Abstract
Antisense radiopharmaceuticals could be used to image gene expression in the brain in vivo, should these polar molecules be made transportable through the blood–brain barrier. The present studies describe an antisense imaging agent comprised of an iodinated peptide nucleic acid (PNA) conjugated to a monoclonal antibody to the rat transferrin receptor by using avidin–biotin technology. The PNA was a 16-mer antisense to the sequence around the methionine initiation codon of the luciferase mRNA. C6 rat glioma cells were permanently transfected with a luciferase expression plasmid, and C6 experimental brain tumors were developed in adult rats. The expression of the luciferase transgene in the tumors in vivo was confirmed by measurement of luciferase enzyme activity in the tumor extract. The [125I]PNA conjugate was injected intravenously in anesthetized animals with brain tumors and killed 2 h later for frozen sectioning of brain and film autoradiography. No image of the luciferase gene expression was obtained after the administration of either the unconjugated antiluciferase PNA or a PNA conjugate that was antisense to the mRNA of a viral transcript. In contrast, tumors were imaged in all rats administered the [125I]PNA that was antisense to the luciferase sequence and was conjugated to the targeting antibody. In conclusion, these studies demonstrate gene expression in the brain in vivo can be imaged with antisense radiopharmaceuticals that are conjugated to a brain drug-targeting system.
Keywords: blood–brain barrier, transferrin receptor, monoclonal antibody, peptide nucleic acid
The availability of the human genome sequence will accelerate the pace of the discovery of pathologic genes that cause cancer or chronic disease in brain and other organs, and in vivo gene imaging technology is needed. Gene expression is imaged in vitro with antisense technology based on the complementary hybridization of an antisense agent with a target mRNA sequence. However, the extension of antisense technology to imaging gene expression in vivo is limited by several factors including rapid metabolism in vivo, toxicity, and poor transport of antisense agents across biological membranes (1). Antisense imaging of gene expression in the brain is particularly difficult because of the presence of the blood–brain barrier (BBB).
Potential antisense imaging agents include phosphodiester (PO) oligodeoxynucleotides (ODN), phosphorothioate (PS)-ODNs, or peptide nucleic acids (PNA). PO-ODNs have been radiolabeled as antisense imaging agents, but these molecules are rapidly degraded in vivo by 3′ exonucleases and endonucleases (2). PS-ODNs are more metabolically stable, but these agents are neurotoxic (3, 4), probably because of the avidity of PS-ODNs for binding multiple cellular proteins (5), and are strongly bound by plasma proteins (6). In addition, PS-ODNs activate RNase H (7). Formation of the duplex between the PS-ODN antisense radiopharmaceutical and the target mRNA would lead to degradation of the target transcript, which is not desired in a diagnostic modality. The PNAs have a polyamide backbone (8) and are not susceptible to degradation by nucleases (9). PNAs do not activate RNase H and would appear to be an ideal antisense imaging agent, particularly because the melting point of nucleic acid duplexes formed by PNAs is much higher than with PO- or PS-ODNs (8). However, PNAs do not cross cell membranes in general and do not cross the BBB (10). Therefore, the successful imaging of gene expression in vivo will require the development of a brain drug-targeting technology that can be applied to antisense radiopharmaceuticals.
Antisense agents such as PNAs can be made transportable through the BBB with the use of the chimeric peptide technology, as described previously (10). In this approach, the drug that is normally not transported through the BBB is biotinylated and then bound to a conjugate of streptavidin (SA) and a brain drug-targeting vector (11). The latter is a ligand such as a peptide or peptidomimetic mAb that undergoes receptor-mediated transcytosis (RMT) through the BBB in vivo by virtue of binding to one of several endogenous peptide receptor systems on the brain capillary endothelium, which forms the BBB in vivo. Transferrin or transferrin receptor (TfR) peptidomimetic mAbs, such as the mouse OX26 mAb to the rat TfR, undergo RMT through the BBB in rats in vivo (11). In the present studies, the conjugate of the OX26 mAb and SA is used and is designated OX26/SA or SA-OX26. The antisense imaging agent is a PNA, which hybridizes to the region around the methionine initiation codon of the luciferase mRNA. C6 rat glioma cells were permanently transfected with a luciferase expression plasmid (Fig. 1A), and the luciferase transgene was expressed in C6 experimental brain tumors in adult rats. The specific expression of the luciferase transgene in the brain tumors was imaged after the i.v. injection of the imaging agent in rats with brain tumors expressing the luciferase gene in vivo.
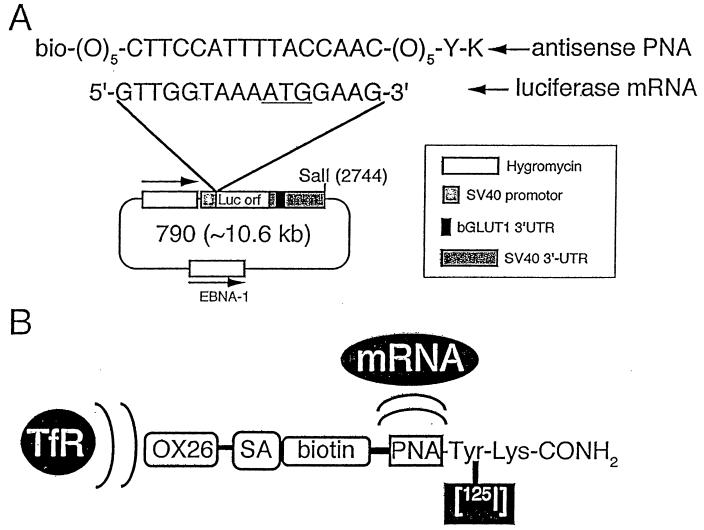
Figure 1.
(A) The sequence of the antiluciferase antisense PNA is shown along with the biotin (bio) moiety at the amino terminus and the tyrosine (Y) and lysine (K) moiety at the carboxy terminus. There are five linkers (O) at both the near carboxy and near amino termini. The sequence of luciferase mRNA around the methionine initiation codon (ATG) is shown. The luciferase (Luc) ORF (orf) was subcloned into a eukaryotic expression plasmid, designated clone 790, which was described previously (12). The hygromycin selection gene, the SV40 promoter, the SV40 3′ UTR, and the Epstein–Barr nuclear antigen (EBNA)-1 gene are shown. The SV40 3′ UTR contains 200 nucleotides from the bovine (b) GLUT1 glucose transporter mRNA, which optimizes gene expression through mRNA stabilization (12). (B) Antisense imaging agent is comprised of the OX26 mAb to the rat TfR, which is linked to SA, which binds the monobiotinylated PNA antisense agent. The PNA contains a Tyr and Lys residue at the amidated carboxy terminus to enable radiolabeling with 125-iodine on the Tyr residue or with 111-indium on the Lys residue.
Experimental Procedures
Conjugate Synthesis.
The sequence of the antiluciferase PNA is shown in Fig. 1A and was synthesized by Applied Biosystems and contained a biotin at the amino terminus followed by 5 linkers, followed by the 16-mer nucleic acid sequence, followed by another 5 linkers, followed by a tyrosine and lysine residue and an amidated carboxy terminus. Each of the five linkers is comprised of -NH(CH2)2-O-(CH2)2-O-CH2-CO-, which are incorporated in the PNA synthesis by the manufacturer. The calculated molecular mass of the PNA was 6,193 kDa, and the observed molecular mass of the PNA was 6,193 kDa, as determined by mass spectrometry. A control PNA that should not hybridize to any transcripts in brain was prepared with a sequence antisense to the rev gene of the HIV. The anti-rev PNA had the following nucleic acid sequence: CTCCGCTTCTTCCTGCCA, and has been described previously (10). Either PNA was radioiodinated with 125-iodine and chloramine T, as described previously (10), to a specific activity of 75–90 μCi/μg and a trichloroacetic acid (TCA) precipitability of 95–98%. The conjugate of the OX26 mAb and recombinant SA was prepared as previously described by using a stable thioether linkage (10).
Luciferase Expression Plasmid.
C6 glioma cells were stably transfected with the luciferase gene by using clone 790, which has been described previously (12), and these transfected cells are designated C6–790. Clone 790 contains a 200-bp fragment of the 3′ untranslated region (UTR) of the Glut1 glucose transporter mRNA, corresponding to nucleotides 2100–2300 of the GLUT1 mRNA sequence, and this was inserted within the luciferase mRNA 3′ UTR. The insertion of the GLUT1 3′ UTR element into the SV40 3′ UTR maximizes luciferase gene expression in C6 glioma cells by stabilizing the mRNA (12).
Experimental Brain Tumors.
The C6–790 cells were implanted in the caudate–putamen nucleus of male CD Fischer 344 rats (Harlan Breeders, Indianapolis, IN), weighing 250–275 g under stereotaxic guidance, as described previously (13). The animals were examined 14 days later. To confirm expression of the luciferase transgene in the tumor in vivo, tumor extracts were prepared in Promega lysis buffer, and luciferase enzyme activity was measured with luciferin as substrate (Promega) by using a Berthold (Nashua, NH) luminometer, as described previously (12). A luciferase standard curve was also assayed, and the enzyme activity was expressed as picograms of luciferase equivalent per milligram of tissue protein. The C6–790 cells were grown in tissue culture as described previously (12), and these cells were also extracted in Promega lysis buffer for measurement of luciferase activity in the cells grown in tissue culture before implantation in brain. Control experiments were performed with C6 cells described previously (13), and these cells were used to develop brain tumors exactly as described above, except these tumor cells were not transfected with the luciferase transgene.
Autoradiography in Tumor-Bearing Rats.
Fourteen days after implantation of C6–790 cells, the rats were anesthetized with ketamine/xylazine for i.v. injection of brain-imaging agents, as described previously (13). Each rat received 100 μCi of PNA labeled with [125I], and 3 groups of rats were studied: Group A rats received antiluciferase PNA conjugated to OX26/SA; group B received antiluciferase PNA without conjugation to the brain targeting system; and group C received anti-rev PNA conjugated to OX26/SA. In these studies, each rat received 0.2 nmol of PNA and 40 μg (0.2 nmol) of OX26/SA. Each rat was also administered 20 mg of l-tyrosine and 2 mg of sodium iodide i.p. 15 min before the study to block brain uptake of radiolabeled metabolites such as [125I]tyrosine or iodide. The animals were decapitated 2 h after i.v. injection of the isotope, the brain was rapidly removed, cut into coronal slabs, immediately frozen in powdered dry ice, and tissue blocks were stored at −70°C. Cryostat sections of 15-μm thickness were prepared on a Bright cryostat and mounted on glass cover slips, which were then exposed to Reflection blue film with intensifying screens (Dupont/NEN). X-ray films were exposed at −70°C for 3 days, followed by development for 1 minute in Kodak developer and fixation for 5 min in Kodak fixer. The x-ray film was scanned in a UMAX flatbed scanner with transparency adapter, cropped in Adobe (Adobe Systems, Mountain View, CA) photoshop 5.5 on a G4 Power Macintosh, and images were colorized with NIH image software. After autoradiography, the glass cover slips were stained with Mayer's hematoxylin to visualize the tumor, and these specimens were subsequently scanned and imaged. All brain scans or all autopsy stains were scanned and colorized simultaneously.
Although the autoradiography was performed on frozen sections of brain, the imaging of gene expression was performed in vivo, because the radiolabeled antisense imaging agent was administered in vivo and was not applied to tissue sections in vitro.
Pharmacokinetics and Organ Uptake in Nontumor-Bearing Rats.
Either the unconjugated antiluciferase PNA or the antiluciferase PNA conjugated to OX26/SA was injected intravenously into ketamine/xylazine-anesthetized adult male Sprague–Dawley rats (270–300 g) by using methods described previously (10). The dose of radioactivity in these experiments was 5 μCi/rat of PNA (0.02 nmol) and 20 μg/rat of OX26/SA (0.1 nmol).
RNase Protection Assay.
The luciferase RNase protection assay demonstrated specific hybridization of the antiluciferase PNA to the target luciferase mRNA despite conjugation to the OX26/SA vector. These methods were identical to those described previously (10). The luciferase RNA was prepared with a luciferase transcription plasmid, designated clone 760, which has been described previously (14). The sense RNA was synthesized with T7 RNA polymerase after linearization of the plasmid with EcoRI. The transcribed RNA was radiolabeled with [32P-α]ATP, and the correct size of the radiolabeled transcribed sense RNA was determined by agarose/formaldehyde gel electrophoresis followed by film autoradiography. For the RNase protection assay, 0.5 pmol of biotinylated luciferase PNA with or without conjugation to 10 pmol of OX26/SA was added to 105 cpm of 32P-labeled sense luciferase RNA (8 fmol) in 3 μl buffer (50 mM NaCl/5 mM Tris, pH 8/0.5 mM EDTA) and annealed for 30 min at 56°C. Then 15 units of RNase T1 and 0.4 units of RNase A were added to samples in 15 μl of RNase digestion buffer III (Ambion, Austin, TX). RNA fragments were analyzed by 7 M urea/20% PAGE and autoradiography, as described previously (10). Labeled RNA and PNA were heat denatured for 2 min at 95°C and then incubated on ice for 2 min immediately before the experiment or conjugation to OX26/SA.
Uptake and Pulse–Chase Experiments in Cultured C6 and C6–790 Cells.
The uptake of 125I-labeled anti-rev or antiluciferase PNA was investigated in C6–790 and C6 cells in the presence or absence of OX26SA. Cells were grown in 24-well dishes and incubated with 5 μCi/ml (12 nM) [125I]PNA with and without OX26SA (1:1 molar ratio) for 2, 4, 6, or 24 h. The monolayers were then washed 3 times in 2.5 ml cold PBS (10 mM phosphate buffer, pH 7.2/150 mM NaCl) and lysed with 250 μl reporter lysis buffer (Promega), as previously described (12). Aliquots of samples (100 μl) and standards (10 μl) were precipitated with TCA, and the percent of medium PNA that was taken up into the TCA-precipitable cellular fraction was measured and expressed either as nanograms PNA per milligram protein or percentage uptake per milligram protein. Twenty-microliter lysate aliquots were also resolved by SDS in a 12% gel. Gels were fixed in 50% MeOH and 10% acetic acid solution for 30 min, incubated in 7% MeOH, 7% acetic acid and 1% glycerol for 5 min, and dried before autoradiography with Kodak BioMax film and intensifying screens. For the pulse–chase study, either control C6 or C6–790 cells were incubated as described above with 10 μCi/ml (24 nM) [125I]antiluciferase PNA conjugated to OX26SA for 24 h. The medium was discarded and fresh medium added that contained no additional radiolabeled PNA. TCA-precipitable cellular radioactivity was then measured at 0, 2, 6, or 24 h of incubation.
Results
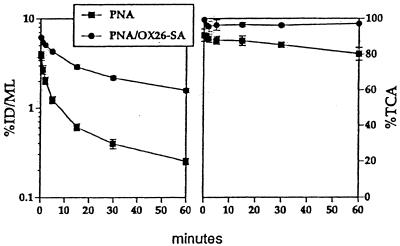
The [125I]antiluciferase PNA, with or without conjugation to the OX26/SA drug-targeting system, was injected intravenously into adult Sprague–Dawley rats. The profile of plasma radioactivity for the unconjugated PNA or for the PNA conjugate is shown in Fig. 2. The plasma clearance of the unconjugated PNA and of the PNA conjugate was 7.2 ± 0.4 and 1.1 ± 0.1 ml/min/kg, respectively, and the plasma area under the concentration curve (AUC) was inversely related to the plasma clearance. The delayed plasma clearance of the PNA conjugate was paralleled by an increase in metabolic stability as reflected by the high percentage of plasma radioactivity that was precipitable by TCA for at least 60 min after i.v. injection of the PNA conjugate (Fig. 2).
Figure 2.
(Left) Plasma radioactivity, expressed as percent of ID/ml, is plotted vs. time after i.v. injection for the unconjugated PNA and the PNA conjugate, designated PNA/OX26-SA. Data are mean ± SE (n = three rats). (Right) The percent of plasma radioactivity that is TCA precipitable is shown.
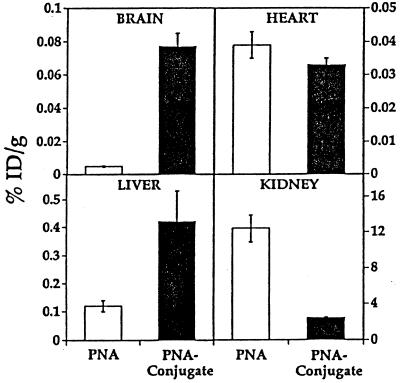
Organ uptake of the radiolabeled PNA or PNA conjugate was measured 60 min after the i.v. injection, and these data are shown in Fig. 3. There was no measurable transport of the unconjugated PNA into brain. However, there was an increase in brain uptake of the PNA after conjugation to the OX26/SA drug-targeting system, and this level of brain uptake, 0.08% of injected dose (ID) per gram of brain, is in excess of the brain uptake of a neuroactive small molecule such as morphine (11). There was no specific targeting of the PNA to heart (Fig. 3), because conjugation of the PNA restricts membrane permeability in heart in parallel with an increase in the plasma AUC (Fig. 2), and these have offsetting effects on the percent of ID per gram (11). There was increased uptake of the PNA conjugate in liver (Fig. 2) because of the expression of the transferrin receptor on hepatocytes in vivo. There was a decrease in the renal uptake of the PNA conjugate, because conjugation of the PNA to the OX26/SA vector, which has a molecular mass of 200,000 kDa, effectively restricts glomerular filtration of the 6,000-kDa PNA.
Figure 3.
The percent of ID delivered per gram of tissue is shown for brain, heart, liver, and kidney. Data are mean ± SE (n = three rats per group). The radiolabeled PNA was injected in one of two forms: (i) unconjugated, which is designated PNA in the figure, and (ii) as a conjugate of the PNA and the OX26/SA targeting system, which is designated PNA conjugate. Mean ± SE (n = three rats).
The ability of the PNA to hybridize to the target mRNA after conjugation to the OX26/SA drug-targeting system was demonstrated by an RNase A/T1 protection assay. Both the unconjugated and the PNA conjugate hybridized to the luciferase sense RNA and resulted in protection of 16-mer RNA fragments. These results indicated that conjugation of the antiluciferase PNA to the OX26/SA drug-targeting system did not impair the hybridization of the PNA to the target mRNA.
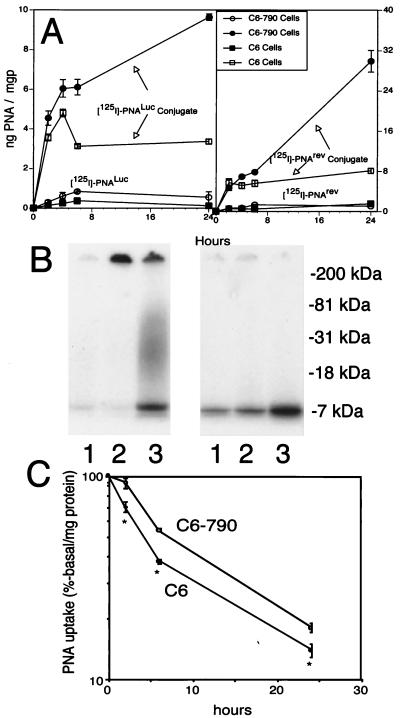
The uptake of either the unconjugated anti-rev PNA or the unconjugated antiluciferase PNA by either the C6 cells or the C6–790 cells was negligible (Fig. 4A). However, either PNA was taken by these cell lines after conjugation to OX26/SA (Fig. 4A). By 24 h of incubation, the [125I]anti-rev PNA was metabolized, and the [125I]tyrosine was recycled into cellular proteins as shown by the SDS/PAGE (Fig. 4B Left). However, the [125I]antiluciferase PNA was metabolically stable during the 24-h incubation period, as no radioactivity incorporated into cellular proteins was detected (Fig. 4B Right). The metabolic stability of the [125I]antiluciferase PNA enabled further pulse–chase experiments, and these showed preferential retention of the [125I]antiluciferase PNA in the C6–790 cells, compared with the C6 cells lacking the luciferase mRNA (Fig. 4C).
Figure 4.
(A) The uptake in either C6 cells or C6–790 cells is plotted against time for four different [125I] tracers: the unconjugated antiluciferase PNA (designated PNALuc), the unconjugated anti-rev PNA (designated PNArev), the PNALuc conjugated to OX26/SA, and the PNArev conjugated to OX26/SA. Data are mean ± SE (n = three rats) for each time point. (B) Film autoradiography after SDS/PAGE of C6–790 cell extracts obtained at 4 (lane 1), 6 (lane 2), or 24 (lane 3) h incubation with either the PNArev conjugate (Left) or the PNALuc conjugate (Right). (C) Pulse–chase experiment showing rate of loss of radioactivity from the cellular TCA precipitable fraction during a second 24-h period after the C6 cells or C6–790 cells were pulsed during an initial 24-h incubation with the labeled PNALuc conjugate. Data are mean ± SE (n = three dishes) for each time point. The radioactivity was significantly higher in the C6–790 cells at all time points (* indicates P < 0.01, Student's t test).
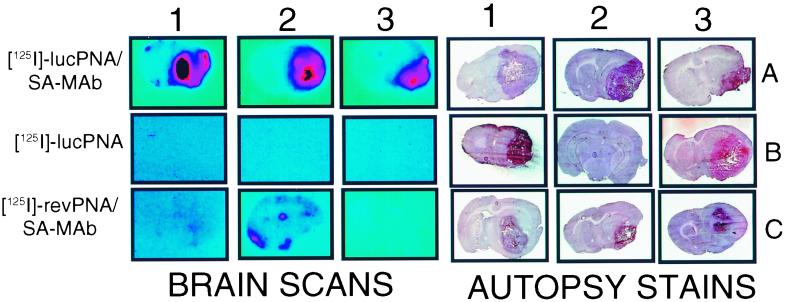
The brain scans and autopsy stains for three different groups of adult Fischer rats bearing the C6–790 gliomas are shown in Fig. 5. The luciferase activity in the tumor extract and in the C6–790 cells in tissue culture was 204 ± 66 and 76 ± 2 pg equivalent per milligram of tissue protein, respectively, indicating the luciferase transgene was fully expressed in the experimental tumor in vivo. Group A rats received the radiolabeled antiluciferase PNA conjugated to the OX26/SA drug-targeting system, which is designated SA-mAb in Fig. 5. Group B rats received the antiluciferase PNA without conjugation to the drug-targeting system. Group C rats received the anti-rev antisense PNA that was conjugated to the OX26/SA drug-targeting system. All rats formed medium to large tumors with the exception of rat 2 in group B, as shown by the autopsy stains (Fig. 5). There was no imaging of either normal brain or brain tumor in the group B rats after i.v. injection of the luciferase PNA without conjugation to the drug-targeting system, because the PNA does not cross the BBB in either normal brain or in the tumor. Conversely, there was imaging of luciferase gene expression in the brain tumor in all group A rats after i.v. injection of the luciferase PNA conjugated to the drug-targeting system. The size of the tumor imaged with the antisense radiopharmaceutical was comparable to the size of the tumor shown on the autopsy stain (Fig. 5). In contrast, there was no imaging of the tumors after conjugation of the rev antisense PNA to the drug-targeting system as shown in the group C rats (Fig. 5). In further control experiments, C6 cells not transfected with the luciferase transgene (13) were grown as experimental tumors in 10 rats. At 14 days after implantation, 5 rats received 100 μCi each of the unconjugated [125I]antiluciferase PNA, and 5 rats received 100 μCi of 125I-labeled antiluciferase PNA conjugated to the OX26/SA drug-targeting system. Brains were removed at 2 h, frozen sections prepared, and film autoradiography performed with the same methods used for the studies shown in Fig. 5. The brain sections were scanned in parallel with the sections from the group A rats (Fig. 5), and no measurable radioactivity was detectable in these control C6 tumors with either the unconjugated or conjugated antiluciferase PNA.
Figure 5.
Brain scans (Left) and autopsy stains (Right) are shown for three groups of rats designated A, B, and C. Group A rats received an i.v. injection of the [125I] antiluciferase PNA bound to the conjugate of the OX26 mAb and SA, which is designated SA-mAb. Group B rats received [125I]antiluciferase PNA without conjugation to SA-OX26. Group C rats received an i.v. injection of [125I]anti-rev PNA bound to the SA-OX26 conjugate, which is designated SA-mAb.
Discussion
These studies are consistent with the following conclusions. First, it is not possible to image gene expression in the brain in vivo with an unconjugated antisense radiopharmaceutical, because these molecules do not cross the BBB in vivo. Second, antisense imaging of gene expression in the brain in vivo is possible if a BBB drug-targeting technology is used. The development of an antisense imaging agent for in vivo applications requires the merger of antisense technology and drug-targeting technology.
The antisense imaging agent is comprised of four domains (Fig. 1B). The first domain is the peptidomimetic mAb that targets the TfR, which is expressed on both the BBB and the tumor cell membrane (13). Transport through both of these membranes is required because the target of the antisense imaging agent, the luciferase mRNA, is localized in the cytoplasm of the tumor cells. The TfR is expressed on brain cells (15) and on C6 glioma cells (13), and the data in Fig. 4A show the increased uptake of the PNA conjugate by C6 cells. The data in Fig. 3 show increased transport across the BBB of the PNA after conjugation to the targeting vector. Therefore, the targeting system enables transport of the PNA across both the BBB and the C6 tumor cell membrane. The second part of the imaging agent is the linker domain, comprised of the SA moiety, which is attached to the mAb through a stable thioether linkage, and the biotin moiety, which is incorporated at the amino terminus of the PNA, as shown in Fig. 1B. The third domain of the antisense imaging agent is the radionuclide. At the carboxy terminus of the PNA, there are tyrosine (Y) and lysine (K) residues to enable radiolabeling with either 125-iodine or 111-indium, respectively. In the present study, the PNA was radiolabeled on the tyrosine residue with 125-iodine. The carboxy terminus of the PNA is amidated to enhance resistance to carboxypeptidases. The fourth domain of the imaging agent is the antisense sequence of the PNA which hybridizes to the target mRNA (Fig. 1). The RNase protection assay demonstrates hybridization of the PNA to the target mRNA despite conjugation to the drug-targeting vector.
The experimental model used in these studies is a C6 glioma brain tumor that expresses the luciferase gene in vivo (Results). The C6 cells were permanently transfected with the luciferase gene, and these cells produce high levels of the luciferase mRNA. The abundance of the luciferase mRNA in these cells is comparable to that of the actin mRNA (12, 16). This high expression of the luciferase mRNA is caused by the insertion of a cis element derived from the Glut1 glucose transporter mRNA 3′ UTR into the luciferase mRNA 3′ UTR. This modification greatly stabilizes the luciferase mRNA and augments the cellular level of the transcript (12). Therefore, the gene targeted in the present studies is expressed at high levels, and this expression is exclusive to the tumor cell with no luciferase gene expression in other cells of brain. These factors contribute to the marked differences in imaging the tumor vs. normal brain by using the PNA conjugate (Fig. 5).
The brain scans in Fig. 5 show that the tumor expressing the luciferase transgene is not imaged with a PNA radiopharmaceutical that is not conjugated to a brain drug-targeting system (group B, Fig. 5). These findings corroborate the brain uptake measurements in control rats with the antiluciferase PNA (Fig. 3) and previous studies showing no transport of a PNA across the BBB (10). Antisense molecules are highly charged and form extensive hydrogen bonding in aqueous solution, which restricts the transport across the endothelial plasma membranes forming the BBB in vivo (11). Tyler et al. (17) report that unconjugated PNAs do cross the BBB in vivo. In this study, the uptake of the PNA by rat brain was measured with a gel shift analysis of extracts of saline-perfused brain. However, this report shows the brain uptake of the PNA is <0.0001% ID/g (17), which is a level of brain uptake comparable to that of sucrose, a molecule that traverses the BBB at the lower limit of detection (11). In contrast, the brain uptake of the PNA conjugate is 3 logarithm orders of magnitude greater (Fig. 3), and this higher brain uptake enables imaging of gene expression in vivo (Fig. 5).
A radiolabeled PS-ODN has been reported to cross the BBB to enable imaging of gene expression for glial fibrillary acidic protein in experimental brain tumors in rats (18). However, this study actually uses a drug-targeting technology, because the [11C]PS-ODN 25-mer included cholesterol conjugated at the 5′ terminus. The addition of a cholesterol moiety to ODNs increases cellular uptake in tissue culture (19, 20). The conjugation of cholesterol to drugs is a “lipidization” drug-targeting strategy. The problem with this approach is that the cholesterol adduct is soluble only in organic solvents, and the i.v. administration of these solvents can cause solvent-mediated disruption of the BBB. In studies with the 5′ cholesterol-[11C]PS-ODN, the conjugate was solubilized in dichloromethane before i.v. injection (18). Dichloromethane is a solvent that is neurotoxic (21).
The imaging of gene expression in brain requires the use of an antisense agent with the correct sequence, as the brain tumor was not imaged with an anti-rev PNA conjugated to SA-OX26 (group C, Fig. 5). The brain uptake of the anti-rev PNA conjugated to SA-OX26 is higher than the brain uptake of a PNA administered without conjugation to the targeting system (10). However, the differential brain uptake between the conjugated PNA (group C, Fig. 5) and the unconjugated PNA (group B, Fig. 5) is not observed with the film autoradiography, because the brain radioactivity in either case is below the limits of detection. The exposure of the film to the brain sections was limited to 3 days (Experimental Procedures), because this duration was sufficient to image the region of interest, which was the brain tumor. The absence of tumor imaging in the group C rats (Fig. 5) shows that tumor imaging with the targeted antiluciferase PNA is not derived from binding of the anti-TfR mAb to the tumor cells and is not derived from leakiness of the C6 glioma. Similar findings were made with imaging of brain tumors with peptide radiopharmaceuticals conjugated to the SA-OX26 targeting system. In these studies, radiolabeled human epidermal growth factor (EGF) was conjugated to OX26 and administered to rats with C6 experimental tumors. However, no imaging of the tumor was observed, because the C6 cells did not express the EGF receptor (13). In another study, the EGF peptide radiopharmaceutical that was conjugated to OX26 was administered to nude rats bearing U87 human glial brain tumors that did overexpress the human EGF receptor (22). In this model, the brain tumor was clearly imaged compared with normal brain (22), similar to the result of the present study (Fig. 5). The imaging of structures within the brain with peptide or antisense radiopharmaceuticals requires that two conditions be met. First, the radiopharmaceutical must be enabled to traverse the BBB and/or brain cell membrane so that the radiopharmaceutical can access the target. Second, the region of interest must overexpress the target receptor, in the case of a peptide radiopharmaceutical, or the target mRNA in the case of an antisense radiopharmaceutical. Binding of the radiopharmaceutical to the target receptor or mRNA within the region of interest causes a sequestration of the radioactivity in that region. The selective sequestration of the antiluciferase PNA conjugate by the C6–790 cells is shown in Fig. 4C. The difference between the rate of efflux of the PNA from the C6–790 cells and the control C6 cells in tissue culture is not large. This is because the total number of PNA molecules taken up by the cells in the conjugate form is very high and is greatly in excess of the amount of luciferase mRNA. After 24 h of incubation in cell culture, there is 10 ng PNA per milligram of protein (Fig. 4A). This is equivalent to 1.2 × 106 PNA molecules per cell, given 106 cells per milligram of protein. Therefore, the number of PNA molecules inside the cell is at least 100-fold greater than the number of luciferase mRNA molecules. However, the ratio of PNA/mRNA molecules is much lower in vivo. Given a brain uptake of 0.08% ID/g (Fig. 3), and assuming 100 mg protein per gram of brain and 106 mg protein per cell, then there are only about 900 PNA molecules per cell in vivo. This number most likely approximates the luciferase mRNA copy number in the tumor cells. The approximation of the number of PNA molecules per cell by the number of target mRNA molecules in vivo enables the selective sequestration of the labeled PNA in the target cell in vivo. This accounts for the high signal-to-noise ratio in the tumor relative to normal brain (Fig. 5, group A).
Imaging gene expression with antisense radiopharmaceuticals requires that the imaging agent traverse three membranes in series: the BBB, the brain-target cell membrane, and the intracellular endosomal membrane. PNAs are able to traverse the endosomal membrane once the PNA is taken up by the cell (23). Moreover, the endosomal membrane may be a more formidable barrier in cultured cells than in vivo. Recent studies have shown that 85-nm pegylated immunoliposomes are able to enter the cytoplasm after transport across the BBB and the neuronal plasma membrane. This was inferred from the finding of active β-galactosidase gene expression in brain after the i.v. injection of this exogenous gene (24).
The brain drug-targeting technology described in these studies in rats uses peptidomimetic mAbs that bind endogenous BBB peptide receptor systems (11). The OX26 mAb and transferrin bind to different sites on the BBB TfR, and very large doses, 190 mg/kg of OX26 mAb, are required to inhibit brain uptake of circulating transferrin (25). The dose of OX26 mAb used in these imaging studies is 160 μg/kg (Experimental Procedures), which is 3 logarithm orders of magnitude lower than the mAb dose that inhibits endogenous transport (25). The brain drug-targeting technology used in these experiments in rats could be adapted to the imaging of brain gene expression in humans. In this case, the human insulin receptor (HIR) mAb, which is up to 10-fold more active in primates than the TfR mAb (11), would be used as the BBB-targeting agent. The insulin receptor is also widely expressed on brain cells (26). A genetically engineered chimeric HIR mAb has been prepared, has the same affinity for the HIR as the original murine HIR mAb (27), and could be used to target antisense imaging agents across the BBB in humans.
Abbreviations
- BBB
blood–brain barrier
- TfR
transferrin receptor
- ODN
oligodeoxynucleotide
- PS
phosphorothioate
- PNA
peptide nucleic acid
- TCA
trichloroacetic acid
- ID
injected dose
- SA
streptavidin
- UTR
untranslated region
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250332397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250332397
References
- 1.Hnatowich D J. J Nucleic Med. 1999;40:693–703. [PubMed] [Google Scholar]
- 2.Kang Y-S, Boado R J, Pardridge W M. Drug Metab Dispos. 1995;23:55–59. [PubMed] [Google Scholar]
- 3.Whitesell L, Geselowitz D, Chavany C, Fahmy B, Walbridge S, Alger J R, Neckers L M. Proc Natl Acad Sci USA. 1993;90:4665–4669. doi: 10.1073/pnas.90.10.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wojcik W J, Swoveland P, Zhang X, Vanguri P. J Pharmacol Exp Ther. 1996;278:404–410. [PubMed] [Google Scholar]
- 5.Rockwell P, O'Connor W J, King K, Goldstein N I, Zhang L M, Stein C A. Proc Natl Acad Sci USA. 1997;94:6523–6528. doi: 10.1073/pnas.94.12.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu D, Boado R J, Pardridge W M. J Pharmacol Exp Ther. 1996;276:206–211. [PubMed] [Google Scholar]
- 7.Crooke S T. FASEB J. 1993;7:533–539. doi: 10.1096/fasebj.7.6.7682523. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen P E, Egholm M, Buchardt O. Bioconjugate Chem. 1994;5:3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- 9.Demidov V V, Potaman V N, Frank-Kamenetskii M D, Egholm M, Buchard O, Sonnichsen S H, Nielsen P E. Biochem Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 10.Pardridge W M, Boado R J, Kang Y-S. Proc Natl Acad Sci USA. 1995;92:5592–5596. doi: 10.1073/pnas.92.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardridge W M J. J Cereb Blood Flow Metab. 1997;17:713–731. doi: 10.1097/00004647-199707000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Boado R J, Pardridge W M. Mol Brain Res. 1998;59:109–113. doi: 10.1016/s0169-328x(98)00134-x. [DOI] [PubMed] [Google Scholar]
- 13.Kurihara A, Deguchi Y, Pardridge W M. Bioconjugate Chem. 1999;10:502–511. doi: 10.1021/bc980123x. [DOI] [PubMed] [Google Scholar]
- 14.Tsukamoto H, Boado R J, Pardridge W M. J Neurochem. 1997;68:2587–2592. doi: 10.1046/j.1471-4159.1997.68062587.x. [DOI] [PubMed] [Google Scholar]
- 15.Mash D C, Pablo J, Flynn D D, Efange S M N, Weiner W J. J Neurochem. 1990;55:1972–1979. doi: 10.1111/j.1471-4159.1990.tb05784.x. [DOI] [PubMed] [Google Scholar]
- 16.Boado R J, Li J Y, Nagaya M, Zhang C, Pardridge W M. Proc Natl Acad Sci USA. 1999;96:12079–12084. doi: 10.1073/pnas.96.21.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyler B M, Jansen K, McCormick D J, Douglas C L, Boules M, Stewart J A, Zhao L, Lacy B, Cusack B, Fauq A, Richelson E. Proc Natl Acad Sci USA. 1999;96:7053–7058. doi: 10.1073/pnas.96.12.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobori N, Imahori Y, Mineura K, Ueda S, Fujii R. NeuroReport. 1999;10:2971–2974. doi: 10.1097/00001756-199909290-00018. [DOI] [PubMed] [Google Scholar]
- 19.de Smidt P C, Doan T L, de Falco S, van Berkel T J C. Nucleic Acids Res. 1991;19:4695–4700. doi: 10.1093/nar/19.17.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieg A M, Tonkinson J, Matson S, Zhao Q, Saxon M, Zhang L-M, Bhanja U, Yakubov L, Stein C A. Proc Natl Acad Sci USA. 1993;90:1048–1052. doi: 10.1073/pnas.90.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebert C S, Mattecucci M M, Pryor G T. Pharmacol Biochem Behav. 1990;36:351–356. doi: 10.1016/0091-3057(90)90416-f. [DOI] [PubMed] [Google Scholar]
- 22.Kurihara A, Pardridge W M. Cancer Res. 1999;54:6159–6163. [PubMed] [Google Scholar]
- 23.Chinnery P F, Taylor R W, Diekert K, Lill R, Turnbull D M, Lightowlers R N. Gene Ther. 1999;6:1919–1928. doi: 10.1038/sj.gt.3301061. [DOI] [PubMed] [Google Scholar]
- 24.Shi N, Pardridge W M. Proc Natl Acad Sci USA. 2000;97:7567–7572. doi: 10.1073/pnas.130187497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda F, Raja K B, Simpson R J, Trowbridge I S, Bradbury M W B. J Neurochem. 1993;60:106–113. doi: 10.1111/j.1471-4159.1993.tb05828.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhao W, Chen H, Xu H, Moore E, Meiri N, Quon M J, Alkon D L. J Biol Chem. 1999;274:34893–34902. doi: 10.1074/jbc.274.49.34893. [DOI] [PubMed] [Google Scholar]
- 27.Coloma M J, Lee H J, Kurihara A, Landaw E M, Boado R J, Morrison S L, Pardridge W M. Pharm Res. 2000;17:266–274. doi: 10.1023/a:1007592720793. [DOI] [PubMed] [Google Scholar]