Abstract
To investigate the importance of the rabies virus (RV) glycoprotein (G) in protection against rabies, we constructed a recombinant RV (rRV) in which the RV G ecto- and transmembrane domains were replaced with the corresponding regions of vesicular stomatitis virus (VSV) glycoprotein (rRV-VSV-G). We were able to recover rRV-VSV-G and found that particle production was equal to rRV. However, the budding of the chimeric virus was delayed and infectious titers were reduced 10-fold compared with the parental rRV strain containing RV G. Biochemical analysis showed equal replication rates of both viruses, and similar amounts of wild-type and chimeric G were present in the respective viral particles. Additional studies were performed to determine whether the immune response against rRV-VSV-G was sufficient to protect against rabies. Mice were primed with rRV or rRV-VSV-G and challenged with a pathogenic strain of RV 12 days later. Similar immune responses against the internal viral proteins of both viruses indicated successful infection. All mice receiving the rRV vaccine survived the challenge, whereas immunization with rRV-VSV-G did not induce protection. The results confirm the crucial role of RV G in an RV vaccine.
Rabies virus (RV), a negative-stranded RNA virus, is a member of the genus Lyssavirus within the family Rhabdoviridae. Five structural proteins, including the nucleoprotein (N), phosphoprotein, matrix protein (M), glycoprotein (G), and RNA-dependent RNA polymerase (L), are encoded by the 12-kb viral genome. G, the only protein exposed on the surface of the viral particle, is the mediator of both binding to cellular receptors and entry into host cells. Being a highly immunogenic protein, virus-neutralizing antibodies that are induced against this protein (1, 2) protect against RV infection (3). Furthermore, the RV G induces cytotoxic T lymphocytes (4) and T helper cells (5). Although RV G seems to be the major determinant of protection from RV infection, some researchers have reported that it is not essential in a rabies vaccine. Immunization of mice and raccoons with RV ribonucleoprotein (RNP) in complete Freund's adjuvant resulted in protection (6). Furthermore, administration of N alone or via a viral vector expressing RV N has resulted in protection against lethal challenge with RV (7–9). On the other hand, others have reported only partial protection after immunization with RV N and suggest that the immunization reduces the incubation period and clinical symptoms of rabies (10). Hence, the question of the importance of RV G alone or in combination with N in protection remains. Further clarification of this debate will help to develop more effective vaccines.
The development of an effective recovery system for the rescue of infectious RV from cDNA has allowed for direct manipulations of the RV genome and the production of genetically modified recombinant viruses. By using this technology, RVs expressing stable heterologous proteins have been rescued. Foreign genes such as CAT (11), CD4, and CXCR4 (12) or HIV-1 gp160 (13) have been inserted into the RV genome, in addition to the five standard genes. One advantage of rhabdovirus-based vectors is the possibility of incorporating foreign glycoproteins into viral particles. For RV, this process requires the replacement of the cytoplasmic domain of the foreign glycoprotein with the corresponding fragment of RV G (12, 14). This process seems to be different for vesicular stomatitis virus (VSV), another rhabdovirus. Incorporation of foreign glycoproteins into VSV particles only depends on the expression level of the glycoproteins and does not require any modification (15–17). Additionally, VSV even incorporates its own glycoprotein containing a foreign cytoplasmic domain as efficiently as wild-type VSV G protein (18).
To investigate the importance of RV G in inducing protection from RV infection, we developed a chimeric virus in which the RV G ecto- and transmembrane domains were replaced with those of VSV glycoprotein. The G of VSV, another rhabdovirus from the genus Vesiculovirus, was chosen for several reasons. First, both glycoproteins form homotrimers on the cellular surface (19–21). Second, no crossreactivity to antibodies against G exists between the species. As a result, the two viruses will be as similar as possible, thereby inhibiting any overlap in immune responses specific for RV G. This chimeric virus will allow us to analyze the importance of RV G in protection against a pathogenic RV infection in the context of the other four RV proteins.
As we report here, a recombinant RV (rRV) containing a chimeric VSV/RV G (cVSV-G) instead of RV G was constructed and recovered. This chimeric virus grew to high titers but 10-fold lower than wild-type RV. In contrast to foreign glycoproteins containing the RV G cytoplasmic tail, the chimeric G protein was incorporated as efficiently as wild-type RV protein. Recombinant virions were produced at similar levels as wild-type particles, but infectivity was reduced 10-fold. Mice were immunized with rRV or rRV-VSV-G virus, but protection against pathogenic RV challenge was induced only by rRV priming. We therefore conclude that RV G is an essential component of an effective RV vaccine.
Materials and Methods
Plasmid Construction.
PCRs were performed with VENT polymerase (New England Biolabs) to minimize introduction of sequence errors. Two unique restriction sites were introduced into the previously described RV cDNA pSAD-L16 (22) upstream of RV G (SmaI) and downstream of the ψ (NheI) by using site-directed mutagenesis (GeneEditor, Promega) and primer RP11 5′-CCT CAA AAG ACC CCG GGA AAG ATG GTT CCT CAG 3′ (SmaI, bold) and RP1 5′-GCC GTC CTT TCA ACG ATC CAA GTC TCG CGA TTT TGC TAG CTT ATA AAG TGC TGG GTC ATC TAA GC-3′ (NheI, bold). The resulting plasmid was entitled pSN and infectious virus recovered with pSN was designated rRV. To construct an RV containing the ecto- and transmembrane domains of VSV G fused to the cytoplasmic tail of RV G, the cytoplasmic tail of RV G was PCR amplified from pSN with the primers RP8 5′-CCT CTA GAT TAC AGT CTG GTC TCA CCC CC 3′ (XbaI, bold) and RP29 5′-CCC GGG TTA ACA GAA GAG TCA ATC GAT CAG AAC 3′ (HpaI, bold). The ecto- and transmembrane domains of VSV G gene were amplified from pVSV-XN1 (23) with the primers RP33 5′-TTA AGT TAA CCA AGA ATA GTC CAA TGA-3′ (HpaI, bold) and RP34 5′-TCT CGA GCC CGG GAC TAT GAA GTG CCT TTT GTA C-3′ (XbaI, bold). Both PCR products were digested with HpaI and ligated. The ligation products were PCR reamplified with the primers RP8 and RP34, and the PCR product was cloned by using the XmaI and XbaI sites into pSN that had been digested with XmaI and NheI. The resulting plasmid was designated pSN-VSV-G, and the infectious virus recovered by using pSN-VSV-G was designated rRV-VSV-G.
Recovery of Infectious RVs from cDNA.
BSR-T7 cells, which constitutively express T7 RNA polymerase (24), were transfected as described (25) with pSN or pSN-VSV-G along with 2.5 μg of pTIT-N, 1.25 μg of pTIT-P, 1.25 μg of pTIT-L, and 1 μg of pTIT-G plasmids (25) by using a calcium phosphate transfection kit (Stratagene). Supernatants were transferred to BSR cells 3 days later. The cells were analyzed 3 days after for infectious RV by immunostaining with an anti-RV N monoclonal antibody.
Immunofluorescence Assays.
To ensure expression of RV G and VSV G by rRV and rRV-VSV-G, respectively, BSR cells (BHK clones) were plated on coverslips in a 6-well plate. The cells were infected with rRV or rRV-VSV-G at a multiplicity of infection (moi) of 0.1. Supernatants were removed 48 h later, and the cells were washed twice with 1× PBS and fixed as noted below. For internal staining, cells were fixed with 80% (vol/vol) acetone at 4°C. FITC-labeled antibody against RV N (Centocor) was added and incubated at 37°C for 30 min. For surface staining, cells were fixed at 4°C with 3% (vol/vol) paraformaldehyde, blocked with PBS containing 1% BSA and 10 mM glycine for 1 h at room temperature, and incubated with either two monoclonal anti-VSV G antibodies (I1, I4; ref. 23) diluted 1:200 or a polyclonal antibody directed against RV G, diluted 1:400. After incubation for another 30 min, secondary antibodies were added [RV G: 1:100 donkey anti-rabbit FITC (Jackson ImmunoResearch) VSV G: 1:100 donkey anti-mouse FITC (Jackson ImmunoResearch)] and analyzed by fluorescent microscopy.
One-Step Growth Curve.
BSR cells were grown to ≈70% confluency in 60-mm dishes. Cells were infected at an moi of 3 focus-forming units (ffu). After incubation at 37°C for 1 h, cells were washed four times with PBS to remove all unabsorbed virus. DMEM [3 ml 10% (vol/vol) FBS] was added. 100 μl of culture supernatants was removed at each time point (8, 16, 24, 48, and 72 h). Infectious titers of supernatants were determined on BSR cells in duplicate.
Northern Blot Analysis.
BSR cells (5 × 106) were plated in a 6-well plate and infected the next day with rRV or rRV-VSV-G with an moi of 1. Total RNA was isolated from each culture 48 h later with an RNA isolation kit (Qiagen). Blots were performed as described (26). RV N specific probes were synthesized by digesting pTIT-N (25) with NcoI/PstI and the N gene fragment was isolated. pSPBN-IG was digested with XbaI/HpaI and the 1.5-kb band was used as a VSV G probe. To create the ψ + L probe, pSN was digested with NheI/StuI and the 1,500 kb-band was isolated.
Virus Incorporation Assays.
BSR cells were grown to ≈70% confluency in 6-well plates. Cells were infected with rRV or rRV-VSV-G at an moi of 50 overnight. DMEM containing 0.3% BSA and lacking methionine were used to replace the original cell media of the first well per virus, after which 250 μCi (1 Ci = 37 GBq) [35S]methionine was added. Media were removed from the cells 24 h later, transferred to microcentrifuge tubes, and stored at 4°C. The procedure was repeated for the remaining wells at 48 and 72 h. Virions were purified twice by ultracentrifugation over 20% sucrose. Virus pellets were resuspended in 100 μl PBS, and viral proteins were separated by SDS/PAGE. Viral proteins were quantified by PhosphorImaging.
Endoglycosidase H (Endo H) Assays.
BSR cells were plated at a density of 2 × 106 in 35-mm plates and infected 24 h later with rRV-VSV-G at an moi of 2. Cells were washed 16 h later and labeled with methionine-negative media (DMEM, 0.3% BSA) containing 100μCi [35S]methionine for 10 min and chased for 0, 30, 40, 50, 60, 90, or 120 min. Cells were lysed with lysis buffer [50 mM Tris, pH 7.4/150 mM NaCl/1% Nonidet P-40/1× protease inhibitor mixture (Sigma)]. The cell debris was pelleted and the protein suspension was transferred to a microcentrifuge tube. Endo H (New England Biolabs) digestions were completed for 4 h, and 1 μl of a rabbit antibody directed against the RV G cytoplasmic domain was added to each sample. After overnight incubation at 4°C, Protein A-agarose (GIBCO/BRL) was added, incubated for 4 h at 4°C, and washed three times with RIPA buffer (1% Nonidet P-40/1% deoxycholate/0.1% SDS/150 mM NaCl/10 mM Tris⋅HCl, pH 7.4). After denaturing at 95°C for 2 min, the proteins were separated by SDS/PAGE (10%) and analyzed by a PhosphorImager (Molecular Dynamics).
Immunization and Challenge.
Two groups of 10 BALB/c female mice aged 6–8 weeks were immunized i.m. with 1 × 106 ffu of rRV or 1 × 106 ffu of rRV-VSV-G, and another group of 10 mice served as a control. Nine days later, all mice were bled. All mice were challenged 12 days after immunization with 5.5 × 106 ffu of RV strain CVS-N2C (27) i.m. and evaluated daily for 4 weeks for clinical signs of rabies.
ELISA.
ELISAs were performed as described (13) by using RV G protein or RV RNP (28) as the target antigen. Briefly, 200 ng of antigen (RV G or RNP) was diluted in 10 ml of coating buffer (50 mM Na2CO3, pH 9.6) and used to coat 96-well plates (Maxisorp plates, Nunc) overnight at 4°C. Plates were blocked with 5% milk powder in PBS for 1 h then washed three times. Sera dilutions were added as indicated (see Fig. 7). Plates were incubated for 1 h at room temperature and washed three times, and the secondary antibody (goat anti-mouse IgG horseradish peroxidase, Jackson ImmunoResearch Laboratories) was added (1:5,000). After 30 min at 37°C, the plates were washed, and 200 μl of OPD substrate (Sigma) was added and incubated at room temperature for 15 min. The reaction was stopped by adding 50 μl of 3 M H2SO4. Optical density was determined at 490 nm on a DuPont kinetic microplate reader.
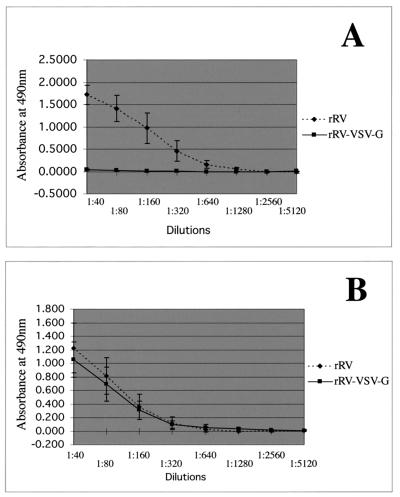
Figure 7.
ELISA analysis of mouse sera against RV RNP and RV G. Sera were collected 9 days after priming from all mice and analyzed for reactivity against RV G (A) or RV RNP (B). Sera from each mouse were run in duplicate. Graphs represent the average of 10 mice per group with the error bars representing standard deviations.
Results
Construction and Expression of Recombinant Viruses.
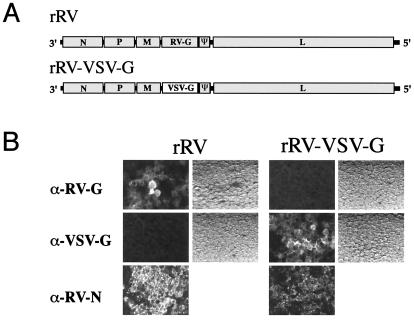
The goal of this study was to investigate further the role of the RV glycoprotein in protection against RV challenge. Having the ability to recover infectious RV from cDNA opens the possibility to recover genetically similar RVs that are only distinguished by their viral glycoprotein. We chose to introduce a portion of the VSV G gene because both viruses have a similar structure and life cycle but are serologically unrelated. First, we developed an RV vaccine strain (SAD B19; ref. 29)-based vector (pSAD L16; ref. 22) by introducing two unique restriction sites (SmaI/NheI) flanking the coding region of RVG (Fig. 1A). The plasmid was designated pSN. Because studies indicated that the RV G cytoplasmic tail is required for incorporation of a foreign glycoprotein into budding RV, we used a PCR strategy to construct a chimeric RV/VSV G that contains the VSV ecto- and transmembrane domains fused to the RV G cytoplasmic tail. The PCR product was used to replace the gene coding for the RV G gene within pSN, resulting in the plasmid pSN-VSV-G.
Figure 1.
(A) Genome of rRV and rRV-VSV-G. RV SAD L16 was modified to contain a SmaI site upstream of the RV G gene and a NheI site upstream of the ψ region. rRV-VSV-G was created by replacing the RV G ecto- and transmembrane domains with those of VSV G by using a newly created HpaI site. (B) Expression of RV G and VSV G by rRVs. BSR cells were infected with rRV or rRV-VSV-G (moi of 0.1) and immunostained 48 h later with anti-RV-G, anti-VSV-G, or anti-RV-N antibody. Light field pictures are shown for anti-RV-G and anti-VSV-G.
Pilot studies indicated that wild-type VSV efficiently infects BSR cells. This suggested that we should be able to recover both viruses by standard RV-recovery protocols. As expected, 50% of the cultures transfected with pSN or pSN-VSV-G produced rRV or rRV-VSV-G, respectively. Next, we evaluated the rRVs derived from these cultures for expression of their respective glycoproteins by immunostaining (Fig. 1B). Cells were infected with rRV or rRV-VSV-G and immunostained for detection of RV G and VSV G on the cell surface (Fig. 1B). Additionally, cells were stained internally for RV N as a control. As expected, rRV-infected cells stained positive for RV N and RV G expression but not VSV G. rRV-VSV-G cultures were positive for RV N as well as VSV G, but not for RV G. These results indicate that both glycoproteins, wild type and chimeric, are being expressed on the surface of infected cells.
Replication Kinetics of Recombinant Viruses.
While growing stocks of each virus, we noticed the infectious titer of rRV-VSV-G was typically lower than that of rRV. To examine the replication rates of both viruses, a one-step growth curve was performed. BSR cells were infected with rRV or rRV-VSV-G at an moi of 3 to allow synchronous infection of all cells and were washed extensively 1 h later to remove any unabsorbed virus. Fresh medium was added back and small aliquots of supernatant were taken at 8, 16, 24, 48, and 72 h after infection. Viral titers were determined at the indicated time points. As shown in Fig. 2, rRV-VSV-G replicated at a rate nearly 1 log less than rRV. However, the chimeric virus remains capable of growing to high titers, 107, which is sufficient for a live virus vaccine approach.
Figure 2.
One-step growth curves of rRV and rRV-VSV-G. BSR cells were infected with an moi of 10 with either rRV (♦) or rRV-VSV-G (▪). Supernatant (100 μl) was taken at 8, 16, 24, 48, and 72 h after infection and titered in duplicate.
Transcription of rRVs.
The lower infectious titers and replication kinetics of rRV-VSV-G in comparison to rRV could be caused by alterations in the transcription of rRV-VSV-G. To examine the transcription rate of the glycoprotein genes of each virus, we infected BSR cells with rRV or rRV-VSV-G at an moi of 1. Total RNA was isolated 48 h after infection, and Northern blot analysis was performed with three different probes specific for RV N, VSV G, or RV-ψ + L. As shown in Fig. 3A, the ratio of genomic RNA to N mRNA is approximately the same for both viruses. Hybridization with a VSV-G-specific probe confirmed the transcription of cVSV-G by rRV-VSV-G (Fig. 3B). The amounts of G transcripts could be directly compared by using a probe specific for the entire ψ gene, the 3′-noncoding region in the RV G mRNA that both viruses still contain, and a small fragment of the 5′ terminus of RV L. We found that the transcription rates of the RV G and the cVSV-G were analogous (Fig. 3C). Because viral gene transcription of rRV and rRV-VSV-G are equivalent, the lower titer of rRV-VSV-G does not result from a lower transcription rate of the G mRNA.
Figure 3.
Northern blot analysis of rRVs. BSR cells were infected with rRV or rRV-VSV-G (moi of 1), and total RNA was isolated 24 h later. RNA was separated by electrophoresis and transferred to a nylon membrane that was hybridized with a probe specific for RV-N (A), VSV-G (B), or a cDNA fragment spanning the entire RV-ψ and a portion of the RV-L gene (ψ + L) (C). G/L = RV G + L bicistronic RNA, L = RV L, G = glycoprotein, N = RV-N.
Comparison of Viral Spread.
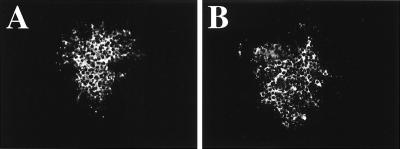
To determine whether the lower infectious titer of rRV-VSV-G was caused by a decrease in the ability of the virus to spread to neighboring cells, BSR cells were infected at an moi of 0.1 with rRV or rRV-VSV-G. Cells were fixed 2 days later and immunostained with a monoclonal antibody specific for RV N. Foci size was examined and was similar for the two viruses (Fig. 4), suggesting that the lower titer of rRV-VSV-G is not caused by slower cell-to-cell spread.
Figure 4.
Comparison of foci size between rRV and rRV-VSV-G. BSR cells were infected with rRV and rRV-VSV-G (moi of 0.1). Cells were fixed 48 h after infection and immunostained with an RV-N-specific antibody. (A) rRV. (B) rRV-VSV-G.
Composition of Recombinant Virions.
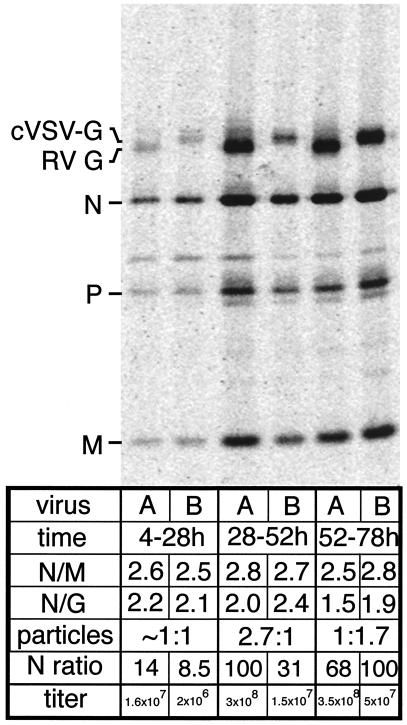
Studies have indicated that the cytoplasmic domain of RV G is required for incorporation of a foreign glycoprotein into budding RV particles. Deletion of this domain from RV G resulted in a 6-fold reduced production of RV particles. To analyze whether the lower titer of rRV-VSV-G is caused by a reduced production of virions, a reduced infectivity of the viral particles, or both, we designed an experiment which enabled us to determine both parameters. BSR cells were infected with rRV or rRV-VSV-G (moi of 50) and labeled with [35S]methionine for 24 h over a time course of 4–28, 28–52, or 52–78 h. Infectious titers from each time point were determined, and virions from supernatants were purified over 20% (vol/vol) sucrose. Viral proteins were separated by SDS/PAGE and quantified with a PhosphorImager. As shown in Fig. 5, the N/M ratio is similar for both rRV and rRV-VSV-G, suggesting that the composition of the recombinant virions is not affected by cVSV-G. Surprisingly, the ratio of N/G, after correcting for the different methionine contents, was also constant for rRV and rRV-VSV-G, indicating that both Gs are efficiently incorporated. This could be due in part to the similar structure and function of the Gs.
Figure 5.
Particle production and infectious titers of rRV and rRV-VSV-G. BSR cells were infected with rRV (A) or rRV-VSV-G (B) at an moi of 50 and labeled with [35S]methionine for 24 h over 4–28, 28–52, or 52–78 h. Labeled virions were purified twice over 20% (vol/vol) sucrose and disrupted. Then the viral proteins were separated by SDS/PAGE. Gels were analyzed by PhosphorImaging, and viral proteins were quantified. The ratio of N protein to M protein (N/M) per virus was determined along with the ratios of N protein to G protein (N/G). The lane with the highest amount of N protein was determined for each virus and set to equal 100 (N ratio lane) and the other values were calculated in proportion to 100. The N ratios of rRV and rRV-VSV-G were compared and estimated in the particles row. A portion of the initial supernatant was used to determine the infectious titer at each time point (titer).
To determine virus production over time, we analyzed the amount of virus particles produced during each time point. The highest value of N protein per virus was set to 100 and the other two time points were calculated in proportion to this standard (Fig. 5, N ratio). rRV and rRV-VSV-G both produced few particles during the first 24 h of infection. However, rRV increased particle production rapidly during the next 24 h, whereas rRV-VSV-G peaked during the final 24 h of infection. We also determined the infectious titers from each time point for both viruses and compared it to the amount of viral particles. The titers of rRV-VSV-G were ≈10-fold less than rRV at each time point. Of note, the total amount of virions produced over 72 h was similar for both viruses, but the lower infectious titer of rRV-VSV-G suggests that 10-fold more particles of rRV-VSV-G are noninfectious as compared with rRV virions.
Glycoprotein Processing.
One possible explanation for the delay in budding of rRV-VSV-G is a slower transport of cVSV-G to the cell surface. Wild-type VSV G contains two N-linked glycosylation sites (30), whereas wild-type RV G contains 3–4 putative sites (31), although each one is not always used (32, 33). The difference in both composition and glycosylation sites of the glycoproteins may cause a fluctuation in time required for the protein to be transported through the secretory pathway. To investigate the transport and processing of rRV-VSV-G glycoprotein, we monitored the accumulation of endo H-resistant oligosaccharides. rRV-VSV-G-infected cells were pulse-labeled with [35S]methionine for 10 min, chased for 0, 30, 40, 50, 60, 90, or 120 min, lysed, and subjected to endo H or mock digestion. The viral glycoproteins were then extracted by using immunoprecipitation with an RV G tail-specific antibody. As can been seen in Fig. 6, ≈50% of the oligosaccharides on the cVSV-G of rRV-VSV-G become endo H resistant within 50 min. A half-time of 50 min for H-resistant sugars has been reported for several RV G proteins (20, 34, 35). These results indicate that the delayed budding of rRV-VSV-G is not caused by a slower transport of cVSV-G.
Figure 6.
Transport kinetics of cVSV-G. Infected cells were pulsed with [35S]methionine for 10 min and chased for 0, 30, 40, 50, 60, 90, or 120 min. Cell extracts were incubated with or without endo H for 4 h at 37°C and immunoprecipitated with an RV G endoplasmic tail-specific antibody. Proteins were separated by SDS/PAGE and analyzed by PhosphorImaging. Endo H + or − indicates with or without endo H.
Immunization of Mice with rRV or rRV-VSV-G.
As described above, the chimeric virus, rRV-VSV-G, is similar to wild-type RV, but each has a serologically distinct glycoprotein. Therefore, we will be able to answer the question of the importance of the RV glycoprotein in protection. Because rRV and rRV-VSV-G contain the same internal proteins, protection studies that use these two viruses for immunization against a highly neurotropic challenge of RV will indicate the significance of the immune response directed toward G after priming.
Two groups of 10 BALB/c mice were immunized i.m. in the hind leg with 106 ffu of rRV or rRV-VSV-G. Another group of 10 mice was not infected and served as a control for challenge. All mice were bled 9 days after immunization, and their sera were analyzed by ELISA to detect specific humoral responses against RV RNP and RV G (Fig. 7). As expected, RV-G-specific antibodies were detected only in sera of mice immunized with rRV (Fig. 7A). However, both groups of mice had similar humoral responses specific for RV RNP (Fig. 7B). This result indicated that both viruses were able to infect and replicate similarly in the immunized mice consequently creating an immune response against the internal proteins of RV.
To analyze whether the induced immune response was sufficient to protect the mice from infection with a pathogenic RV, a challenge experiment was performed. All mice were infected 12 days after immunization i.m. in the hind leg with 5.5 × 106 ffu of CVS-N2C (27), a highly pathogenic strain of RV. Mice were evaluated for clinical signs of rabies for 4 weeks (Table 1). As expected, all mice receiving one immunization of rRV were completely protected. However, nonimmunized mice and mice immunized once with rRV-VSV-G were not protected, and all animals died by 8 days after challenge. There was no statistically significant difference in survival time between nonimmunized and mice immunized with rRV-VSV-G. These data clearly indicate that RV G protein is required for protection against a highly pathogenic RV challenge. Although an immune response was detected against the internal proteins of both viruses, it was not sufficient to confer protection against this particular challenge virus.
Table 1.
Survival rates of mice primed with rRV or rRV-VSV-G and challenged with a highly pathogenic rabies virus
Mice were challenged with 5.5 × 106 ffu of a highly pathogenic strain of rabies virus, CVS-N2C (27).
10 mice were primed i.m. with 1 × 106 ffu rRV.
10 mice were primed i.m. with 1 × 106 ffu rRV-VSV-G.
Discussion
The present study describes the generation of an RV, rRV-VSV-G, in which the RV G gene has been replaced with a chimeric G gene encoding the ecto- and transmembrane domains of VSV G and the cytoplasmic domain of RV G. The data presented herein demonstrate that rRV-VSV-G buds efficiently and incorporates similar levels of the cVSV-G glycoprotein compared with wild-type RV.
Reports have indicated that foreign glycoproteins expressed from rhabdoviruses are efficiently incorporated into rhabdovirus particles. In the case of VSV, it has been shown that no specific amino acid sequence is required to promote incorporation of most foreign viral and cellular membrane proteins into VSV virions. These proteins were incorporated by their expression level rather than a specific signal located within the transmembrane or cytoplasmic domain of VSV G (15, 16).
In contrast to VSV, a signal sequence located within the cytoplasmic tail of RV G is always required to incorporate a foreign membrane protein into RV particles (12, 14). An RV lacking the cytoplasmic domain of RV G displayed a different phenotype than wild-type RV as indicated by lower viral titers, 6-fold reduced particle production, and a reduced amount of RV G on virus particles (34). As shown by Mebatsion et al. (34, 36), the RV G cytoplasmic tail specifically interacts with the internal viral proteins, probably M. In experiments with RVs expressing a chimeric HIV-1 envelope protein (gp160) in which we replaced the cytoplasmic tail with that of RV G, we have noticed a 5% rate of incorporation compared with RV G, whereas wild-type HIV-1 gp160 was not detected (H.D.F. and M.J.S., unpublished data). Our rRV-VSV-G containing the chimeric VSV/RV protein produced different results. Whereas rRV-VSV-G had reduced titers like the RV G tail-deleted RV, incorporation of the chimeric VSV/RV-G protein into virions and particle production of rRV-VSV-G were similar to rRV. One explanation why cVSV-G is incorporated so efficiently may be the structural similarity between the two rhabdoviral G proteins. Both glycoproteins form homotrimers on the cellular surface and have the same function in the viral life cycle (19–21), and therefore, the ectodomain of VSV may efficiently substitute for the RV domain.
Consequently, why is the budding of rRV-VSV-G delayed compared with RV? The Northern blot analysis indicated equal levels of mRNA encoding the glycoproteins and an endo H assay indicated a similar transport rate to the Golgi for both cVSV-G and RV G. These results suggest that the later onset of virion production for rRV-VSV-G is not caused by a lower transcription rate of the cVSV-G mRNA or slower transport kinetics of the VSV/RV protein. One possible explanation for slower budding of rRV-VSV-G virions is that less of the chimeric VSV/RV protein is produced within the infected cells, or the protein is less stable.
As described above, we were able to create a virus differing only in the glycoprotein gene. Several studies have analyzed the effects of the immune responses against the internal RV proteins on protection from RV challenge. Dietzschold et al. (6) reported that liposomes containing RNPs conferred no protection in mice against lethal intracerebral challenge with RV, but mice and raccoons resisted lethal peripheral challenge with homologous or heterologous RV strains after immunization with RNPs in complete Freund's adjuvant. Priming with a recombinant vaccinia virus expressing RV N protein protected five of seven dogs against a challenge with a street RV, but three of the five dogs developed VNA after challenge, indicating a protective immune response induced by the challenge virus (10). The mechanism by which internal RV proteins induce protection is not well understood. It was proposed that priming with RV N protein induces antiviral cytokines, the production of virus-neutralizing antibodies from CD4+ T cells, or the induction of RV-specific cytotoxic T lymphocytes, but the exact mechanism is still unknown (6, 9).
Published results are all based on other viral vectors used for expression of RV proteins or the administration of native or rRV proteins. The possibility to recover genetically manipulated RV from cDNA enabled us to analyze the immune response against two very similar RVs, just different in their single-surface glycoprotein G. As described above, we detected a similar humoral response against the internal RV proteins in sera from rRV- and rRV-VSV-G-primed mice. Because a single inoculation with killed RV does not induce detectable immune responses against RV RNP by ELISA (C.A.S. and M.J.S., unpublished data), the response must have resulted from replicating RV, indicating similar replication rates of both viruses after the infection. Only the sera of rRV infected mice reacted positively against RV G. As the uninfected control group, all rRV-VSV-G-primed mice failed to survive a peripheral challenge with a pathogenic RV strain. Of note, a less pathogenic challenge virus might have resulted in protection, but the minimal requirement for an RV vaccine is certainly an effective immune response against a highly pathogenic RV strain. Taken together, these data indicate that expression of RV G protein is a requirement for an effective RV-based vaccine.
We also can apply these results to our approach with RV-based vectors as vaccines for other viral diseases such as HIV-1. We described an rRV expressing HIV-1 gp160 in addition to the other five RV proteins. The same vector containing cVSV-G instead of RV G should not be neutralized by a humoral response against RV G, thus permitting several viral life cycles to boost the immune responses against the expressed foreign proteins.
Acknowledgments
This study was supported by National Institutes of Health Grants AI45097 (to B.D.) and AI44340 (to M.J.S.), by internal Thomas Jefferson University funds (to M.J.S.), and by the Center for Human Virology.
Abbreviations
- RV
rabies virus
- G
glycoprotein
- rRV
recombinant RV
- VSV
vesicular stomatitis virus
- N
nucleoprotein
- M
matrix protein
- L
RNA-dependent RNA polymerase
- RNP
ribonucleoprotein
- cVSV-G
chimeric VSV/RV G
- moi
multiplicity of infection
- ffu
focus-forming units
- endo H
endoglycosidase H
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011510698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011510698
References
- 1.Cox J H, Dietzschold B, Schneider L G. Infect Immun. 1977;16:754–759. doi: 10.1128/iai.16.3.754-759.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiktor T J, Gyorgy E, Schlumberger D, Sokol F, Koprowski H. J Immunol. 1973;110:269–276. [PubMed] [Google Scholar]
- 3.Perrin P, Thibodeau L, Sureau P. Vaccine. 1985;3:325–332. doi: 10.1016/s0264-410x(85)90224-5. [DOI] [PubMed] [Google Scholar]
- 4.Macfarlan R I, Dietzschold B, Koprowski H. Mol Immunol. 1986;23:733–741. doi: 10.1016/0161-5890(86)90084-2. [DOI] [PubMed] [Google Scholar]
- 5.Celis E, Ou D, Dietzschold B, Koprowski H. J Virol. 1988;62:3128–3134. doi: 10.1128/jvi.62.9.3128-3134.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzschold B, Wang H H, Rupprecht C E, Celis E, Tollis M, Ertl H, Heber-Katz E, Koprowski H. Proc Natl Acad Sci USA. 1987;84:9165–9169. doi: 10.1073/pnas.84.24.9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii H, Takita-Sonoda Y, Mifune K, Hirai K, Nishizono A, Mannen K. J Gen Virol. 1994;75:1339–1344. doi: 10.1099/0022-1317-75-6-1339. [DOI] [PubMed] [Google Scholar]
- 8.Sumner J W, Fekadu M, Shaddock J H, Esposito J J, Bellini W J. Virology. 1991;183:703–710. doi: 10.1016/0042-6822(91)90999-r. [DOI] [PubMed] [Google Scholar]
- 9.Fu Z F, Dietzschold B, Schumacher C L, Wunner W H, Ertl H C, Koprowski H. Proc Natl Acad Sci USA. 1991;88:2001–2005. doi: 10.1073/pnas.88.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fekadu M, Sumner J W, Shaddock J H, Sanderlin D W, Baer G M. J Virol. 1992;66:2601–2604. doi: 10.1128/jvi.66.5.2601-2604.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mebatsion T, Schnell M J, Cox J H, Finke S, Conzelmann K-K. Proc Natl Acad Sci USA. 1996;93:7310–7314. doi: 10.1073/pnas.93.14.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mebatsion T, Finke S, Weiland F, Conzelmann K-K. Cell. 1997;90:841–847. doi: 10.1016/s0092-8674(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 13.Schnell M J, Foley H D, Siler C A, McGettigan J P, Dietzschold B, Pomerantz R J. Proc Natl Acad Sci USA. 2000;97:3544–3549. doi: 10.1073/pnas.050589197. . (First Published March 7, 2000; 10.1073/pnas.050589197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mebatsion T, Conzelmann K-K. Proc Natl Acad Sci USA. 1996;93:11366–11370. doi: 10.1073/pnas.93.21.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kretzschmar E, Buonocore L, Schnell M J, Rose J K. J Virol. 1997;71:5982–5989. doi: 10.1128/jvi.71.8.5982-5989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnell M J, Buonocore L, Kretzschmar E, Johnson E, Rose J K. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell M J, Johnson J E, Buonocore L, Rose J K. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 18.Schnell M J, Buonocore L, Boritz E, Ghosh H P, Chernish R, Rose J K. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudin Y, Ruigrok R W, Tuffereau C, Knossow M, Flamand A. Virology. 1992;187:627–632. doi: 10.1016/0042-6822(92)90465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitt M, Buonocore L, Prehaud C, Rose J K. Virology. 1991;185:681–688. doi: 10.1016/0042-6822(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 21.Doms R W, Keller D S, Helenius A, Balch W E. J Cell Biol. 1987;105:1957–1969. doi: 10.1083/jcb.105.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnell M J, Mebatsion T, Conzelmann K-K. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnell M J, Buonocore L, Whitt M A, Rose J K. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchholz U J, Finke S, Conzelmann K-K. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finke S, Conzelmann K-K. J Virol. 1999;73:3818–3825. doi: 10.1128/jvi.73.5.3818-3825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conzelmann K-K, Schnell M. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morimoto K, Hooper D C, Carbaugh H, Fu Z F, Koprowski H, Dietzschold B. Proc Natl Acad Sci USA. 1998;95:3152–3156. doi: 10.1073/pnas.95.6.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider L G, Dietzschold B, Dierks R E, Matthaeus W, Enzmann P J, Strohmaier K. J Virol. 1973;11:748–755. doi: 10.1128/jvi.11.5.748-755.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conzelmann K-K, Cox J H, Schneider L G, Thiel H J. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 30.Rose J K, Gallione C J. J Virol. 1981;39:519–528. doi: 10.1128/jvi.39.2.519-528.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietzschold B. J Virol. 1977;23:286–293. doi: 10.1128/jvi.23.2.286-293.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shakin-Eshleman S H, Remaley A T, Eshleman J R, Wunner W H, Spitalnik S L. J Biol Chem. 1992;267:10690–10698. [PubMed] [Google Scholar]
- 33.Wunner W H, Dietzschold B, Smith C L, Lafon M, Golub E. Virology. 1985;140:1–12. doi: 10.1016/0042-6822(85)90440-4. [DOI] [PubMed] [Google Scholar]
- 34.Mebatsion T, Konig M, Conzelmann K-K. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 35.Mebatsion T, Schnell M J, Conzelmann K-K. J Virol. 1995;69:1444–1451. doi: 10.1128/jvi.69.3.1444-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mebatsion T, Weiland F, Conzelmann K-K. J Virol. 1999;73:242–250. doi: 10.1128/jvi.73.1.242-250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]