Abstract
Macrophages have long been regarded as the main target encountered by Salmonella typhimurium, a Gram-negative facultative intracellular pathogen that invades the intestinal mucosa. S. typhimurium, however, are first internalized by dendritic cells. To gain new insights into the interactions between Salmonella and the dendritic cells, we compared the fate of wild-type S. typhimurium and the virulence-attenuated PhoP constitutive (PhoPc) strain. The PhoPc strain is impaired for entry and survival in mammalian cells and is poorly processed by macrophages for antigen presentation on MHC class II molecules. Here, we show that bone marrow-derived dendritic cells can similarly process and present a foreign antigen expressed by the invasive wild-type and the attenuated PhoPc S. typhimurium. This property correlates with equivalent entry and survival efficiencies of both strains in dendritic cells. In addition, Salmonella strains mutated in mgtCB, sseC, and orfL genes required for macrophage survival showed no defect in survival in dendritic cells. Together, these results indicate that uptake of Salmonella by dendritic cells and subsequent antigen processing and presentation do not depend on virulence factors important in macrophages.
Salmonella typhimurium is a Gram-negative bacterium that causes a self-limiting gastroenteritis in humans, but a typhoid-like systemic infection in mice (1). Hosts are infected after ingestion of contaminated food or water. The bacteria then survive the acidic pH of the stomach, penetrate the gut barrier via the specialized M cells, and colonize the Peyer's patches (2, 3). Subsequently, they spread into the draining mesenteric lymph nodes and disseminate via the bloodstream to the spleen, liver, and bone marrow, where they replicate in cellular niches, the macrophages of the reticuloendothelial system (4).
Macrophages are thought to be central to the dissemination of Salmonella but also to initiate the immune reaction. Most of the experiments supporting this model, and in particular the presentation of antigens expressed by recombinant S. typhimurium, have been conducted in macrophage cell lines or in bone marrow-derived macrophages cultured in vitro. Antigens produced by wild-type Salmonella are efficiently processed and presented by macrophages on MHC class II molecules (5). Different attenuated strains were analyzed as well, among which was the PhoPc mutant of S. typhimurium. The PhoP/Q system controls the coordinated expression of virulence genes involved in invasion and intracellular survival. The PhoPc bacteria have impaired ability to enter into mammalian cells, especially epithelial cells, and to survive in macrophages (6–9). Antigens expressed by these bacteria are poorly presented to T cells by macrophages, compared with antigens expressed by the wild-type bacteria (5). In mice however, the strain is highly immunogenic via different mucosal routes of immunization (8, 10, 11). Thus, the in vitro results on antigen presentation in macrophages do not correlate with in vivo immune responses for the PhoPc strain.
The role of macrophages as antigen-presenting cells at the site of entry of Salmonella in the Peyer's patches has been questioned. Light has recently been shed on the major role of dendritic cells (DCs) in the capture and presentation of antigens in the Peyer's patches (12–14). These cells are located just below the epithelium associated with lymphoid follicles, close to M cells, and are ideally placed to sample antigens. In a previous study, we have shown that DCs, and not macrophages, internalize S. typhimurium early after oral administration to mice or injection in a ligated loop of intestine (15). Both wild-type and attenuated strains were efficiently taken up by DCs in vivo. Therefore, we postulated that the discrepancies mentioned above for the PhoPc bacteria between in vitro assays and the immunization experiments in mice could be explained by differences in uptake of bacteria by DCs versus macrophages and/or by a different fate of the bacteria in these two cell types. To test this hypothesis, we investigated the interactions between bone marrow-derived DCs and the attenuated PhoPc strain or its wild-type counterpart. We found that attenuated and wild-type bacteria behave similarly in DCs in terms of entry, survival, and antigen presentation, in contrast to what had been reported in macrophages. The interactions of S. typhimurium with DCs may be preponderant over macrophages in vivo, an observation that has strong implications for the design and screening of new vaccine strains.
Materials and Methods
Antibodies and Reagents.
Grade VII Ovalbumin (OVA) was purchased from Sigma. Monoclonal antibodies against Thy-1 (AT83, rat IgM), CD45R (B220, rat IgM), CD11b/Mac 1 (M1/70, rat IgG2b), and MHC class II I-Ad molecules [11.54.3, rat IgG2b (15, 16)] were produced as supernatants and used as such or after concentration in a Labscale TFF system (Millipore). Biotinylated anti-CD40 antibodies were a kind gift of Hans Acha-Orbea (Biochemistry Institute, Epalinges, Switzerland). Biotinylated antibodies against CD80, CD86, and CD11c were from PharMingen, streptavidin coupled to phycoerythrin was from Serotec, anti-OVA rabbit antibodies were from Chemicon International (Temecula; CA), and anti-rabbit Ig coupled to peroxidase was from Amersham Pharmacia.
Salmonella Strains.
All strains are derived from the virulent strain ATCC14028. S. typhimurium CS022 or PhoPc, a kind gift of John Mekalanos (Harvard Medical School, Boston), contains a point mutation, pho-24, in the phoQ gene (8). S. typhimurium EG9527 (6) was a kind gift of Eduardo Groisman (Washington University School of Medicine, St. Louis). BA1501 contains a MudJ insertion in SPI-4 in orfL as in MS2097 (17) and was kindly provided by Brian Ahmer (Ohio State University, Columbus, OH). The ATCC14028 derivative strain SIN6 (sseC∷aphT) was obtained by phage P22HT105/1-int-mediated transduction (18) with HH104 (19), kindly provided by David Holden (Imperial College School of Medicine, London), as donor strain. The GFP-expressing S. typhimurium harbor the pKKGFP plasmid and have been described in ref. 15. The strains producing OVA were obtained by electroporation of Salmonella (10) with the pKKOVA plasmid, a pKK233.3 expression vector containing the ova cDNA (20) and kindly provided by Noboyuki Takahashi (Kyoto University, Uji, Japan). In all experiments, bacteria were grown overnight at 37°C with shaking in LB broth, then subcultured without shaking for 4–5 h in LB containing 300 mM NaCl. These culture conditions have been shown to increase S. typhimurium invasiveness (21, 22). Ampicillin was added at 100 μg/ml to grow recombinant bacteria. The absorbance at 600 nm of the bacterial suspensions was used to determine the multiplicity of infection, by estimating that 109 bacteria per ml give an A600 of 1. The inoculum dose was then calculated by plating serial dilutions onto LB agar plates.
Dot Blot.
Bacteria were lysed with B-Per (Pierce). Serial dilutions of bacterial lysates and purified OVA were spotted on a nitrocellulose membrane (Schleicher & Schuell) by using a Dot-Blot device (Bio-Rad), and immunoblot was performed with anti-OVA antibodies followed by horseradish peroxidase-coupled anti-rabbit antibodies, revealed by enhanced chemiluminescence according to the manufacturer's instructions (Amersham Pharmacia) and film exposure (Kodak). The film was scanned and analyzed with the nih image software.
Bone Marrow Cultures.
Bone marrow-derived DCs and macrophages were cultivated from 6- to 8-week-old female BALB/c (H-2d) mice femoral and tibial bones, as described by Inaba et al. (23). Briefly, the cell suspensions were depleted in red blood cells, and in T and B cells by incubation with anti-Thy-1 and anti-CD45R antibodies and rabbit complement (Saxon Europe, Newmarket, Suffolk, U.K.) for 45 min in an incubator at 37°C. They were then plated overnight in culture-treated Petri dishes (Nunc). The next day, nonadherent cells were collected and cultivated for 5 days without subculture in the presence of 10 ng/ml of rGM-CSF (PharMingen or Immugenex, Los Angeles, CA) in complete RPMI medium 1640 [containing 5% FCS (Myoclone superplus), 2 mM L-glutamine, 10 mM Hepes, and 1 mM sodium pyruvate (all from GIBCO/BRL)]. After 5 days, nonadherent DCs were separated from any debris by ficoll purification. The adherent macrophages were physically scrapped, washed, and plated in new wells. Both the macrophage and the DC fractions were infected with bacteria a maximum of 3 h after separation to avoid further maturation due to subculture, as described in refs. 23 and 24. The cells' phenotype was then analyzed by a fluorescence-activated cell sorter (FACScan, Becton Dickinson) by using antibodies specific for various surface markers.
Presentation of Antigens Expressed by Bacteria.
Antigen-presenting cells (APCs) were irradiated at 700 cGy and seeded at 2 × 105 cells per condition in 100 μl of complete RPMI in duplicate wells of a 96-well flat-bottom tissue culture plate (macrophages) or in 5 ml tubes (DCs). They were infected with bacteria at a multiplicity of infection of 25 for 2.5 h and then washed four times. DCs were then transferred to 96 wells. CD4+ T cells recognizing an I-Ad + OVA323–339 complex were purified from the lymph nodes of the DO11.10 transgenic mice (25) generated by Kenneth Murphy and kindly provided by Manfred Kopf (Basel Institute for Immunology, Basel, Switzerland), by using magnetic beads coated with anti-CD4 antibodies (miniMACS, Miltenyi Biotec, Auburn, CA). Purity of the T cell population was routinely 98%, as assessed by FACS analysis for the CD4 marker. To each well were added 105 T cells in 100 μl of complete medium containing 100 μg/ml of gentamicin (GIBCO/BRL), so that the final concentration of gentamicin in the wells is 50 μg/ml. After 72 h of coculture, the cells were pulsed for 16 h with 1 μCi (1 Ci = 37 GBq) per well of [3H]thymidine (Amersham Pharmacia, 20 Ci/mmol). The cells content was harvested on fiberglass filters and [3H]thymidine incorporation was measured in a Betaplate counter (1205, Wallac, Gaithersburg, MD).
Plating Assay.
DCs were transferred to 5 ml tubes (5.105 cells per tube in 1 ml of complete RPMI medium) and infected at a multiplicity of infection of 50. After a 30 min incubation at 37°C in 5% CO2, the cells were washed twice with sterile PBS, once with complete RPMI medium supplemented with 50 μg/ml of gentamicin, and then incubated in the latter medium for the indicated times. At each time point, cells were washed twice with PBS and lysed with 0.5% Triton X-100 in PBS (Fluka). The number of viable bacteria present at each time point was determined by plating serial dilutions on LB agar plates.
Binding Assay.
Cells and GFP-expressing bacteria were chilled on ice for 15 min before contact. Cells were incubated with bacteria for 30 min at 4°C and then washed 4 times with cold PBS. The binding of GFP-expressing bacteria to cells was analyzed by FACS using an FSC/SSC gate that excludes free bacteria.
Electron Microscopy.
DCs were incubated with bacteria for the indicated times in an incubator at 37°C. The cells were washed and then prepared for transmission electron microscopy as described previously (15). For scanning electron microscopy, DCs were infected in tubes and then transferred onto poly-L-lysine-coated glass coverslips. Samples were fixed and dehydrated as for transmission electron microscopy, then critical-point dried in CO2, sputter-coated to produce 15 nm gold coating, and examined by using a JSM 6300 F JEOL microscope (Centre de Microscopie Electronique, Lausanne, Switzerland) at an accelerating voltage of 5 kV.
Statistical Analysis.
The measure of bacterial survival was analyzed by Student's t test. Probabilities (P) of 0.05 or less were considered significant.
Results
Dendritic Cells but Not Macrophages Efficiently Present Antigens Expressed by an Attenuated S. typhimurium Strain.
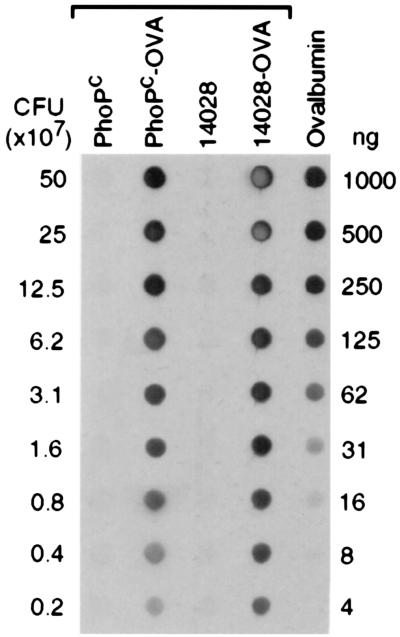
To analyze the presentation of antigens expressed by Salmonella in DCs, we used OVA as a model antigen. OVA was constitutively expressed in the cytosol of the attenuated S. typhimurium PhoPc and the virulent strain ATCC14028. The amount of OVA produced was quantified by dot blot (Fig. 1). Densitometric analysis and comparison with a standard curve of purified OVA allowed us to calculate that 106 PhoPc and ATCC14028 bacteria contained 15 and 25 ng of OVA, respectively.
Figure 1.
Dot blot analysis of OVA production by recombinant Salmonella. Strains ATCC14028, ATCC14028(pKKOVA), PhoPc, and PhoPc(pKKOVA) were grown in conditions described in Material and Methods. Serial dilutions of bacterial lysates were analyzed by dot blotting onto a nitrocellulose membrane. Dilutions of soluble OVA were blotted to generate a standard curve. The membrane was reacted with anti-OVA antibodies.
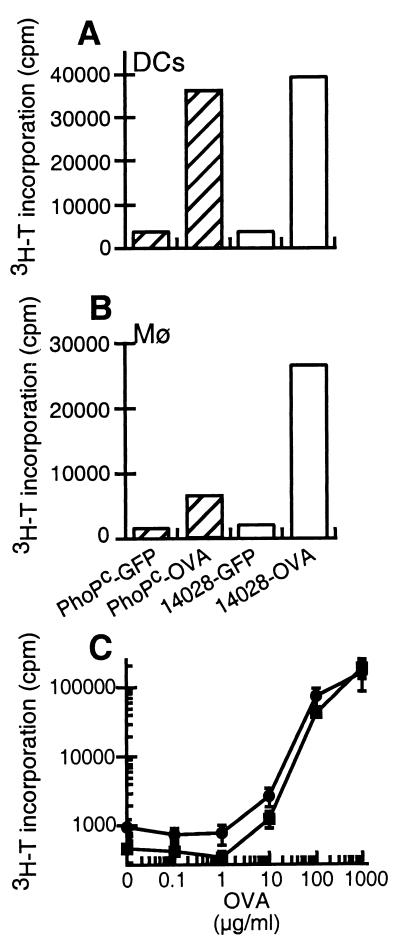
The ability of bone marrow-derived DCs to process and present antigens expressed by the recombinant bacteria was assessed by measuring the proliferation of naive CD4+ T lymphocytes purified from DO11.10 transgenic mice, which express a T cell receptor specific for OVA323–339 presented on I-Ad molecules. OVA expressed by attenuated bacteria was presented by DCs as efficiently as OVA produced by the wild-type bacteria (Fig. 2A). The activation of DO11.10 T lymphocytes was specific, because incubation of DCs with bacteria expressing GFP as a control did not lead to T cell proliferation. As described previously, the attenuated PhoPc bacteria were poorly processed by macrophages, in contrast to wild-type bacteria (Fig. 2B and ref. 5). In addition, both cell types presented the soluble model antigen with similar efficiency when incubated with various concentrations of soluble OVA (Fig. 2C). This indicates that the macrophages have the capacity to present antigens on MHCII molecules. Nevertheless, only DCs were able to present antigens expressed by the PhoPc bacteria.
Figure 2.
Differential presentation of OVA-producing S. typhimurium PhoPc by DCs and macrophages. (A and B) A representative experiment of at least two independent experiments performed in duplicate. The difference between the duplicates was <15%. Bone-marrow-derived DCs (A) or macrophages (B) were irradiated and incubated with S. typhimurium PhoPc (hatched bars) or the virulent ATCC14028 (open bars) strain expressing OVA or GFP as a control, at a multiplicity of infection of 25. (C) The bone-marrow-derived DCs (squares) or macrophages (circles) were incubated with purified OVA at various concentrations. After 2.5 h, the cells were washed and incubated with gentamicin to kill extracellular bacteria. Naive CD4+ transgenic T cells specific for OVA323–339 + IAd were added. Proliferation of T cells was measured after 90 h by 3H-T incorporation. The mean ± SD of four values is plotted.
We next compared the level of proliferation obtained after incubation of antigen-presenting cells with OVA-expressing bacteria or soluble OVA. Incubation of macrophages or DCs with OVA-expressing bacteria led to comparable T cell proliferation as incubation with 75 μg/ml of soluble OVA. The number of recombinant bacteria added to presenting cells produced an equivalent of 0.75 μg/ml of OVA (Fig. 1). Accordingly, the presentation of antigens derived from bacteria was at least 100 times more efficient than the presentation of soluble antigens.
S. typhimurium Induce Phenotypic Maturation of DCs.
Because only DCs processed foreign antigens expressed by PhoPc Salmonella and presented them to naive T cells, we restricted our study to DCs. In response to various stimuli, including inflammatory cytokines and viral or microbial components, DCs undergo a maturation process reflected by the up-regulation of costimulatory molecules or chemokine receptors that potentiates their ability to activate T cells (26). The efficient presentation of OVA expressed by S. typhimurium as compared with soluble antigen could be due to such an adjuvant activity of the bacteria on DCs. Therefore, we tested whether phenotypic changes characteristic of maturation occurred on DCs incubated with bacteria.
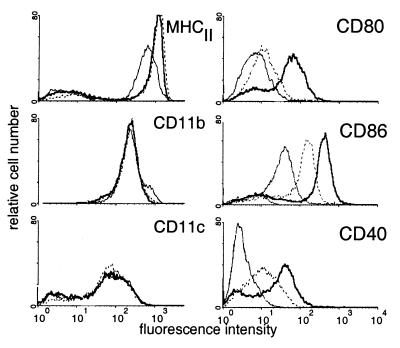
The phenotype of uninfected DCs was first analyzed by FACS. As shown in Fig. 3, the bone marrow-derived DCs expressed MHC class II molecules, the CD11c and CD11b markers typical of myeloid DCs, and low levels of CD80 and CD86 costimulatory molecules. No CD40 expression could be detected. This phenotype is similar to that of the DCs present in the subepithelial dome of the Peyer's patches (14), which internalize S. typhimurium in vivo (15). DCs incubated for 3 h with either tissue culture medium alone, bacteria, or soluble OVA, washed extensively, and cultured overnight at 37°C were analyzed by FACS as described above. After an overnight culture in medium, the DCs spontaneously matured further as described (24) and exhibited increased levels of MHCII, CD80, CD86, and CD40 molecules on their surface. Incubation of the cells with OVA did not induce any change as compared with medium alone (data not shown). In contrast, the CD80, CD86, or CD40 costimulatory molecules were up-regulated in DCs that were in contact with either the PhoPc strain (Fig. 3) or wild-type S. typhimurium (data not shown). The marked increase in the expression of costimulatory molecules on DCs after incubation with bacteria may explain, at least in part, the efficiency of presentation to naive T cells of antigens expressed by the bacteria as compared with soluble antigens.
Figure 3.
Up-regulation of costimulatory molecules on DCs by S. typhimurium. Bone marrow-derived DCs were analyzed before any treatment by flow cytometry (thin line). The cells were then incubated for 3 h in medium alone (dotted line) or with S. typhimurium PhoPc (bold line), washed, further cultivated for 18 h, and then analyzed. The mean fluorescence obtained after staining with secondary antibodies alone was <10. Results are representative of five different experiments.
Dendritic Cells Efficiently Internalize Noninvasive S. typhimurium PhoPc.
To better understand the capacity of dendritic cells to present antigens expressed by S. typhimurium PhoPc, we investigated internalization and subsequent intracellular survival of the bacteria in DCs.
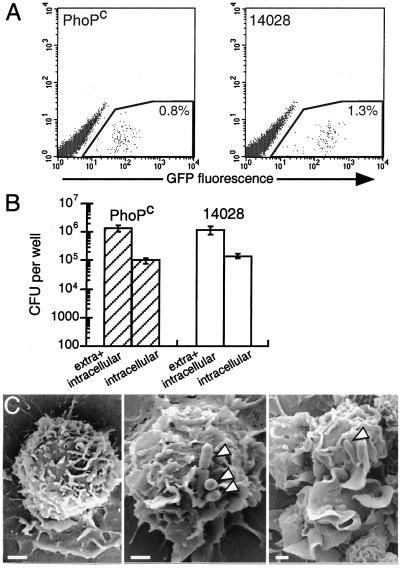
In the attenuated PhoPc bacteria, the genes involved in cell invasion encoded by the Salmonella pathogenicity island-1 (SPI-1) are constitutively repressed (27). We analyzed the effect of this mutation on the entry of Salmonella in DCs. Bacteria were grown in conditions that enhance the invasiveness of the wild-type Salmonella. We first studied the adhesion of bacteria to DCs. To this end, we measured the binding at 4°C to DCs of GFP-producing PhoPc and wild-type Salmonella. The cells were analyzed by flow cytometry, which allows the cells rendered fluorescent by adherent bacteria to be distinguished from the free bacteria. Fig. 4A shows that the attenuated PhoPc bacteria were able to bind to DCs at 4°C as efficiently as the wild-type bacteria.
Figure 4.
Entry of S. typhimurium in DCs. (A) Adhesion of S. typhimurium to DCs. S. typhimurium PhoPc and wild-type expressing GFP were incubated at 4°C for 30 min with DCs. After washing, the cell suspension was analyzed by flow cytometry. The proportion of cells associated with the fluorescent bacteria is indicated in the gated region. The figure shows a representative experiment of three independent experiments performed. (B) Internalization of S. typhimurium in DCs. S. typhimurium PhoPc (hatched bars) and wild-type (open bars) strains were incubated for 30 min with DCs. After washing, the cells were lysed and plated onto agar to count the number of extracellular and internalized bacteria, or incubated for 30 min with gentamicin to kill extracellular bacteria, lysed, and plated onto agar to count the internalized bacteria. The figure shows the mean ± SD of four independent experiments performed in duplicate. (C) Scanning electron micrographs of DCs not treated (Left) or incubated for 3 min with S. typhimurium PhoPc (Middle), or wild-type (Right). Arrowheads point to bacteria. Bar = 1 μm.
Lowering the temperature to 4°C inhibits the bacterial internalization and any energy-dependent processes required for binding. Therefore, we quantified the adhesion and entry of Salmonella into DCs at 37°C. The DCs were incubated with attenuated PhoPc and wild-type bacteria, lysed, and the lysate then plated onto agar to quantify cell-associated bacteria, including adherent and internalized bacteria. Alternatively, gentamicin was added to DCs to kill the extracellular bacteria, and the internalized bacteria were counted (Fig. 4B). The number of wild-type and PhoPc S. typhimurium associated with DCs after infection was similar. The proportion of cell-associated bacteria that were internalized by DCs was not different for the attenuated PhoPc and the wild-type bacteria (11 ± 5% vs. 11 ± 3%, n = 5).
S. typhimurium invades eukaryotic cells by a trigger mechanism reminiscent of macropinocytosis, inducing ruffles and cytoskeletal modifications by injecting bacterial toxins into the host cytosol via type III secretion systems (28, 29). To further analyze the entry of S. typhimurium PhoPc into DCs, DCs were incubated with wild-type or attenuated bacteria for 3 min, immediately fixed, and processed for scanning electron microscopy. Compared with noninfected cells (Fig. 4C Left), DCs incubated with bacteria exhibited surface modifications with membrane extensions (Fig. 4C Middle and Right). This membrane ruffling occurred on the whole cell population, including cells that were apparently not in contact with bacteria (data not shown). Ruffling was observed with wild-type bacteria and, to a lesser extent, with PhoPc strains.
Taken together, these results suggest that the binding to and entry into DCs of S. typhimurium involves a mechanism that is independent of the PhoP/Q regulated genes.
Attenuated S. typhimurium Survive in DCs.
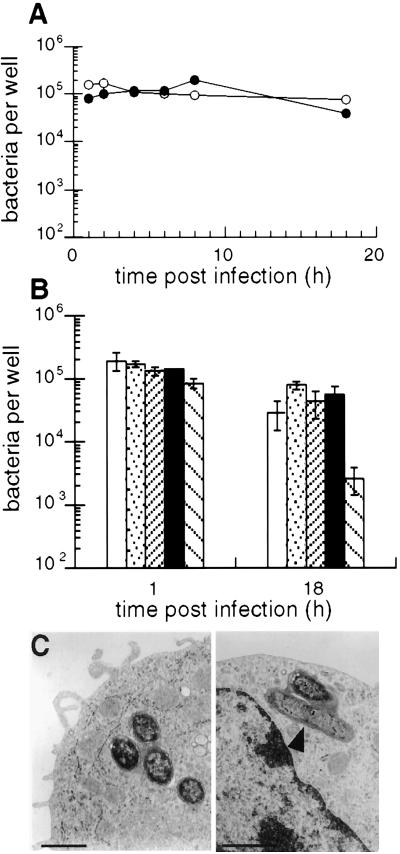
The PhoPc strain was initially described as attenuated for survival in macrophages in vitro (8). Therefore, we analyzed the kinetics of survival of the PhoPc bacteria in DCs. To this end, infected cells were lysed after different incubation times, and live bacteria were counted after plating on agar (Fig. 5A). The number of live bacteria found in DCs at the different time points, as well as 18 h after internalization, was comparable for wild-type and PhoPc S. typhimurium. The proportion of bacteria that survived was not significantly different (P = 0.069) for attenuated bacteria (33 ± 11%, n = 4) as compared with their wild-type counterparts (47 ± 7%, n = 4). Therefore, the phoP locus is not required for survival in DCs. To extend this study to other loci, we analyzed mutants in SPI-2 (sseC), SPI-3 (mgtCB), and SPI-4 (orfL) that are impaired in macrophage survival (refs. 17, 19, and 30 and data not shown). In DCs (Fig. 5B), these mutants showed no defect in survival, in contrast to an E. coli laboratory strain (P = 0.029). This indicates that virulence factors required for Salmonella to survive in macrophages are not necessary in DCs, but suggests that other factors absent from E. coli may be involved.
Figure 5.
Intracellular survival of S. typhimurium in bone marrow-derived DCs. (A) Kinetics of survival in DCs. Cells were infected for 30 min with the attenuated PhoPc (filled circles) and virulent (open circles) strains of Salmonella, washed, and incubated in gentamicin. The number of viable bacteria at different incubation times was counted after plating the cells onto agar. The figure shows a representative experiment of four independent experiments performed in duplicate. (B) Overnight survival of wild-type (open bars), SIN6 (dotted bars), EG9527 (close hatching), BA1501 (filled bars), and JM105 (spaced hatching) in dendritic cells was measured as described in A. Mean ± SD of two independent experiments performed in duplicate out of four performed. (C) Transmission electron micrographs of intracellular PhoPc (Left) and wild-type (Right) Salmonella at 4 h postinfection. Bar = 250 nm.
Next, we examined internalized bacteria by transmission electron microscopy (Fig. 5C). Both PhoPc and wild-type bacteria were located in membrane-bound tight vacuoles. Some bacteria showed signs of degradation, whereas others were in the process of division. These observations are consistent with the survival of the bacteria in DCs on the one hand, and the processing for antigen presentation on the other.
Discussion
The present study was aimed at dissecting the various steps involved in binding, uptake, and antigen presentation of recombinant S. typhimurium by dendritic cells. To mimic physiological conditions, we used bone marrow-derived DCs phenotypically close to the myeloid DC subset present in the dome of Peyer's patches and encountered by Salmonella in vivo. In addition, to analyze antigen presentation, we followed the activation of naive T lymphocytes. With this model, we show here for the first time that DCs, in contrast to macrophages, internalize and process Salmonella for antigen presentation independently of the PhoP-regulated virulence factors.
Entry of S. typhimurium into host cells, especially the epithelial cells of the gut, depends on virulence factors encoded by the SPI-1 as well as other loci (31–33). In the PhoPc bacteria, the PhoP/Q system that controls the expression of virulence genes by sensing the environment is constitutively turned on. In turn, the SPI-1 genes are down-regulated (32, 34). The data reported here indicate that the phoP/Q locus is not required for the entry of Salmonella into DCs. One explanation of our results is that constitutive macropinocytosis, a hallmark of DCs (35, 36), allows the bacteria to be internalized independently of the virulence factors required for invasion of other cell types. Such a mechanism implies that, in DCs, adhesion is the key step that drives uptake. We report here that the binding to, and uptake of PhoPc by DCs is as efficient as that of wild-type Salmonella, a prerequisite for a similar loading of the cells for antigen presentation.
Attenuated PhoPc and wild-type S. typhimurium also exhibited the same survival in bone marrow-derived DCs. Garcia del Portillo et al. (37) recently reported that the S. typhimurium PhoP− strain, impaired in macrophage survival, survived in a transformed dendritic cell line. We extended our analysis to other strains impaired in macrophage survival and mutated in loci of various pathogenicity islands. Altogether, the results indicate for the first time that macrophage-specific virulence genes are not required for intracellular survival of Salmonella in DCs. The intracellular compartment that harbors Salmonella may be different in DCs or macrophages. This is supported by the observation that S. typhimurium resides in a vacuole lacking lysosomal markers like the lysosomal associated membrane protein-1 (Lamp-1) in the DC line (37), whereas in macrophages the bacteria containing vacuoles are Lamp-1-positive (38, 39). Thus, Salmonella ends up in a different intracellular compartment in DCs compared with macrophages, suggesting that the bacteria do not require in DCs the program of gene expression necessary for survival in macrophages. This would facilitate the survival of attenuated bacteria that lack this program.
The interaction of S. typhimurium with DCs is relevant to the in vivo situation. First, DCs, but not macrophages, are enriched at the site of entry of Salmonella in the Peyer's patches (2, 13). The recruitment of macrophages and polymorphonuclear cells that provide a first niche of infection is triggered by IL-8. This proinflammatory chemokine is released by the epithelial cells following interaction with wild-type S. typhimurium (40, 41). In contrast to wild-type Salmonella, the attenuated PhoPc bacteria are impaired in their ability to induce IL-8 production by epithelial cells (42). Consistent with this, PhoPc did not trigger an inflammatory reaction with massive influx of macrophages in the mouse Peyer's patches (S. Hopkins and J.-P. Kraehenbuhl, unpublished observations). Because no macrophages are recruited by the PhoPc strain, DCs remain the main target cell type below the follicle-associated epithelium of the Peyer's patches, the preferred portal of entry of Salmonella. We have previously identified DCs as the first Peyer's patch cell type to internalize wild-type and attenuated S. typhimurium in mice (15). The role of DCs in the uptake of bacteria may not be limited to gut-associated lymphoid tissues; they may also be involved in direct uptake of bacteria through the epithelium of the villi. Recently, CD18+ cells were shown to take up noninvasive Salmonella in the intestine (43). These CD18+ leukocytes could represent DCs. Interactions of Salmonella with DCs occur also in lymphoid tissues distant from the intestine. Bacteria have been found in mouse splenic DCs after oral administration of a S. typhimurium aroA mutant (44) or i.p. injection of Salmonella dublin (45).
Our results suggest that interaction of Salmonella, and in particular noninflammatory vaccine strains, with DCs plays an essential role in triggering an immune response, whereas macrophages participate in the clearance of the bacteria. This view is in agreement with the dual role played by macrophages in pathogenesis and the control of murine Salmonella infection as proposed by Wijburg et al. (46). By using a macrophage-specific depletion, these authors showed that macrophages are essential for the development of infection by virulent Salmonella. On the other hand, macrophages are responsible for the clearance of virulence-attenuated bacteria, but are not required for the induction of protective immunity after vaccination with an attenuated strain. We further propose DCs as the antigen-presenting cells important for the induction of an immune response during vaccination. According to current models, DCs are activated on interaction with bacterial or viral products. As a consequence of antigen deposition and inflammation, they differentiate, acquire a mature phenotype, and migrate to draining lymph nodes where they then stimulate T cells (26, 47). We show here that DCs infected in vitro with S. typhimurium mature and express costimulatory molecules that are crucial to activate naive CD4+ T lymphocytes. This is consistent with previous reports showing that the lipopolysaccharide from other Gram-negative bacteria induce the maturation of DCs (35, 48). The up-regulation of costimulatory molecules may explain why recombinant Salmonella are more efficient than the soluble antigen to induce T cell proliferation. Processing and presentation of antigens derived from wild-type S. typhimurium has already been reported in bone marrow-derived DCs (49), as well as the induction of cytokine release by splenic DCs infected with wild-type S. dublin (45). We show here that an attenuated strain is efficiently processed for antigen presentation by DCs, but not macrophages. This efficiency can be correlated with the efficient entry of the noninvasive bacteria in DCs.
Attenuated strains of S. typhimurium are attractive candidates as vectors for vaccination, because they target the intestinal DCs and initiate a mucosal immune response (50). There is now a rationale to design S. typhimurium strains efficient in mucosal immunization by screening mutants that would be unable to induce an inflammatory reaction and to survive in macrophages, but which retain the capacity to enter, survive in, and be presented by DCs.
Acknowledgments
We are grateful to Dr. S. Hopkins for her contribution to the initial experiments. We thank Drs. N. Takahashi, K. Murphy, M. Kopf, H. Acha-Orbea, B. Ahmer, D. Holden, E. Groisman, and J. Mekalanos for the generous gift of reagents, mice, or bacteria; Franco Ardizzoni (Centre de Microscopie Electronique, Lausanne) and Jeannine Bamat for expert assistance with scanning and transmission electron microscopy, respectively; Marcel Allegrini for photography; and Dr. Lucy Hathaway for reading the manuscript. This work was supported by grants from the the Swiss National Science Foundation (31–56936-99) and the Swiss League against Cancer (SKL 635–2—1998). F.N. holds a long-term European Molecular Biology Organization fellowship.
Abbreviations
- PhoPc
PhoP constitutive
- DCs
dendritic cells
- OVA
ovalbumin
- FACS
fluorescence-activated cell sorter
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darwin K H, Miller V L. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones B D, Ghori N, Falkow S. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen V B, Harty J T, Jones B D. Infect Immun. 1998;66:3758–3766. doi: 10.1128/iai.66.8.3758-3766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter-Dahlfors A, Buchan A M J, Finlay B B. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wick M J, Harding C V, Twesten N J, Normark S J, Pfeiffer J D. Mol Microbiol. 1995;16:465–476. doi: 10.1111/j.1365-2958.1995.tb02411.x. [DOI] [PubMed] [Google Scholar]
- 6.Garcia Véscovi E, Soncini F C, Groisman E A. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 7.Gunn J S, Miller S I. J Bacteriol. 1996;178:6369–6373. doi: 10.1128/jb.178.21.6369-6373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller S I, Mekalanos J J. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpuche-Aranda C M, Racoosin E L, Swanson J A, Miller S I. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopkins S, Kraehenbuhl J-P, Schodel F, Potts A, Peterson D, de Grandi P, Nardelli-Haefliger D. Infect Immun. 1995;63:3279–3286. doi: 10.1128/iai.63.9.3279-3286.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benyacoub J, Hopkins S, Potts A, Kelly S, Kraehenbuhl J-P, Curtiss R, III, De Grandi P, Nardelli-Haefliger D. Infect Immun. 1999;67:3674–3679. doi: 10.1128/iai.67.7.3674-3679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruedl C, Rieser C, Bock G, Wick G, Wolf G. Eur J Immunol. 1996;26:1801–1806. doi: 10.1002/eji.1830260821. [DOI] [PubMed] [Google Scholar]
- 13.Kelsall B L, Strober W. J Exp Med. 1996;183:237–247. doi: 10.1084/jem.183.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki A, Kelsall B L. J Exp Med. 2000;191:1381–1393. doi: 10.1084/jem.191.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins S, Niedergang F, Corthésy-Theulaz I, Kraehenbuhl J-P. Cell Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-5822.2000.00035.x. [DOI] [PubMed] [Google Scholar]
- 16.Wall K A, Lorber M I, Loken M R, McClatchey S, Fitch F W. J Immunol. 1983;131:1056–1064. [PubMed] [Google Scholar]
- 17.Fields P I, Swanson R V, Haidaris C G, Heffron F. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsolis R, Heffron F. Methods Cell Biol. 1994. 79–106. [DOI] [PubMed] [Google Scholar]
- 19.Shea J E, Beuzon C R, Gleeson C, Mundy R, Holden D W. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi N, Orita T, Hirose M. Gene. 1995;161:211–216. doi: 10.1016/0378-1119(95)00234-w. [DOI] [PubMed] [Google Scholar]
- 21.Lee C A, Falkow S. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galan J E, Curtiss R., III Infect Immun. 1990;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman R M. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inaba K, Turley S, Iyoda T, Yamaide F, Shimoyama S, Reis e Sousa C, Germain R N, Mellman I, Steinman R M. J Exp Med. 2000;191:927–936. doi: 10.1084/jem.191.6.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy K M, Heimberger A B, Loh D Y. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 26.Banchereau J, Steinman R M. Nature (London) 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 27.Pegues D A, Hantman M J, Behlau I, Miller S I. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 28.Francis C L, Ryan T A, Jones B D, Smith S J, Falkow S. Nature (London) 1993;364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 29.Galàn J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 30.Blanc-Potard A-B, Groisman E A. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallis T S, Galyov E E. Mol Microbiol. 2000;36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- 32.Galàn J E. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 33.Marcus S L, Brumell J H, Pfeifer C G, Finlay B B. Microb Infect. 2000;2:145–156. doi: 10.1016/s1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 34.Miller S I. Mol Microbiol. 1991;5:2073–2078. doi: 10.1111/j.1365-2958.1991.tb02135.x. [DOI] [PubMed] [Google Scholar]
- 35.Sallusto F, Cella M, Danieli C, Lanzavecchia A. J Exp Med. 1995;182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norbury C C, Chambers B J, Prescott A R, Ljunggren H-G, Watts C. Eur J Immunol. 1997;27:280–288. doi: 10.1002/eji.1830270141. [DOI] [PubMed] [Google Scholar]
- 37.Garcia del Portillo F, Jungnitz H, Rohde M, Guzman C A. Infect Immun. 2000;68:2985–2991. doi: 10.1128/iai.68.5.2985-2991.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh Y-K, Alpuche-Aranda C, Berthiaume E, Jinks T, Miller S I, Swanson J A. Infect Immun. 1996;64:3877–3883. doi: 10.1128/iai.64.9.3877-3883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mills S D, Finlay B B. Eur J Cell Biol. 1998;77:35–47. doi: 10.1016/S0171-9335(98)80100-3. [DOI] [PubMed] [Google Scholar]
- 40.Eckmann L, Kagnoff M F, Fierer J. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung H C, Eckmann L, Yang S-K, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff M F. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gewirtz A T, Siber A M, Madara J L, McCormick B A. Infect Immun. 1999;67:608–617. doi: 10.1128/iai.67.2.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasquez-Torres A, Jones-Carson J, Baumler A J, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks W T, Fang F C. Nature (London) 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 44.Paglia P, Medina E, Arioli I, Guzman C A, Colombo M P. Blood. 1998;92:3172–3176. [PubMed] [Google Scholar]
- 45.Marriott I, Hammond T G, Thomas E K, Bost K L. Eur J Immunol. 1999;29:1107–1115. doi: 10.1002/(SICI)1521-4141(199904)29:04<1107::AID-IMMU1107>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 46.Wijburg O L C, Simmons C P, van Rooijen N, Strugnell R A. Eur J Immunol. 2000;30:944–953. doi: 10.1002/1521-4141(200003)30:3<944::AID-IMMU944>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 47.Reis e Sousa C, Sher A, Kaye P. Curr Op Immunol. 1999;11:392–399. doi: 10.1016/S0952-7915(99)80066-1. [DOI] [PubMed] [Google Scholar]
- 48.Winzler C, Rovere P, Rescigno M, Granucci F, Penna G, Adorini L, Zimmermann V S, Davoust J, Ricciardi-Castagnoli P. J Exp Med. 1997;185:317–328. doi: 10.1084/jem.185.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svensson M, Stockinger B, Wick M J. J Immunol. 1997;158:4229–4236. [PubMed] [Google Scholar]
- 50.Sirard J-C, Niedergang F, Kraehenbuhl J-P. Immunol Rev. 1999;171:5–26. doi: 10.1111/j.1600-065x.1999.tb01340.x. [DOI] [PubMed] [Google Scholar]