Abstract
The calcium- and calmodulin-dependent protein phosphatase calcineurin has been implicated in the transduction of signals that control the hypertrophy of cardiac muscle and slow fiber gene expression in skeletal muscle. To identify proteins that mediate the effects of calcineurin on striated muscles, we used the calcineurin catalytic subunit in a two-hybrid screen for cardiac calcineurin-interacting proteins. From this screen, we discovered a member of a novel family of calcineurin-interacting proteins, termed calsarcins, which tether calcineurin to α-actinin at the z-line of the sarcomere of cardiac and skeletal muscle cells. Calsarcin-1 and calsarcin-2 are expressed in developing cardiac and skeletal muscle during embryogenesis, but calsarcin-1 is expressed specifically in adult cardiac and slow-twitch skeletal muscle, whereas calsarcin-2 is restricted to fast skeletal muscle. Calsarcins represent a novel family of sarcomeric proteins that link calcineurin with the contractile apparatus, thereby potentially coupling muscle activity to calcineurin activation.
Calcineurin is a calcium/calmodulin-dependent serine, threonine phosphatase that plays an important role in transducing calcium-dependent signals in a variety of cell types (1). Calcineurin exists as a heterodimer comprising a catalytic A subunit (CnA) that contains a calmodulin-binding site and an autoinhibitory domain and a regulatory B-subunit that binds calcium. Activation of calcineurin in vivo occurs in response to sustained, low-amplitude calcium transients evoked by ligand binding to cell surface receptors (2, 3). The functions of calcineurin have been studied most extensively in T lymphocytes, where calcineurin transduces immunogenic signals in response to T cell receptor activation. Activation of the immune response by calcineurin is coupled to dephosphorylation of the nuclear factor of activated T cells (NFAT) family of transcription factors, which results in their translocation to the nucleus, where they associate combinatorially with other transcription factors to activate transcription through composite DNA sequence elements (4). Calcineurin also associates with and dephosphorylates a variety of other proteins, many of which regulate calcium homeostasis (1, 5).
Recently, calcineurin has been shown to have a profound influence on the properties of striated muscle cells. In cardiac muscle, calcineurin is activated by hypertrophic agonists, as well as by alterations in sarcomere function, which are known to alter calcium handling (3, 6). Overexpression of a constitutively active form of calcineurin in hearts of transgenic mice is also sufficient to induce cardiac hypertrophy that progresses to heart failure and sudden death (3). Conversely, inhibition of calcineurin activity with cyclosporine A (CsA) can block hypertrophic growth of cardiomyocytes in response to a variety of intrinsic and extrinsic stimuli (reviewed in ref. 7). In skeletal muscle, calcineurin activation has been shown to be necessary for hypertrophic growth in response to insulin-like growth factor-1, which can mobilize intracellular calcium (8). Calcineurin has also been demonstrated to stimulate the slow-twitch phenotype of skeletal muscle, which is dependent on sustained calcium signaling (9, 10).
Although the principal transcriptional targets for calcineurin signaling appear to be similar in diverse tissues, it remains to be determined whether calcineurin signaling may be specialized in different cell types through the involvement of tissue-specific substrates or binding proteins. Given the differences in duration and amplitude of calcium transients in T cells and striated muscle cells and the significantly different subcellular organization of these cell types, it seems likely that the calcineurin pathway may be modulated in a tissue-specific fashion.
In an effort to identify potential cardiac-specific regulators of calcineurin, we conducted a yeast-two hybrid screen, using the CnA subunit as bait. Here, we describe a novel family of striated muscle-specific calcineurin-interacting proteins called calsarcins. Calsarcins interact and colocalize with the z-disk protein α-actinin in vitro and in vivo and thereby tether calcineurin to the sarcomere of cardiac and skeletal muscle. These properties of calsarcins suggest an important role for these proteins in modulating the function and substrate specificity of calcineurin in striated muscle cells.
Materials and Methods
Yeast Two-Hybrid Screens.
A full-length mouse CnA-α cDNA, fused to the GAL4 DNA binding domain, was used as bait in a two-hybrid screen of approximately 1.5 × 106 clones of a human heart cDNA library (CLONTECH), as described (3). From this screen, we identified a cDNA encoding calsarcin-1. Additional two-hybrid screens of the same cDNA library were performed, using calsarcin-1 and calsarcin-2 as bait.
Northern Blot Analysis.
Northern blots of RNA from human and mouse multiple tissues (CLONTECH) as well as from C2C12 cell extracts were performed as described (11).
Generation of Calsarcin Antiserum and Western Blots.
A rabbit antiserum was generated against the complete ORF of calsarcin-1 fused in-frame with glutathione S-transferase. IgG was purified from rabbit serum and used for Western blotting and immunostaining.
Radioactive In Situ Hybridization.
RNA probes corresponding to the sense and antisense strands of calsarcin-1 and calsarcin-2 cDNAs were prepared by using T7 and T3 RNA polymerase (Roche Molecular Biochemicals) and 35S-labeled UTP. Sections of mouse embryos and adult hindlimbs were subjected to in situ hybridization, as described (12).
Cell Culture, Transfections, and Immunoprecipitations.
Cos-7 cells were maintained in DMEM containing 10% FBS. Cells (2 × 105) were transfected with 1 μg of expression plasmids for full-length and truncated forms of calsarcin-1 and calsarcin-2, CnA, and α-actinin-2, using FuGENE 6 reagent (Roche Molecular Biochemicals). Calsarcin peptides were fused with a N-terminal hemagglutinin-epitope or a C-terminal Myc-epitope, α-actinin-2 was fused with N-terminal Myc- or FLAG-epitopes, and CnA constructs were fused with a N-terminal FLAG epitope. Forty-eight hours after transfection, cells were harvested in ELB buffer, containing 50 mM Hepes (pH 7.0), 250 mM NaCl, 5 mM EDTA, 0.1% NP-40, 1 mM DTT, 1 mM PMSF, and protease inhibitors (Complete; Roche Molecular Biochemicals). Cells were briefly sonicated, and debris was removed by centrifugation. Tagged proteins were immunoprecipitated for 2–3 h at 4°C, using protein A/G agarose and 1 μg of the appropriate antibody (monoclonal anti-FLAG, monoclonal anti-Myc, and polyclonal anti-Myc). Subsequently, the pellet was washed with ELB buffer, subjected to SDS/PAGE, transferred to polyvinylidene membranes, and immunoblotted using anti-FLAG, anti-Myc, or anti-hemagglutinin-antibodies, respectively.
Immunostaining.
The subcellular localization of calsarcin-1, α-actinin, and CnA was determined in neonatal rat cardiomyocytes, which were harvested and maintained as described (3). Immunostaining was performed as described (11). The following antibodies were used: anti-calsarcin-1, anti-sarcomeric α-actinin (Sigma), anti-CnA (Sigma and Transduction Laboratories, Lexington, KY); secondary antibodies: anti-mouse/rabbit, Texas red, and FITC-labeled (Vector Laboratories), respectively. Cryosections of mouse heart and skeletal muscle were fixed in 3.7% formalin for 3 min, permeabilized in 0.3% Triton X-100 for 5 min, and subsequently stained as described above.
Mapping of Calsarcin-1 Interaction Domains.
Several N- and C-terminal truncations of calsarcin-1 were fused in-frame with the GAL4 DNA-binding domain in vector pAS1. CnA and α-actinin were fused with the GAL4 transactivation domain in the two-hybrid vector pACT2. Because both full-length and constitutively active CnA displayed background β-galactosidase activity when transfected alone, a mutated CnA lacking enzymatic activity (13) was used in subsequent experiments and did not display any background signal. Calsarcin-1 constructs were cotransformed with CnA, α-actinin, or pACT2 (as negative control) and grown on appropriate selective medium for 3 days. β-Galactosidase activity was determined with filter-lift assays as described (14) and monitored for 1–4 h. Because several C-terminal truncations of calsarcin-1 exhibited β-galactosidase activity when cotransformed with pACT2, complementary coimmunoprecipitation experiments were performed to further define calsarcin's interaction domains for CnA and α-actinin, as described above.
Results
Cloning and Characterization of Calsarcin-1 and Calsarcin-2 cDNA.
The CnA subunit was fused to the DNA-binding domain of yeast GAL4 and used as bait in a two-hybrid screen of an adult human heart cDNA library. One of the positive clones identified in the screen contained a 2,422-bp cDNA with an ORF encoding a novel 264-aa protein (Fig. 1). We named this protein calsarcin-1 (calcineurin-associated sarcomeric protein-1, GenBank accession no. AY013295). The interaction of calsarcin-1 with CnA was confirmed by retransformation of both cDNAs in yeast, followed by β-galactosidase assays.
Figure 1.
Protein sequence alignment of human calsarcin-1 and calsarcin-2. The deduced amino acid sequences of human calsarcin-1 and calsarcin-2 are shown. Conserved amino acids are highlighted.
Several cardiac- and muscle-derived human and mouse expressed sequence tags matched the sequence of the calsarcin-1 cDNA and contained in-frame termination codons 5′ of the putative translation initiation codon, suggesting that the complete ORF was contained in the original cDNA. Using public expressed sequence tag database information, overlapping sequences for the mouse homolog of calsarcin-1 were also identified (GenBank accession no. AY013296). Sequence comparison revealed 88% amino acid identity between the mouse and human sequences. Expressed sequence tag searches also identified a related cDNA encoding a protein we named calsarcin-2 (GenBank accession nos. AY013297 and AY013298) (Fig. 1).
Calsarcin-1 and calsarcin-2 show the highest homology toward their amino and carboxyl termini, whereas the intervening amino acids are less well conserved (Fig. 1). blast searches with both protein sequences did not reveal any significant homology to known proteins.
Muscle-Specific Expression of Calsarcin-1 and Calsarcin-2.
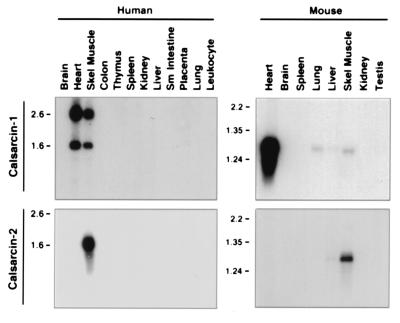
Northern blot analysis of human and mouse tissues revealed a highly striated muscle-specific expression pattern of calsarcin-1 and calsarcin-2 (Fig. 2). Calsarcin-1 is specifically expressed in heart and skeletal muscle, with two mRNAs of 1.6 and 2.6 kb in human tissues, and only a single transcript of approximately 1.3 kb in mouse. Faint expression of calsarcin-1 was also detected in mouse lung and liver. Calsarcin-2 transcripts of 1.6 kb and 1.3 kb were detected exclusively in adult human and mouse skeletal muscle, respectively. The relative difference in expression level of calsarcin-1 between human and mouse skeletal muscle may reflect differences in slow- versus fast-twitch fiber composition of the muscle tissue (see below).
Figure 2.
Northern blot analysis of calsarcin-1 and calsarcin-2 in adult human and mouse tissues. Calsarcin transcripts were detected by Northern analysis of the indicated human and mouse tissues. Calsarcin-1 mRNA is predominantly detected in heart and skeletal muscle, whereas the calsarcin-2 transcript was detected in skeletal muscle of both species.
Developmental Time Course of Calsarcin-1 and Calsarcin-2 Expression.
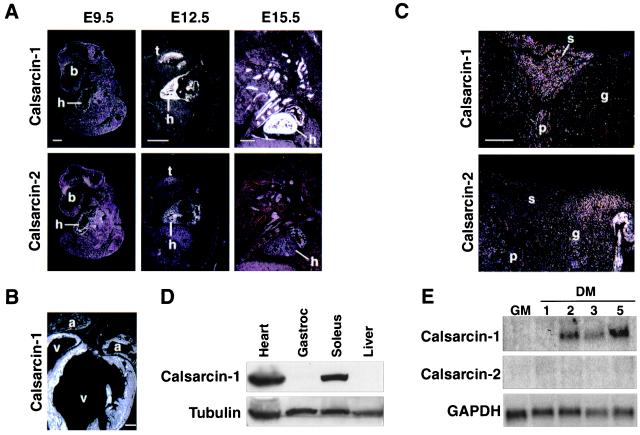
Embryonic expression of calsarcin was determined by radioactive in situ hybridizations on mouse embryo sections (Fig. 3A). At embryonic day (E) 9.5, relatively weak expression of calsarcin-1 was observed in the heart, whereas at E12.5 and E15.5, intense signals were detected in both cardiac and skeletal muscle tissue. In contrast, adjacent sections from the same embryo probed with calsarcin-2 displayed significant cardiac expression at E9.5, which was still detectable at E12.5. Low-level expression of calsarcin-2 in skeletal muscle of the tongue was also visible at this stage. At E15.5, cardiac expression of calsarcin-2 was down-regulated and was only weakly detected in the atria, whereas skeletal muscle expression became more robust. Expression of calsarcin-1 in all cardiac chambers persisted through adulthood (Fig. 3B). Thus, calsarcin-1 is expressed in all striated muscle tissues throughout development, whereas calsarcin-2 is transiently expressed in the heart during early embryogenesis and later becomes restricted to skeletal muscle.
Figure 3.
Developmental expression of calsarcin-1 and calsarcin-2. (A) Calsarcin-1 and calsarcin-2 transcripts were detected by radioactive in situ hybridization of mouse embryo sagittal sections at the embryonic time points indicated above each set of panels. b, brain; h, heart; t, tongue. (B) Calsarcin-1 transcripts were detected by radioactive in situ hybridization of a frontal section of an adult mouse heart. Transcripts are detected throughout the atria (a) and ventricles (v). (C) Calsarcin transcripts were detected by radioactive in situ hybridization of sections through adult mouse hindlimb muscle. Calsarcin-1 transcripts are localized to the soleus (s) and plantaris (p), whereas calsarcin-2 transcripts are localized to the gastrocnemius (g). (D) Calsarcin-1 and α-tubulin protein expression was detected by Western blot analysis of extracts from the indicated tissues. (E) Calsarcin-1 transcripts were detected by Northern analysis of RNA from C2 cells in growth medium (GM) or differentiation medium (DM) for the indicated days. (Scale bar, 500 μm.)
Fiber Type Specificity of Calsarcin-1 and Calsarcin-2 Expression in Skeletal Muscle.
To determine whether calsarcins might exhibit fiber type specificity of expression in skeletal muscle, we performed in situ hybridizations with sections of adult mouse hindlimb, using calsarcin-1 and calsarcin-2 probes (Fig. 3C). Calsarcin-1 expression was localized to the soleus and plantaris muscles, which are composed predominantly of slow-twitch fibers. In contrast, calsarcin-2 expression was enriched in gastrocnemius, which is primarily a fast-twitch muscle type.
Western blots of various tissue extracts using calsarcin-1 antiserum revealed a single 32-kDa protein in heart and soleus muscle (Fig. 3D). No expression was detected in liver or other nonmuscle tissues. The calsarcin-1 antiserum did not recognize recombinant calsarcin-2 in extracts derived from transfected Cos cells, indicating no significant cross-reactivity of the antiserum (data not shown). Only faint expression of calsarcin-1 protein could be detected in extract derived from gastrocnemius (Fig. 3D), confirming the slow fiber-restricted expression of calsarcin-1.
Calsarcin-1 transcripts were up-regulated during differentiation of the C2 skeletal muscle cell line, after transfer of proliferating myoblasts to differentiation medium (Fig. 3E). In contrast, calsarcin-2 expression was undetectable in C2 cells.
Calsarcins Interact with CnA and α-Actinin.
To begin to investigate the functions of calsarcins, we performed two-hybrid screens of the adult heart cDNA library, using calsarcin-1 and calsarcin-2 fused to the GAL4 DNA binding domain as bait. The GAL4-calsarcin-1 bait identified more than 50 clones of α-actinin-2 and one clone of α-actinin-3. A less extensive screen with the GAL4-calsarcin-2 bait yielded two positive clones, α-actinin-2 and the muscle-specific isoform of filamin.
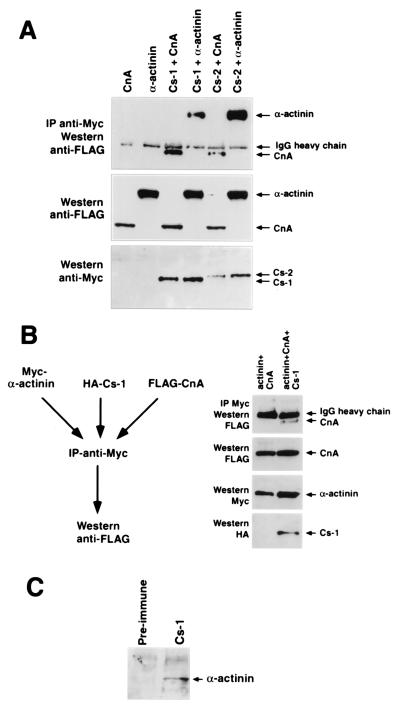
The interaction of calsarcin-1 and calsarcin-2 with CnA and α-actinin was further tested by coimmunoprecipitation of epitope-tagged proteins in transfected Cos cells and of the native proteins from neonatal cardiomyocytes. As shown in Fig. 4A, C-terminal Myc-tagged calsarcin-1 immunoprecipitated FLAG-tagged CnA and α-actinin. Catalytic activity of calcineurin is not required for the calsarcin interaction, as demonstrated by the ability of calsarcin-1 to immunoprecipitate a catalytically inactive CnA mutant (data not shown). Using a triple-immunoprecipitation approach with Myc-tagged α-actinin, hemagglutinin-tagged calsarcin, and FLAG-tagged calcineurin (Fig. 4B), we demonstrated that CnA could only be precipitated by Myc-α-actinin in the presence of calsarcin-1, indicating a trimeric complex. In addition, α-actinin could also be coimmunoprecipitated with native calsarcin-1 from cardiomyocyte extracts (Fig. 4C).
Figure 4.
Coimmunoprecipitation of calsarcins with calcineurin and α-actinin. (A) Cos cells were transiently transfected with expression vectors encoding FLAG-CnA, FLAG-α-actinin-2, or Myc-calsarcin-1/-2 (Cs-1/-2), and immunoprecipitations were performed. (Top) Anti-FLAG immunoblot of anti-Myc immunoprecipitates demonstrates the association of CnA and α-actinin with calsarcins. IgG heavy chain is also recognized by the secondary antibody. (Middle) Anti-FLAG immunoblot of cell extracts to demonstrate the presence of CnA and α-actinin. (Bottom) Anti-Myc immunoblot of cell extracts to demonstrate the presence of calsarcins. (B) Cos cells were transiently transfected with expression vectors encoding Myc-α-actinin-2, hemagglutinin–calsarcin-1, or FLAG-CnA, and immunoprecipitations were performed with anti-Myc antibody followed by immunoblotting with anti-FLAG antibody. The top panel shows an anti-FLAG immunoblot of anti-Myc immunoprecipitates and demonstrates association of CnA with calsarcin-1. The second panel from the top shows an anti-FLAG immunoblot of cell extracts to demonstrate the presence of CnA. The next panels show an anti-Myc immunoblot to demonstrate the presence of α-actinin and an anti-hemagglutinin immunoblot to demonstrate the presence of calsarcin-1, respectively. (C) Extracts prepared from primary neonatal rat cardiomyocytes were immunoprecipitated with anti-calsarcin-1 antibody or preimmune serum and analyzed by immunoblotting with anti-α-actinin antibody. α-Actinin is specifically immunoprecipitated with anti-calsarcin-1.
Mapping of Interaction Domains.
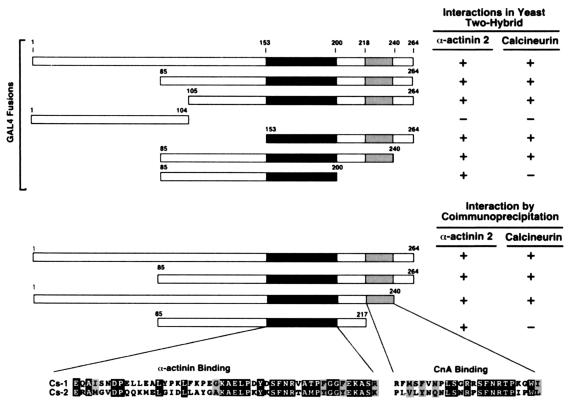
N- and C-terminal truncations of calsarcin-1 were used to characterize the CnA and α-actinin interaction domains. Yeast two-hybrid assays and complementary immunoprecipitation experiments revealed that amino acids 153–200 of calsarcin are necessary for the interaction of calsarcin with α-actinin-2 (Fig. 5A). Twenty-five residues within this region are highly conserved between mouse and human calsarcin-1 and calsarcin-2, suggesting that this might constitute the minimal interaction domain. Because a motif between amino acids 245 and 250 resembles known calcineurin docking sites on NFAT (PxIxIT) and muscle-enriched calcineurin inhibitor protein (PxIxxIT), we tested a C-terminal truncation lacking those residues. However, calsarcin lacking these amino acids was still able to bind CnA, by both two-hybrid assay (GAL4-calsarcin 85–240) and coimmunoprecipitation (Myc-calsarcin-1–240). In contrast, a calsarcin-1 mutant lacking residues 217–264 was unable to bind CnA, implying that residues 217–240 are necessary for binding. Mapping of the interaction domain on CnA revealed that the calsarcin-interacting domain resides within the catalytic region, whereas the calsarcin-1–interacting domain of α-actinin maps to the second and third spectrin-like repeats (data not shown).
Figure 5.
Mapping of calsarcin-, calcineurin-, and α-actinin-interacting domains. N- and C-terminal calsarcin-1 truncations were generated and fused to a Gal4-DNA-binding domain to test their ability to interact with CnA or α-actinin, as assessed by β-galactosidase activity in yeast. Complementary experiments were conducted by coimmunoprecipitation of Myc-tagged calsarcin-1 with FLAG-tagged CnA or α-actinin, respectively. Taken together, amino acids 153–200 appear to be necessary for the interaction with α-actinin, whereas amino acids 217–240 are required for calsarcin's association with CnA.
Calsarcin Colocalizes with Calcineurin and α-Actinin at the z-Line.
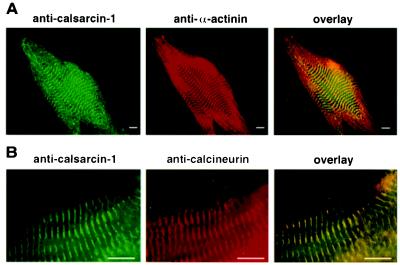
The subcellular localization of calsarcin-1 was determined by immunostaining of neonatal rat cardiomyocytes and cryosections of adult mouse heart and skeletal muscle. As shown in Fig. 6, calsarcin-1 staining was localized to the sarcomere of neonatal cardiomyocytes and overlapped with α-actinin staining, which specifically marks the z-line. A similar staining pattern was observed in sections of adult mouse heart and skeletal muscle (data not shown). Interestingly, calcineurin, detected with an antibody directed against amino acids 247–449 (Transduction Laboratories), was also localized to the z-line, indicating a muscle-specific subcellular localization of the enzyme. The latter finding was confirmed by a second CnA antibody (Sigma; data not shown). CnA staining was also detected in the nucleus, suggesting that CnA is also localized to other subcellular regions (data not shown).
Figure 6.
Subcellular localization of calsarcin-1. Neonatal rat cardiomyocytes were analyzed by immunostaining with calsarcin-1 antiserum and antibodies directed against α-actinin (A) and CnA (B). The overlay indicates that calsarcin-1 colocalizes both with α-actinin and CnA. (Scale bar, 10 μm.)
Discussion
Calsarcins belong to a family of muscle-specific sarcomeric proteins that appear to function as a bridge between calcineurin and α-actinin at the z-line of cardiac and skeletal muscle. Because calcineurin responds to sustained, low-amplitude calcium signals (2) and local calcium concentrations differ dramatically within a cardiomyocyte, calsarcin may serve to localize CnA in the vicinity of a unique intracellular calcium pool, where it can interact with specific upstream activators or downstream substrates.
Association of Calcineurin with Calsarcins.
In addition to serving as a potential docking site for calcineurin on the z-line, calsarcins could act as inhibitors or activators of the enzyme, or they could be substrates for calcineurin dephosphorylation. Preliminary studies indicate that overexpression of calsarcins can inhibit calcineurin activity in vitro (N.F. and E.N.O., unpublished results), although we do not know whether this reflects a direct function of calsarcins or, alternatively, the mislocalization of endogenous calcineurin, resulting in an indirect effect on calcineurin signaling. Interestingly, the muscle-enriched calcineurin inhibitor protein family of calcineurin-interacting proteins, which inhibit calcineurin activity, also show a muscle-enriched expression pattern, although they are not as specific to striated muscle as calsarcin-1 and calsarcin-2 (15, 16). Another calcineurin inhibitor, Cabin/Cain (17, 18), is enriched in brain and testis but is not expressed in heart or skeletal muscle. Thus, despite the wide tissue distribution of calcineurin, its function appears to be regulated by tissue-specific modifiers.
There are numerous examples of intracellular binding proteins that localize signaling molecules to specific sites. Protein kinase A, for example, associates with a family of proteins called AKAPs (A-kinase anchoring proteins; reviewed in ref. 19). Similarly, protein kinase C associates with the PDZ-domain protein PICK1 (20). Interestingly, in neurons AKAP79 has been shown to interact with protein kinase A, as well as with protein kinase C (21) and calcineurin (22, 36), thereby potentially providing a scaffold for integrating multiple signaling pathways. In this regard, it will be interesting to see if calsarcins also bind other signaling molecules. In addition, AKAP79 has been shown to inhibit calcineurin activity and to attenuate cardiomyocyte hypertrophy in vitro (22, 36), although given its brain-enriched expression pattern, it seems unlikely that AKAP79 inhibits cardiac calcineurin activity in vivo. Calcineurin-binding domains have also been identified in several other proteins, including NFATs (4), the viral protein A238L (23), muscle-enriched calcineurin inhibitor protein (15), and Cabin/Cain (1). All of these proteins bind calcineurin through a PxIxIT (or closely related) motif that interacts with a domain N-terminal of the catalytic site of the enzyme. The calcineurin-binding domain of calsarcin does not contain this motif and therefore seems to interact with a different region of calcineurin.
Calsarcin-1 and calsarcin-2 are restricted to slow- and fast-twitch skeletal muscle fibers, respectively, which exhibit distinct contractile, physiologic, and metabolic properties. The slow-twitch phenotype has been shown to depend on chronic motor neuron stimulation that results in sustained elevation of intracellular calcium (reviewed in ref. 7). In contrast, fast-twitch fibers receive intermittent, higher frequency stimulation and exhibit lower intracellular calcium concentrations. Because calcineurin activation favors a slow-twitch phenotype, via the induction of NFAT- and MEF2-dependent genes (24), it will be of interest to determine whether calsarcin isoforms can differentially modulate the function of calcineurin in a fiber type-specific manner, or if calsarcin isoform expression itself is calcineurin-dependent.
Potential Roles of z-Line Proteins in Mechanosignaling.
Calsarcin-1 and calsarcin-2 interact with α-actinin-2 and -3, which are striated muscle-enriched. Our two-hybrid screens also revealed an interaction between calsarcin-2 and the z-line-associated protein filamin. Although we have not yet examined the potential significance of this interaction, it is intriguing that a mutation in myotilin, another muscle-specific interaction partner of filamin (25), causes autosomal dominant limb-girdle muscular dystrophy (26).
α-Actinin plays a key role in maintaining the structural integrity of the sarcomere by crosslinking actin fibers in an antiparallel fashion at the level of the z-line. Mutations in several proteins associated with α-actinin and/or the z-line, including α-actin (27), desmin (28), and dystrophin (29), have been implicated in the development of dilated cardiomyopathy (reviewed in ref. 30). Mice lacking the muscle LIM domain protein, MLP, which also interacts with α-actinin, develop lethal cardiomyopathy (31). Because of its unique position at the interface of the cytoskeleton and the sarcomere, the z-line has been suggested to play a role as a mechanosensor in cardiomyopathic chamber dilation (32). This concept is also attractive in light of the fact that α-actinin interacts with several signaling molecules, such as PKN (33), phospholipase D2 (34), and G protein-coupled receptor kinases (35). In this regard, calcineurin is a potent inducer of myocyte hypertrophy, and it is well established that alterations in sarcomere function can evoke a hypertrophic response. The mechanism that links altered cardiac contractility to the hypertrophic response has been a focus of intense interest but remains largely unknown. It is worth noting that calcineurin activity has been reported to be elevated in hearts of transgenic mice that exhibit sarcomeric dysfunction due to overexpression of the actin-stabilizing protein tropomodulin (6). Localization of calcineurin to the z-line provides a potential mechanism whereby calcineurin could link specific calcium signals to a hypertrophic response.
Although our results demonstrate the colocalization of CnA and α-actinin at the z-line, it should be emphasized that CnA is also located in other regions of striated myocytes, including the nucleus, where it presumably participates in other signal transduction pathways, such as in the activation of NFAT- and MEF2-dependent transcription.
Future Questions.
The importance of calcineurin for the immune response poses challenges to the development of calcineurin inhibitors that specifically modify its activity in striated muscle cells, without associated immunosuppression. In principle, this problem might be overcome by the identification of muscle-specific calcineurin-interacting proteins that influence the activity or subcellular localization of the enzyme or serve as key substrates that mediate its actions on striated muscle cells. Modulation of the interactions between calcineurin and calsarcins might therefore provide an approach to regulating calcineurin activity in a muscle-specific manner.
Acknowledgments
We thank A. Tizenor and J. Page for help with the manuscript, J. A. Spencer for help with C2 cell differentiation experiments, and J. Shelton for preparing tissue sections for in situ hybridizations. This work was supported by grants from the National Institutes of Health and the Texas Advanced Technology Program (to E.N.O.). N.F. was supported by a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft.
Abbreviations
- CnA
calcineurin A
- CsA
cyclosporin A
- E
embryonic day
- NFAT
nuclear factor of activated T-cells
- AKAPs
A-kinase anchoring proteins
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY013295, AY013296, AY013297, and AY013298).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260501097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260501097
References
- 1.Aramburu J, Rao A, Klee C B. Curr Top Cell Regul. 2000;36:237–295. doi: 10.1016/s0070-2137(01)80011-x. [DOI] [PubMed] [Google Scholar]
- 2.Dolmetsch R E, Lewis R E, Goodnow C C, Healy J I. Nature (London) 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 3.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S E, Olson E N. Cell. 1998;17:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 5.Frey N, McKinsey T A, Olson E N. Nat Med. 2000;6:1221–1227. doi: 10.1038/81321. [DOI] [PubMed] [Google Scholar]
- 6.Sussman M A, Welch S, Gude N, Khoury P R, Daniels S R, Kirkpatrick D, Walsh R A, Price R L, Lim H W, Molkentin J D. Am J Pathol. 1999;155:2101–2113. doi: 10.1016/S0002-9440(10)65528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson E N, Williams R S. Cell. 2000;101:689–692. doi: 10.1016/s0092-8674(00)80880-6. [DOI] [PubMed] [Google Scholar]
- 8.Musaro A, McCullagh K J, Naya F J, Olson E N, Rosenthal N. Nature (London) 1999;400:581–585. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- 9.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naya F J, Mercer B, Shelton J, Richardson J A, Williams R S, Olson E N. J Biol Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 11.Spencer J A, Eliazer S, Ilaria R L, Richardson J A, Olson E N. J Cell Biol. 2000;150:771–784. doi: 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Richardson J A, Gan L, Olson E N. Mech Dev. 1998;73:23–32. doi: 10.1016/s0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 13.Shibasaki F, Price E R, Milan D, McKeon F. Nature (London) 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 14.Fields S, Song O. Nature (London) 1989;340:245–247. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 15.Rothermel B, Vega R B, Yang J, Wu H, Bassel-Duby R, Williams R S. J Biol Chem. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- 16.Kingsbury T J, Cunningham K W. Genes Dev. 2000;14:1595–1604. [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L, Youn H-D, Loh C, Stolow M, He W, Liu J O. Immunity. 1998;8:703–711. doi: 10.1016/s1074-7613(00)80575-0. [DOI] [PubMed] [Google Scholar]
- 18.Lai M M, Burnett P E, Wolosker H, Blackshaw S, Snyder S H. J Biol Chem. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- 19.Edwards A S, Scott J D. Curr Opin Cell Biol. 2000;12:217–221. doi: 10.1016/s0955-0674(99)00085-x. [DOI] [PubMed] [Google Scholar]
- 20.Staudinger J, Zhou J, Burgess R, Elledge S J, Olson E N. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klauck T M, Faux M C, Labudda K, Langeberg L K, Jaken S, Scott J D. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 22.Coghlan V M, Perrino B A, Howard M, Langeberg L K, Hicks J B, Gallatin W M, Scott J D. Science. 1995;267:108–112. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 23.Miskin J E, Abrams C C, Goatley L C, Dixon L K. Science. 1998;281:562–565. doi: 10.1126/science.281.5376.562. [DOI] [PubMed] [Google Scholar]
- 24.Wu H, Naya F J, McKinsey T A, Mercer B, Shelton J M, Chin E R, Simard A R, Michel R N, Bassel-Duby R, Olson E N, Williams R S. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Der Ven P F, Wiesner S, Salmikangas P, Auerbach D, Himmel M, Kempa S, Hayes K, Pacholsky D, Taivainen A, Schroder R, et al. J Cell Biol. 2000;151:235–248. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauser M A, Horrigan S K, Salmikangas P, Torian U M, Viles K D, Dancel R, Tim R W, Bartolini L, Gilchrist J M, Stajich J M, et al. Hum Mol Genet. 2000;9:2141–2147. doi: 10.1093/hmg/9.14.2141. [DOI] [PubMed] [Google Scholar]
- 27.Olson T M, Michels V V, Thibodeau S N, Tai Y S, Keating M T. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 28.Li D, Tapscoft T, Gonzales O, Burch P E, Quinones M A, Zoghbi W A, Hill R, Bachinski L L, Mann D L, Roberts R. Circulation. 1999;100:461–464. doi: 10.1161/01.cir.100.5.461. [DOI] [PubMed] [Google Scholar]
- 29.Muntoni F, Cau M, Ganau A, Congiu R, Arvedi G, Mateddu A, Marrosu M G, Cianchetti C, Realdi G, Cao A. N Engl J Med. 1993;329:921–925. doi: 10.1056/NEJM199309233291304. [DOI] [PubMed] [Google Scholar]
- 30.Nichol R L, Frey N, Olson E N. Annu Rev Genomics Hum Genet. 2000;1:179–223. doi: 10.1146/annurev.genom.1.1.179. [DOI] [PubMed] [Google Scholar]
- 31.Arber S, Hunter J J, Ross J, Hongo M, Sansig G, Borg J, Perriard J-C, Chien K R, Caroni P. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 32.Chien K R. Nature (London) 2000;407:227–232. doi: 10.1038/35025196. [DOI] [PubMed] [Google Scholar]
- 33.Mukai H, Toshimori M, Shibata H, Takanaga H, Kitagawa M, Miyahara M, Shimakawa M, Ono Y. J Biol Chem. 1997;272:4740–4746. doi: 10.1074/jbc.272.8.4740. [DOI] [PubMed] [Google Scholar]
- 34.Park J P, Kim J H, Kim Y, Ha S H, Kim J H, Yoo J-S, Du G, Frohman M A, Su P-G, Ryu S H. J Biol Chem. 2000;275:21295–21301. doi: 10.1074/jbc.M002463200. [DOI] [PubMed] [Google Scholar]
- 35.Freeman J L R, Pitcher J A, Xiaolin L, Bennett V, Lefkowitz R J. FEBS Lett. 2000;473:280–284. doi: 10.1016/s0014-5793(00)01543-x. [DOI] [PubMed] [Google Scholar]
- 36.Taigen T, DeWindt L J, Lim H W, Molkentin J D. Proc Natl Acad Sci USA. 2000;97:1196–1201. doi: 10.1073/pnas.97.3.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]