Abstract
Stem cells, which are clonogenic cells with self-renewal and multilineage differentiation properties, have the potential to replace or repair damaged tissue. We have directly isolated clonogenic human central nervous system stem cells (hCNS-SC) from fresh human fetal brain tissue, using antibodies to cell surface markers and fluorescence-activated cell sorting. These hCNS-SC are phenotypically 5F3 (CD133)+, 5E12+, CD34−, CD45−, and CD24−/lo. Single CD133+ CD34− CD45− sorted cells initiated neurosphere cultures, and the progeny of clonogenic cells could differentiate into both neurons and glial cells. Single cells from neurosphere cultures initiated from CD133+ CD34− CD45− cells were again replated as single cells and were able to reestablish neurosphere cultures, demonstrating the self-renewal potential of this highly enriched population. Upon transplantation into brains of immunodeficient neonatal mice, the sorted/expanded hCNS-SC showed potent engraftment, proliferation, migration, and neural differentiation.
Keywords: neural, transplantation, clonogenic, self-renewal, differentiation
Although much is known about the development of the brain during embryonic life, the pathways and lineages that neural precursors take in neurogenesis during development and adult life are only partly understood (1). Until recently, it was unclear whether neurogenesis proceeded like hematopoiesis, i.e., from pluripotent clonogenic stem cells that both self-renew to produce multipotent progenitors and give rise to progressively more committed progeny (2), or if some other developmental hierarchy, not including stem cells, was operative. Recently, long-term cell culture systems have been developed for rodent and human central nervous system (CNS) cells that continuously propagate a heterogeneous population of early neural stem and/or progenitor cells. CNS neural stem/progenitor cells can grow either in monolayers on substrate-coated tissue culture plates (3, 4) or as self-adherent complexes of cells, forming clusters known as neurospheres (5, 6). In both culture systems, these cells maintain the capacity to produce the three main mature cell classes of the CNS: neurons, astrocytes, and oligodendrocytes (6, 7). In vivo analysis of transplanted cultured rodent cells shows engraftment, migration, and site-specific multilineage differentiation in mice and rats (8, 9). Human long-term cultured neurosphere cells show similar engraftment, migration, and site-specific differentiation upon transplantation into immunosuppressed rats (10). These rodent and human studies suggest that the neural cultures support the growth of stem cells and/or progenitors capable of engraftment and differentiation (3–14). Recently, Roy et al. (13) showed that human primitive progenitor cells from the dentate gyrus of adult hippocampus can be selected by transfecting them with the reporter green fluorescent protein gene driven by the nestin enhancer. However, the direct isolation of human neural stem cells from fresh tissues through the identification of cell surface markers and fluorescence-activated cell sorting (FACS) of either the central or peripheral nervous system, to our knowledge, has not yet been reported.
In the past decade, hematopoietic stem and progenitor cells have been identified by using mAbs against cell surface markers for enriching rare subpopulations that are clonal self-renewing and multipotent stem cells (15, 16) or oligopotent progenitors (17, 18). This strategy was used successfully to isolate rat peripheral nervous system stem cells (14) and, as we report here, to identify and isolate a candidate human CNS stem cell (hCNS-SC). The mAb 5F3 recognizes the CD133 antigen.¶ Antibodies to CD133 define a five-transmembrane protein and have been used to enrich for human hematopoietic stem cells (19). Here we show that mAbs 5F3 and the novel mAb, 5E12, detect a distinct subset of human fetal brain (FBr) cells. FACS using these mAbs results in a subset of human CD133+ FBr cells that are capable of neurosphere initiation, self-renewal, and multilineage differentiation at the single-cell level. Another mAb, 8G1, which recognizes human CD24, can further enrich neurosphere initiating cell activity within the CD133+ CD24−/lo fraction. Because the CD133+ cells self-renew, significantly expand in neurosphere cultures, and differentiate in vitro to neurons and glial cells, they are candidate human CNS-SC. Moreover, transplantation of CD133+ sorted/expanded neurosphere cells into the lateral ventricles of newborn nonobese diabetic–severe combined immunodeficient (NOD-SCID) mouse brains resulted in specific engraftment in numerous sites of the brain, which is similar to results shown by Fricker et al. (10). These cells also demonstrated continued self-renewal, migration, and neural differentiation for at least 7 months.
Materials and Methods
Preparation of Human Neural Tissues.
Human fetal spinal cord (FSC) and brain tissues (FBr) were obtained from Advanced Bioscience Resources, in accordance with all state and federal guidelines. Minced FBr tissues were dissociated enzymatically in 0.1% collagenase (Roche Molecular Biochemicals) and 0.1% hyaluronidase (Sigma) at 37°C for 1 h. Dissociated cells were further treated with 0.05% trypsin-0.53 mM EDTA (GIBCO) for 10–15 min to obtain a single cell suspension for cell sorting (≈1–10 × 108 cells per tissue).
Source of mAbs and Staining FBr Cells for Cell Sorting.
mAb 5F3, also known as mAb CD133, identifies the same cell surface antigen recognized by two other mAbs, CD133/1 and CD133/2 (Miltenyi Biotec, Auburn, CA). The dissociated FBr cells were incubated with mAbs against CD45-FITC (Caltag, South San Francisco, CA), CD133/1 CD133/2-PE, and CD34-APC (Becton Dickinson Immunocytometry Systems) for 20–60 min on ice. Washed cells were resuspended in Hanks' balanced salt solution containing 0.5 μg/ml propidium iodide, sorted with a FACS Vantage SE (Becton Dickinson Immunocytometry Systems), and cultured for analysis.
Generation of Novel mAbs.
mAbs to human neural cells were produced by using a previously described decoy immunization strategy (19). BALB/c mice were immunized in the left footpad with decoy human peripheral blood mononuclear cells and in the contralateral footpad with enzyme-dissociated human FBr. The right popliteal lymph node cells were fused with mouse SP2/0 cells. This fusion resulted in 1,900 hybridoma-positive wells. Approximately 180 hybridomas were selected, expanded, and further tested for specific reactivity to surface epitopes on human FBr cells.
Assay for Neurosphere-Initiating Cells (NS-IC).
Sorted FBr cells were cultured either as described (7) or in neurosphere initiation medium consisting of Ex Vivo 15 (BioWhittaker) medium with N2 supplement (GIBCO), fibroblast growth factor-2 (20 ng/ml), epidermal growth factor (20 ng/ml), lymphocyte inhibitory factor (10 ng/ml), neural survival factor-1 (Clonetics, San Diego), and 60 μg/ml N-acetylcysteine (Sigma). Cultures were fed weekly and passaged at ≈2–3 weeks. In some cases, sorted FBr cells were resorted, and different numbers of cells were deposited by the automated cell deposition unit into 96-well plates to evaluate directly the frequency of precursors that initiate neurosphere cultures. Plates were fed weekly, and wells were scored for neurosphere growth at 6–8 weeks. Linear regression analysis of the proportion of negative wells at each cell concentration was used to determine the frequency of NS-IC (20).
Differentiation Conditions.
Neurosphere cells were harvested, dissociated, and cultured with differentiation medium [Ex Vivo-15/N2/N-acetylcysteine/brain-derived neurotrophic factor (10 ng/ml)/glial-derived neurotrophic factor (10 ng/ml)/laminin (1 μg/ml)] on polyornithine-coated chamber slides. After 1–2 weeks, chamber slides were fixed with 4% paraformaldehyde in PBS and stained to detect differentiation into neurons and astrocytes, with mAbs against β-tubulin III (1:200; Sigma) conjugated with Alexa 488 (Molecular Probes) and glial fibrillary acidic protein-Cy3 (GFAP-Cy3) (1:1,000; Sigma).
Transplantation of CD133+ Sorted/Expanded Neurosphere Cells into Neonatal NOD-SCID Mice.
Expanded CD133+ sorted neurosphere cells at passages 6–10 were harvested and gently dissociated with collagenase. Neonatal mice (<24 h after birth) were anesthetized by placing them in ice for 5–10 min. Once cryoanesthetized, the pups were placed on a stereotaxic device and injected with 2 μl of cells (ranging from 105 to 106 cells per injection) into the lateral ventricle. The injected mice were kept for 7 months before they were tested for the engraftment of human cells.
Immunocytochemical Analysis of Transplanted Mouse Brain.
Seven months after transplantation, the injected mice were perfused with 4% paraformaldehyde. The mouse brains were sectioned sagitally at 5-μm thickness for fluorescence microscopic analysis or at 40-μm thickness for confocal microscopy (Bio-Rad MRC1024 UV confocal scanning microscope). To detect human cells in the transplanted mouse brains, sections were stained with mouse mAbs against human nuclei (1:100; Chemicon) or human neural cell adhesion molecule (N-CAM) (1:20; D.W.B., unpublished work), followed by goat anti-mouse IgG conjugated with Alexa 488 (1:1000, Molecular Probes) or conjugated with Cy-3 (1:500; Jackson ImmunoResearch). To stain lineage-specific cell populations, sections were stained with guinea pig anti-GFAP (1:250; Advanced ImmunoChemical, Long Beach, CA), rabbit anti-Ki-67 (1:1,000, Novocastra), and rabbit antityrosine hydroxylase (1:500; Pel-Freeze Biologicals), followed by donkey anti-guinea pig conjugated with Cy-5 (1:250; Jackson ImmunoResearch) or anti-rabbit conjugated with Cy-3 (1:250; Jackson ImmunoResearch).
Results
Search Strategy for CNS-SC Markers.
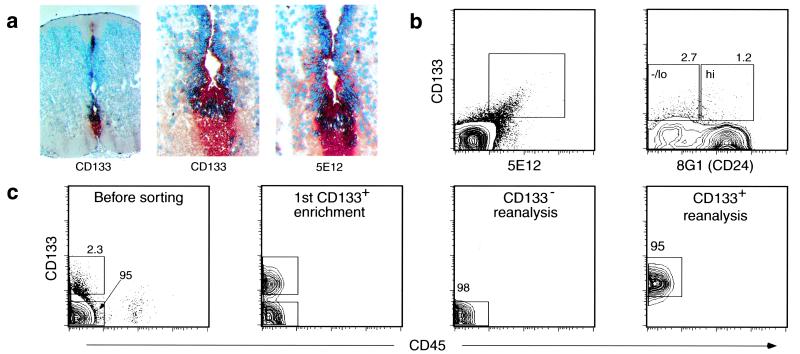
To isolate the human CNS-SC prospectively, we used the approach that had proved successful for the isolation of several stem and progenitor cells, namely, mAb-based cell sorting of candidate regenerative cells followed by placement of the sorted cells into clonogenic assays in vitro and in vivo (2, 15, 16, 21, 22). Because rodent and human neurospheres contain cells of both neuronal and glial lineages (6, 7), and because they contain cell populations capable of engraftment, migration, and multilineage differentiation upon injection into rodent brains (10), we tested the possibility that each neurosphere derives from a clonogenic stem cell. Thus, a cell population that at the single-cell level initiates a neurosphere could be a candidate for hCNS-SC. Initially, we sought a mAb that cleanly separated FBr into two fractions: one fraction that established a neurosphere culture and one that did not. Such candidate hCNS-SC markers should be expressed on only a minor subset of FBr cells. Enzyme-dissociated FBr and long-term cultured neurosphere cells were stained with over 50 known mAbs. CD34 and CD45 were not expressed on the neurosphere cells, whereas they were expressed in FBr on endothelial cells and blood cells, respectively. Antibody screening revealed that another hematopoietic stem cell marker, CD133 (19, 23, 24), was expressed on ∼90–95% of long-term cultured neurosphere cells but only on 1–6% of FBr cells derived from 16- to 20-gestational-week tissue (Fig. 1b). Histochemical analysis of FSC tissue sections with the anti-CD133 mAb 5F3 showed staining of the ventricular zone surrounding the central canal, the roof plate, and the floor plate (Fig. 1a).
Figure 1.
Isolation of human NS-IC from FBr based on cell surface markers. (a) Immunochemical staining of human FSC. Cross sections of human FSC (12 gestational weeks) were fixed with acetone and stained with mAb 5F3 (×10, Left; ×40, Middle) and 5E12 (×40, Right), using peroxidase-labeled secondary antibody with 3-amino-9-ethylcarbazole substrate (red). (b) Flow cytometric analysis of fresh FBr. FBr cells were stained with mAbs as described. A rare subset of FBr cells coexpressed both CD133+ and 5E12+ (Left). The CD133+ FBr cells were further separated based on CD24 expression (Right). Sort regions were set based on isotype controls of FBr. (c) Flow cytometric separation of FBr cells based on CD133 expression. Two cell populations, CD133−CD34−CD45− (CD133−) and CD133+CD34−CD45− (CD133+), were sorted. FACS preenrichment of CD133+ cells followed by a second round of cell sorting resulted in a distinct CD133+ population (second panel). In all subsequent experiments, both CD133− and CD133+ subsets were sorted twice, resulting in CD133− and CD133+ FBr cell fractions with high purity (right two panels).
We also identified two mAbs, 5E12 and 8G1, by generating hybridomas to human FBr cells. The mAb 5E12 costained CD133+ FBr cells (Fig. 1b) and showed a staining pattern similar to that of 5F3 on FSC sections (Fig. 1a). 5E12 recognized a protein of ∼125 kDa, which, based on its tissue distribution pattern and molecular mass, appears to be different from CD133 (D.W.B., unpublished observation) and therefore appears to be novel. In contrast, the mAb 8G1 stains the majority of total FBr cells (80–90%) at high levels; among the CD133+ cells, 8G1 expression is heterogeneous (Fig. 1b) and defines two subsets of CD133+ cells: 8G1hi and 8G1−/lo (Fig. 1b). Immunoprecipitation and blocking studies determined that 8G1 recognizes the human CD24 heat-stable antigen (D.W.B., unpublished observation).
mAbs Against CD133, 5E12, and CD24 Enriched for hCNS-SC.
To test whether hCNS-SC could be isolated from FBr based on the expression of CD133, 5E12, and/or CD24 alone or in combination, two cell populations, CD133−CD34−CD45− (CD133−) and CD133+CD34−CD45− (CD133+), were sorted and cultured to evaluate their NS-IC activity. CD34 and CD45 were used to exclude contaminating vascular and blood cells, respectively, and bulk cultures of sorted CD45+ or CD34+ cells from FBr failed to initiate long-term neurosphere cultures. Fig. 1c shows the analysis of FBr cells stained for CD133 and a reanalysis of the sorted CD133+ and CD133− subsets. Single-cell suspensions of CD133+ cells proliferated and formed small neurospheres as early as 7–10 days after culture initiation. In contrast, the sorted CD133− cells remained as a single-cell suspension, failed to initiate neurospheres, and eventually died. The CD133+-derived neurosphere cultures expanded; the number of CD133+ cells increased by at least 1,000-fold by the fifth passage (n = 5) and were capable of reinitiating neurospheres (see below).
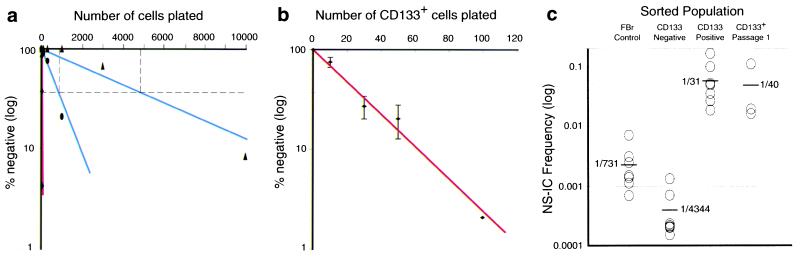
To determine the frequency of NS-IC, unfractionated FBr cells were plated by limiting dilution and were cultured for 6–8 weeks. Fig. 2a shows a representative limiting dilution analysis of control unseparated, CD133+, and CD133− subsets of FBr. Control processed FBr cells contained NS-IC activity at a frequency of 1/880 cells. In the CD133− subset, the NS-IC frequency dropped to 1/4,860 cells. In contrast, NS-IC activity was highly enriched in the CD133+ subset, with a frequency of 1/32 cells (Fig. 2a). The NS-IC frequencies from eight different FBr samples (16–20 gestational weeks) were evaluated and are summarized in Fig. 2 b and c and Table 1. On average, about 1 in 730 unfractionated FBr cells contains NS-IC activity. The CD133− subset, representing >96% of FBr, contained <1 in 4,300 cells with NS-IC. In contrast, NS-IC activity was on average enriched 24-fold in the FBr CD133+ subset with ≈1/31 (range 1/6 to 1/72) cells capable of initiating a neurosphere. Because CD133+ cells represent ∼3.6% of FBr (i.e., 1/28 cells), the enrichment of NS-IC activity virtually paralleled the frequency of CD133+ cells in FBr. Thus, CD133+ cells contained all detectable NS-IC activity in 16-to 20-gestational-week FBr cell suspensions. Similarly, 5E12+ FBr cells are enriched and 5E12− cells are depleted in NS-IC activity (Table 1). As shown in Table 1, the frequency of NS-IC is higher in the CD133+CD24−/lo fraction compared with the CD133+CD24hi subset. Thus, CD133+ cells contained virtually all NS-IC activity but were still heterogeneous; more potent NS-IC activity was enriched in the subset of cells that expressed negative to low levels of CD24. These data suggest that the CD133+CD24hi cells could represent a subset of multipotent progenitor cells, such as the short-term or long-term stem cell subsets defined for the hematopoietic system (16, 25)
Figure 2.
Limiting dilution analyses of CD133+-sorted hCNS-SC cells. (a) A representative limiting dilution analysis for NS-IC activity in FBr and sorted subsets. FBr (●), CD133− CD34− CD45− (▴), and CD133+ CD34− CD45− (⧫, red line) sorted cell populations were plated in a series of limiting cell doses. FBr and CD133− cells (100–10,000 cells per well) and CD133+ cells (10–300 cells per well) were plated into 96-well plates with a FACS–automated cell deposition unit. Cultures were carried for 6–8 weeks, and wells that did not contain neurospheres were scored as negative. A linear regression analysis was used to determine the frequency of NS-IC (20). (b) Neurospheres generated from a single-cell event. CD133+ CD34− CD45− sorted cell populations from eight different FBr samples were plated in a series of limiting cell doses into 96-well plates with a automated cell deposition unit. Results from eight different samples were combined to generate a plot of the number of cells plated vs. log percentage negative wells with SEM (error bars). Linear correlation of log percentage negative wells and number of CD133+ cells plated indicate a neurosphere initiated from a single-hit event. (c) NS-IC frequencies from unsorted, sorted, and cultured FBr cells. NS-IC frequency in FBr control (n = 8), CD133− (n = 8), CD133+ (n = 8) sorted, and CD133+ sorted/expanded (passage 1, n = 3) cell populations was determined. NS-IC frequencies of individual samples were calculated by linear regression analysis. The average NS-IC frequency in a given population is shown alongside the bars.
Table 1.
NS-IC activity in FBr and sorted populations
| Population* | % in brain | NS-IC | NS-IC range |
|---|---|---|---|
| Brain cell control (n = 8) | 100 | 1/731 | 1/139–1435 |
| CD133− sorted (n = 8) | 96 | 1/4,344 | 1/745–4,760 |
| CD133+ sorted (n = 8) | 3.6 | 1/31 | 1/6–1/74 |
| Brain cell control (n = 3) | 100 | 1/1,132 | 1/661–1,435 |
| 5E12− sorted (n = 3) | 97 | 1/12,791 | 1/1,259–35,672 |
| 5E12+ sorted (n = 3) | 2.5 | 1/286 | 1/79–392 |
| Brain cell control (n = 3) | 100 | 1/1,030 | 1/399–1,435 |
| CD133+ CD24−/lo (n = 3) | 1.1 | 1/23 | 1/15–34 |
| CD133+ CD24hi (n = 3) | 1.7 | 1/63 | 1/38–105 |
Limiting dilution assays on the above cell populations were performed and NS-IC frequencies were determined by linear regression analysis.
All sorted populations, except brain cell controls, were also gated for CD34− CD45−, which represent 98–99% of FBr.
Clonal Neurosphere Expansion, Self-Renewal, and Differentiation from Single CD133+-Sorted Cells.
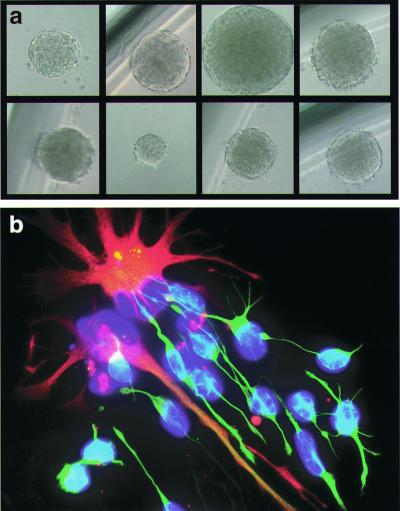
The limiting dilution analysis (Fig. 2b) shows a linear correlation between the number of cells plated and the log percentage negative wells (20, 26), indicating that a neurosphere is generated from single CD133+-sorted cells. To confirm the clonality of neurospheres, single FBr-derived CD133+-sorted cells were directly plated into wells of a 96-well plate with a FACS–automated cell deposition unit. After 8 weeks, about 5–10% of wells contained one neurosphere, confirming that neurospheres are derived from single CD133+-sorted cells (Fig. 3a). When plated in medium with the growth factors, brain-derived neurotrophic factor and glial-derived neurotrophic factor, each of these clonally derived neurospheres differentiated into neurons and astrocytes (Fig. 3b).
Figure 3.
Clonal expansion from single CD133+ cells. (a) Clonal expansion of neural stem/progenitor cells. Neurospheres can be derived from a single sorted CD133+ cell directly isolated from fetal brain or from a single CD133+ cell from sorted/expanded cultured neurospheres. (b) Differentiation capacity of clonally derived neurosphere cells. Progeny of single cell-derived neurospheres can be differentiated into neurons (β-tubulin III, green) and astrocytes (GFAP, red). Nuclei were counterstained with Hoechst 33342 (blue).
We tested NS-IC self-renewal activity by examining the replating activity of single viable cells from the CD133+-sorted/expanded neurosphere cells. Nine of 96 wells had a single neurosphere in one experiment. Cells derived from neurosphere cultures initiated from CD133+-sorted cells could consistently reinitiate the formation of secondary neurospheres, as shown in Fig. 2c. Cells from the primary neurosphere that became CD133− failed to reinitiate secondary neurosphere cultures (data not shown). Together, the data suggest that the mAb against CD133 marks cells that can self-renew.
Potent Engraftment Ability of CD133+ Sorted/Expanded Human Neurosphere Cells.
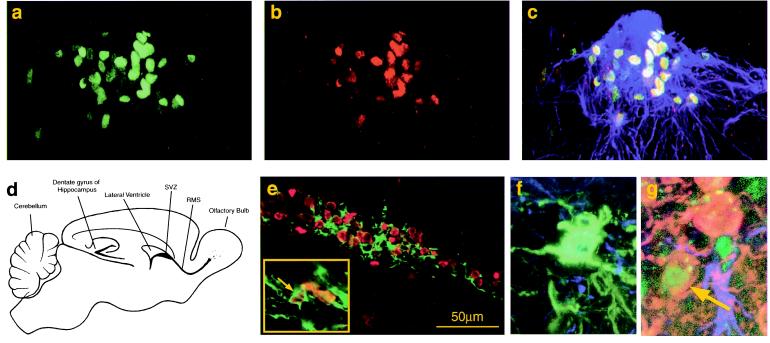
To evaluate in vivo engraftment, migration and the differentiation capacity of sorted/expanded CD133+ hCNS-SC, 105 or 106 cells from CD133+-initiated neurosphere cultures at passage 7–10 were injected into the lateral ventricles of neonatal NOD-SCID mice. Detailed analysis focused particularly on the two sites of the brain previously shown to be sites of active neurogenesis: the subventricular zone (SVZ) of the lateral ventricles and the dentate gyrus of the hippocampus (Figs. 4 and 5). Human cells, detected with an anti-human nuclear antibody, were found throughout the brain of mice transplanted with sorted/expanded human neural cells and were abundant in the SVZ 7 months after injection. Confocal microscopy indicated that most of the human cells were GFAP−, but occasional human GFAP+ cells also were detected (data not shown). Because stem/progenitor cells in the SVZ have been shown to proliferate continuously, it was important to test whether progeny of the transplanted human CD133+ sorted/expanded neurosphere cells were still proliferating in situ and coexpressed the proliferation marker Ki-67, expressed on cells in late G1/S/G2/M phases (27). As shown in Fig. 4 a–c, a cluster of human cells in the SVZ, nested in GFAP+ cells, coexpressed Ki-67; like their mouse counterparts, these cells continued to proliferate 7 months after transplantation in the SVZ.
Figure 4.
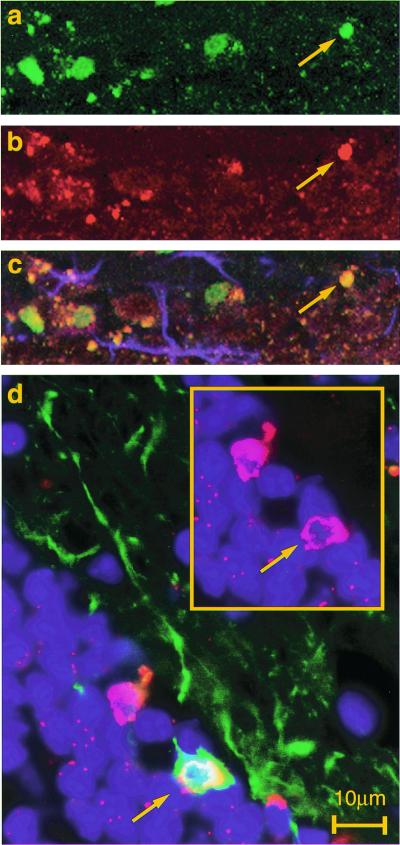
Detection of human cells in the olfactory system. CD133+-sorted/expanded human neurosphere cells were transplanted into the lateral ventricles of neonatal NOD-SCID mice. Engraftment of human cells was analyzed 7 months after transplantation. (a–c) SVZ: detection of proliferating human neural cells in the SVZ. Confocal images are of a sagittal section of the transplanted mouse brain stained with anti-human nuclear antigen (a, green) and Ki-67 (b, red) and GFAP (c, blue). Most of the human nuclear antigen positive cells in the SVZ also coexpressed the proliferation marker Ki-67 (c, merged). (d) Schematic diagram of adult mouse brain. (e) RMS: the array of human nuclear antigen+ cells (red) in the RMS was colocalized with β-tubulin III expression (green). Some of these cells were clearly double positive (arrow). (f and g) Confocal microscopic analysis of the olfactory bulb. Many human cells were distributed in the olfactory bulb, some of which expressed human N-CAM (green) on their neuronal processes (f). Very rare human dopaminergic neurons were detected [i.e., human nuclei (green), tyrosine hydroxylase (red)] in the periglomerular layer (g, arrow).
Figure 5.
Detection of proliferating and differentiating human cells in the dentate gyrus of hippocampus. (a–c) Confocal microscopic analysis of proliferating human neural cells in the subgranular zone of the dentate gyrus. Transplanted mouse brain was stained with anti-human nuclei (a, green), Ki-67 (b, red), and GFAP (c, blue). Some human nuclear antigen+ cells were costained with Ki-67 (c, merged, arrow). (d) Detection of human neurons in the dentate gyrus. The sagittal section of the transplanted mouse brain was stained with anti-human nuclear antigen (red) and anti-β-tubulin III (green). One of two human nuclear antigen-positive cells was also positive for β-tubulin III (arrow).
In the olfactory system of rodents, the progeny of stem/progenitor cells that have proliferated in the SVZ enter the rostral migratory stream (RMS) and migrate to the olfactory bulb (8, 9). These endogenous rodent progenitors, the “chain of neuroblasts” in the RMS, express both β-tubulin III and N-CAM (10, 28). Large numbers of human cells were detected in mice transplanted 7 months previously with CD133+-sorted neurosphere cells, beginning in the SVZ and extending throughout the RMS (Fig. 4 a–c, e). Multiple cells were identified that were double positive for both β-tubulin III and the human nuclear antigen (Fig. 4e, arrow and Inset). In addition, many of these cells in the RMS expressed the human specific marker N-CAM (data not shown).
After migrating through the RMS, the progeny of stem/progenitor cells enter the olfactory bulb and extend toward the olfactory glomerulus to the periglomerular layers (8, 9). The transplanted progeny of human cells distributed into the glomerular as well as the periglomerular layers. Some of these cells expressed human N-CAM, indicating that they were committed to neuronal lineages (Fig. 4f). In a few instances, human dopaminergic neurons, defined by tyrosine hydroxylase expression, were observed (Fig. 4g).
Another critical site where neurogenesis takes place in adult life is the dentate gyrus of the hippocampus (29). We found numerous human cells in the dentate gyrus of the hippocampus. Some of the human cells in the subgranular cell zone coexpressed Ki-67, indicating that they were still able to proliferate 7 months after transplantation (Fig. 5 a–c). Distinct β-tubulin III+ human cells also were detected that had long axonal processes extending toward the hilus of the hippocampus, as expected for developing granular neurons (Fig. 5d). These results indicate that not only do these sorted/expanded hCNS-SC engraft, migrate, continue to proliferate, and differentiate, but, furthermore, their behavior and cell fate are regulated by host cues in a site-specific manner. In no case did the injected cells form tumors even 1 year after transplant into immunodeficient SCID mice.
Discussion
Previously, studies with rodent neural cultures showed that retroviral-marked clones could self-renew and differentiate (30). These cells and rodent CNS-SC monolayer cells or v-myc immortalized rodent neural cells engraft, migrate, and differentiate appropriately upon intracerebral transplantation (9, 31). Here we have shown that human neurosphere cultures are initiated from a rare subset of clonogenic CD133+, 5E12+, CD24−/lo, CD34− CD45− cells that we define as the hCNS-SC. In vitro, these rare cells can initiate neurosphere cultures and differentiate into neurons and glia. It has been reported that the incidence of differentiation to human oligodendrocytes is very rare (32). The rarity of detectable oligodendrocytes could be because stem cells do not readily differentiate toward that lineage, or it could be that the markers of human oligodendrocytes are inadequate both in vivo and in vitro. The latter seems possible, as in vitro cells with oligodendrocyte morphology have been detected in cultures of our human stem cells. More recently, our transplant data suggest that the progeny of human neurosphere cells were found in the fimbria of the hippocampus of engrafted mice, with oligodendrocyte-like morphology. Upon transplantation to the lateral ventricles of NOD-SCID newborn mice, this rare subset of sorted, then expanded, CD133+ neurosphere cells engraft for at least 7 months and give rise to progeny that migrate and differentiate. Some grafted human cells continue to proliferate (i.e., express Ki-67+) in the SVZ and dentate gyrus, where counterpart mouse neural cells undergo the same events.
Rodent neurogeneic cells and ES cells grown in vitro can give rise to neurons and glia (33–35). In addition, Frisen et al. provided evidence that mouse Notch+ ependymal cells also can initiate neurospheres (ref. 36; see ref. 37 for comments). These neurospheres contain multipotent precursors for several nonneural outcomes when transplanted into blastocysts (38). In apparent contradiction, SVZ GFAP+ cells rather than ependymal cells were concluded to be rodent CNS-SC for the RMS-to-olfactory pathway (39, 40). We could not access fresh human adult brain tissue to determine the anatomical location of candidate adult CNS-SC.
Are there other CNS-SC candidates in the human FBr? The data in Fig. 2b and Table 1 demonstrate that there are no other detectable NS-IC cells outside of the CD133+ subset in the human FBr, but this does not mean that there are no other cells in the human fetal or adult brain that have CNS-SC activity, or no other progenitors that might engraft. It is not yet known which cells—stem cells or oligolineage-committed progenitors or even mature cells—engraft best in the brain. The transplant efficiencies of hCNS-SC versus highly enriched progenitors or mature cells in biological and preclinical models have yet to be addressed.
It is striking that the progeny of transplanted hCNS-SC-derived neurosphere cells could be found adjacent to ventricular spaces, and in the hippocampus, cerebral cortex, and the corpus callosum, as well as in the cerebellum, and in the SVZ, along the RMS, and within the olfactory bulb in and near the olfactory glomeruli. Previous experiments by Snyder et al. (31) using v-myc immortalized mouse and human neural cell lines demonstrated this wide (“global”) distribution of cells, and here we show that this distribution is a property not only of myc immortalized cells of murine origin but also of nongenetically modified hCNS-SC that have been considerably expanded in culture.
When analyzed 7–12 months after transplant, these hCNS-SC appeared to still respond to host microenvironmental cues and were not neoplastic. Therefore, these cells are likely to reveal developmental and functional pathways inherent in human neuronal systems not modified by the functions of oncogenes or already neoplastically transformed cell lines (11, 41, 42). The prospective isolation of human CNS-SC provides several opportunities to delineate directly the lineages that derive from cells at a particular stage of differentiation, to obtain a gene expression profile of hCNS-SC, and their immediate downstream progeny in vitro or in vivo, and to test whether specific gene modification of the cells alters their engraftment, migration, differentiation, and/or functional integration. Finally, the prospective isolation of hCNS-SC should allow the transplantation of these cells in mouse analogues of human disease (11, 31) as preclinical tests for their transplantability, differentiation ability, and lack of tumorigenicity.
Acknowledgments
We are grateful to David Anderson, Ben Barres, and Eric Lagasse for stimulating discussions and advice; M. L. Gage for critical review of the manuscript; and Linda Kitabayashi for performing confocal microscopic analysis. We also thank the staff of StemCells, Inc., for their continuous encouragement and support.
Abbreviations
- CNS
central nervous system
- hCNS-SC
human CNS stem cell
- FACS
fluorescence-activated cell sorting
- FBr
fetal brain
- NOD-SCID
nonobese diabetic–severe combined immunodeficient
- FSC
fetal spinal cord
- NS-IC
neurosphere-initiating cells
- GFAP
glial fibrillary acidic protein
- N-CAM
neural cell adhesion molecule
- SVZ
subventricular zone
- RMS
rostral migratory stream
Footnotes
7th Workshop and Conference on Human Leucocyte Differentiation Antigens, Harrogate, U.K., June 20–24, 2000.
References
- 1.Gage F H. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 2.Weissman I L. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 3.Ray J, Peterson D A, Schinstine M, Gage F H. Proc Natl Acad Sci USA. 1993;90:3602–3606. doi: 10.1073/pnas.90.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKay R. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds B A, Tetzlaff W, Weiss S. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss S, Reynolds B A, Vescovi A L, Morshead C, Craig C G, van der Kooy D. Trends Neurosci. 1996;19:387–393. doi: 10.1016/s0166-2236(96)10035-7. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter M K, Cui X, Hu Z Y, Jackson J, Sherman S, Seiger A, Wahlberg L U. Exp Neurol. 1999;158:265–278. doi: 10.1006/exnr.1999.7098. [DOI] [PubMed] [Google Scholar]
- 8.Lois C, Alvarez-Buylla A. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 9.Suhonen J O, Peterson D A, Ray J, Gage F H. Nature (London) 1996;383:624–627. doi: 10.1038/383624a0. [DOI] [PubMed] [Google Scholar]
- 10.Fricker R A, Carpenter M K, Winkler C, Greco C, Gates M A, Bjorklund A. J Neurosci. 1999;19:5990–6005. doi: 10.1523/JNEUROSCI.19-14-05990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flax J D, Aurora S, Yang C, Simonin C, Wills A M, Billinghurst L L, Jendoubi M, Sidman R L, Wolfe J H, Kim S U, et al. Nat Biotechnol. 1998;16:1033–1039. doi: 10.1038/3473. [DOI] [PubMed] [Google Scholar]
- 12.Brustle O, Choudhary K, Karram K, Huttner A, Murray K, Dubois-Dalcq M, McKay R D. Nat Biotechnol. 1998;16:1040–1044. doi: 10.1038/3481. [DOI] [PubMed] [Google Scholar]
- 13.Roy N S, Wang S, Jiang L, Kang J, Benraiss A, Harrison-Restelli C, Fraser R A, Couldwell W T, Kawaguchi A, Okano H, et al. Nat Med. 2000;6:271–277. doi: 10.1038/73119. [DOI] [PubMed] [Google Scholar]
- 14.Morrison S J, White P M, Zock C, Anderson D J. Cell. 1999;96:737–749. doi: 10.1016/s0092-8674(00)80583-8. [DOI] [PubMed] [Google Scholar]
- 15.Spangrude G J, Heimfeld S, Weissman I L. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 16.Morrison S J, Weissman I L. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 17.Kondo M, Weissman I L, Akashi K. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 18.Akashi K, Traver D, Miyamoto T, Weissman I L. Nature (London) 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 19.Yin A H, Miraglia S, Zanjani E D, Almeida-Porada G, Ogawa M, Leary A G, Olweus J, Kearney J, Buck D W. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 20.Lefkovits I, Waldmann H. Limiting Dilution Analysis of Cells of the Immune System. Oxford: Oxford Univ. Press; 1999. [Google Scholar]
- 21.Baum C M, Weissman I L, Tsukamoto A S, Buckle A M, Peault B. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchida N, Weissman I L. J Exp Med. 1992;175:175–184. doi: 10.1084/jem.175.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miraglia S, Godfrey W, Yin A H, Atkins K, Warnke R, Holden J T, Bray R A, Waller E K, Buck D W. Blood. 1997;90:5013–5021. [PubMed] [Google Scholar]
- 24.Corbeil D, Roper K, Hellwig A, Tavian M, Miraglia S, Watt S M, Simmons P J, Peault B, Buck D W, Huttner W B. J Biol Chem. 2000;275:5512–5520. doi: 10.1074/jbc.275.8.5512. [DOI] [PubMed] [Google Scholar]
- 25.Uchida N, Combs J, Chen S, Zanjani E, Hoffman R, Tsukamoto A. Blood. 1996;88:1297–1305. [PubMed] [Google Scholar]
- 26.Visser J W, Bauman J G, Mulder A H, Eliason J F, de Leeuw A M. J Exp Med. 1984;159:1576–1590. doi: 10.1084/jem.159.6.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schluter C, Duchrow M, Wohlenberg C, Becker M H, Key G, Flad H D, Gerdes J. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gage F H, Cristen Y. Isolation, Characterization and Utilization of CNS Stem Cells. Heidelberg: Springer; 1997. [Google Scholar]
- 29.Gage F H, Coates P W, Palmer T D, Kuhn H G, Fisher L J, Suhonen J O, Peterson D A, Suhr S T, Ray J. Proc Natl Acad Sci USA. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer T D, Takahashi J, Gage F H. Mol Cell Neurosci. 1997;8:389–404. doi: 10.1006/mcne.1996.0595. [DOI] [PubMed] [Google Scholar]
- 31.Yandava B D, Billinghurst L L, Snyder E Y. Proc Natl Acad Sci USA. 1999;96:7029–7034. doi: 10.1073/pnas.96.12.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong R J, Svendsen C N. Cell Transplant. 2000;9:139–152. doi: 10.1177/096368970000900202. [DOI] [PubMed] [Google Scholar]
- 33.Brustle O, Jones K N, Learish R D, Karram K, Choudhary K, Wiestler O D, Duncan I D, McKay R D. Science. 1999;285:754–756. doi: 10.1126/science.285.5428.754. [DOI] [PubMed] [Google Scholar]
- 34.McDonald J W, Liu X Z, Qu Y, Liu S, Mickey S K, Turetsky D, Gottlieb D I, Choi D W. Nat Med. 1999;5:1410–1412. doi: 10.1038/70986. [DOI] [PubMed] [Google Scholar]
- 35.Lee S H, Lumelsky N, Studer L, Auerbach J M, McKay R D. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 36.Johansson C B, Momma S, Clarke D L, Risling M, Lendahl U, Frisen J. Cell. 1999;96:25–34. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- 37.Barres B A. Cell. 1999;97:667–670. doi: 10.1016/s0092-8674(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 38.Clarke D L, Johansson C B, Wilbertz J, Veress B, Nilsson E, Karlstrom H, Lendahl U, Frisen J. Science. 2000;288:1660–1663. doi: 10.1126/science.288.5471.1660. [DOI] [PubMed] [Google Scholar]
- 39.Doetsch F, Caille I, Lim D A, Garcia-Verdugo J M, Alvarez-Buylla A. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 40.Chiasson B J, Tropepe V, Morshead C M, van der Kooy D. J Neurosci. 1999;19:4462–4471. doi: 10.1523/JNEUROSCI.19-11-04462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trojanowski J Q, Mantione J R, Lee J H, Seid D P, You T, Inge L J, Lee V M. Exp Neurol. 1993;122:283–294. doi: 10.1006/exnr.1993.1128. [DOI] [PubMed] [Google Scholar]
- 42.Auerbach J M, Eiden M V, McKay R D. Eur J Neurosci. 2000;12:1696–1704. doi: 10.1046/j.1460-9568.2000.00067.x. [DOI] [PubMed] [Google Scholar]