressive course, which appeared in adulthood. Although type 2A fibers appeared to be selectively involved in young individuals, we noted that all muscle fiber types, as assessed by enzyme histochemistry, showed morphological changes in the severely affected muscles of the adult, progressive patients. This phenomenon may possibly be explained by the increased proportion of hybrid muscle fibers, expressing more than one MyHC isoform, which occurs normally in aging (22). An additional possibility would be an increased expression of MyHC-IIa in these individuals with increasing age. Since physical training and inactivity influence MyHC isoform expression (23–25), the type and amount of physical activity may be important factors for the phenotypic expression of the disease in this family.
The morphological features with rimmed vacuoles and filamentous inclusions that appeared in adult progressive patients are of considerable interest in relation to the pathogenesis of rimmed vacuoles in sporadic inclusion body myositis, which is the most common myopathy presenting after 50 years of age (26). A possible link between rimmed vacuoles and MyHC defects could be that degradation of myosin is, at least partly, mediated by proteasomes (27) and that the ATP–ubiquitin–proteasome proteolytic pathway may participate in muscle fiber degeneration in distal myopathy with rimmed vacuoles (28). Increased degradation of myosin, possibly in addition to other genetic factors and aging, may contribute to the formation of rimmed vacuoles.
This paper also presents the genomic sequence for MyHC-IIa. Genetics studies of separate members of the MyHC gene cluster at the cDNA level have been difficult because of the vast homology between the different members, also at the DNA level. We have made use of the increasing sequencing information from the large sequencing projects. The complete raw genomic sequence of the chromosome 17 MyHC gene cluster region has recently been made available on the Internet through the large-scale sequencing efforts of Whitehead Institute/MIT Center for Genome Research. Thus, to study the MyHC-IIa gene we first deduced the exon–intron structure of the gene by comparing the earlier published cDNA sequence with the recently available raw genomic sequence—i.e., through an “in silico” strategy.
Although this is, to our knowledge, the first myopathy shown to be associated with a mutation in a fast MyHC isoform (7), defects in other sarcomeric proteins have recently been shown to cause myopathy. Mutations in the slow α-tropomyosin (TPM3) and nebulin genes are associated with nemaline myopathy (29, 30), mutations in the skeletal muscle α-actin gene are associated with actin myopathy and nemaline myopathy (31), and mutations in the myosin light chain are associated with a myopathy in human heart and skeletal muscle (32). In addition, titin is a candidate gene in tibial muscular dystrophy, another autosomal dominant myopathy with rimmed vacuoles (33). It is likely that other congenital myopathies, possibly variants of “multiminicore” or “minimal change” myopathies in addition to hereditary myopathies with rimmed vacuoles, such as other hereditary inclusion body myopathies and distal myopathies, will turn out to be disorders of fast MyHC isoforms.
Supplementary Material
Acknowledgments
We are grateful for the financial support from the Medical Research Council (Project no. 7122, A.O., and no. 11255, J.W.), Gretchen Olsjös Donationsfond, Linnea och Josef Carlssons Forskningsfond, Göteborgs Barnklinikers Forskningsfond, Swedish Rheumatism Association, and the King Gustaf V 80 Years Anniversary Fund.
Abbreviations
- MyHC
myosin heavy chain
- BAC
bacterial artificial chromosome
- SSCP
single-strand conformation polymorphism
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250289597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250289597
References
- 1.Darin N, Kyllerman M, Wahlström J, Martinsson T, Oldfors A. Ann Neurol. 1998;44:242–248. doi: 10.1002/ana.410440215. [DOI] [PubMed] [Google Scholar]
- 2.Askanas V, Engel W K. Curr Opin Rheumatol. 1998;10:530–542. doi: 10.1097/00002281-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Weiss A, Schiaffino S, Leinwand L A. J Mol Biol. 1999;290:61–75. doi: 10.1006/jmbi.1999.2865. [DOI] [PubMed] [Google Scholar]
- 4.Bonne G, Carrier L, Richard P, Hainque B, Schwartz K. Circ Res. 1998;83:580–593. doi: 10.1161/01.res.83.6.580. [DOI] [PubMed] [Google Scholar]
- 5.Cuda G, Fananapazir L, Zhu W S, Sellers J R, Epstein N D. J Clin Invest. 1993;91:2861–2865. doi: 10.1172/JCI116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fananapazir L, Dalakas M C, Cyran F, Cohn G, Epstein N D. Proc Natl Acad Sci USA. 1993;90:3993–3997. doi: 10.1073/pnas.90.9.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laing N G. Curr Opin Neurol. 1999;12:513–518. doi: 10.1097/00019052-199910000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Martinsson T, Darin N, Kyllerman M, Oldfors A, Hallberg B, Wahlström J. Am J Hum Genet. 1999;64:1420–1426. doi: 10.1086/302375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pette D, Staron R S. Int Rev Cytol. 1997;170:143–223. doi: 10.1016/s0074-7696(08)61622-8. [DOI] [PubMed] [Google Scholar]
- 10.Argov Z, Eisenberg I, Mitrani-Rosenbaum S. Curr Opin Rheumatol. 1998;10:543–547. doi: 10.1097/00002281-199811000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Dubowitz V, Brooke M H. Muscle Biopsy–A Modern Approach. London: Saunders; 1973. [Google Scholar]
- 12.Sjoberg R M, Hallstensson K, Nordling M, Kogner P, Gardellin P, Lubyova B, Onyango P, Weith A, Martinsson T. Int J Oncol. 1996;8:1137–1142. doi: 10.3892/ijo.8.6.1137. [DOI] [PubMed] [Google Scholar]
- 13.Asmussen G, Traub I, Pette D. FEBS Lett. 1993;335:243–245. doi: 10.1016/0014-5793(93)80738-g. [DOI] [PubMed] [Google Scholar]
- 14.Cope M J T, Whisstock J, Rayment I, Kendrick-Jones J. Structure. 1996;4:969–987. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- 15.Rayment I, Rypniewski W R, Schmidt-Base K, Smith R, Tomchick D R, Benning M M, Winkelmann D A, Wesenberg G, Holden H M. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 16.Ruppel K M, Spudich J A. Annu Rev Cell Dev Biol. 1996;12:543–573. doi: 10.1146/annurev.cellbio.12.1.543. [DOI] [PubMed] [Google Scholar]
- 17.Houdusse A, Kalabokis V N, Himmel D, Szent-Györgyi A G, Cohen C. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 18.Acakpo-Satchivi L J, Edelmann W, Sartorius C, Lu B D, Wahr P A, Watkins S C, Metzger J M, Leinwand L, Kucherlapati R. J Cell Biol. 1997;139:1219–1229. doi: 10.1083/jcb.139.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker K D, Gottshall K R, Hickey R, Perriard J C, Chien K R. J Cell Biol. 1997;137:131–140. doi: 10.1083/jcb.137.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roopnarine O, Leinwand L A. Biophys J. 1998;75:3023–3030. doi: 10.1016/S0006-3495(98)77743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiaffino S, Reggiani C. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 22.Andersen J L, Terzis G, Kryger A. Muscle Nerve. 1999;22:449–454. doi: 10.1002/(sici)1097-4598(199904)22:4<449::aid-mus4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Andersen J L, Klitgaard H, Bangsbo J, Saltin B. Acta Physiol Scand. 1994;150:21–26. doi: 10.1111/j.1748-1716.1994.tb09655.x. [DOI] [PubMed] [Google Scholar]
- 24.Andersen J L, Gruschy-Knudsen T, Sandri C, Larsson L, Schiaffino S. J Appl Physiol. 1999;86:455–460. doi: 10.1152/jappl.1999.86.2.455. [DOI] [PubMed] [Google Scholar]
- 25.Williamson D L, Godard M P, Porter D A, Costill D L, Trappe S W. J Appl Physiol. 2000;88:627–633. doi: 10.1152/jappl.2000.88.2.627. [DOI] [PubMed] [Google Scholar]
- 26.Oldfors A, Lindberg C. Curr Opin Neurol. 1999;12:527–533. doi: 10.1097/00019052-199910000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Eble D M, Spragia M L, Ferguson A G, Samarel A M. Cell Tissue Res. 1999;296:541–548. doi: 10.1007/s004410051315. [DOI] [PubMed] [Google Scholar]
- 28.Kumamoto T, Fujimoto S, Nagao S, Masuda T, Sugihara R, Ueyama H, Tsuda T. Intern Med. 1998;37:746–752. doi: 10.2169/internalmedicine.37.746. [DOI] [PubMed] [Google Scholar]
- 29.Pelin K, Hilpela P, Donner K, Sewry C, Akkari P A, Wilton S D, Wattanasirichaigoon D, Bang M L, Centner T, Hanefeld F, et al. Proc Natl Acad Sci USA. 1999;96:2305–2310. doi: 10.1073/pnas.96.5.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laing N G, Wilton S D, Akkari P A, Dorosz S, Boundy K, Kneebone C, Blumbergs P, White S, Watkins H, Love D R, Haan E. Nat Genet. 1995;9:75–79. doi: 10.1038/ng0195-75. [DOI] [PubMed] [Google Scholar]
- 31.Nowak K J, Wattanasirichaigoon D, Goebel H H, Wilce M, Pelin K, Donner K, Jacob R L, Hubner C, Oexle K, Anderson J R, et al. Nat Genet. 1999;23:208–212. doi: 10.1038/13837. [DOI] [PubMed] [Google Scholar]
- 32.Poetter K, Jiang H, Hassanzadeh S, Master S R, Chang A, Dalakas M C, Rayment I, Sellers J R, Fananapazir L, Epstein N D. Nat Genet. 1996;13:63–69. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 33.Haravuori H, Makela-Bengs P, Udd B, Partanen J, Pulkkinen L, Somer H, Peltonen L. Am J Hum Genet. 1998;62:620–626. doi: 10.1086/301752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.