Abstract
MaizeGDB is the Maize Genetics and Genomics Database. Available at MaizeGDB are diverse data that support maize research including maps, gene product information, loci and their various alleles, phenotypes (both naturally occurring and as a result of directed mutagenesis), stocks, sequences, molecular markers, references and contact information for maize researchers worldwide. Also available through MaizeGDB are various community support service bulletin boards including the Editorial Board's list of high-impact papers, information about the Annual Maize Genetics Conference and the Jobs board where employment opportunities are posted. Reported here are data updates, improvements to interfaces and changes to standard operating procedures that have been made during the past 2 years. MaizeGDB is freely available and can be accessed online at http://www.maizegdb.org.
INTRODUCTION
Maize (Zea mays ssp. mays) has long been the number one production crop in the United States, and in 2001 it also became number one in the world. As maize is economically important and also serves as a model organism for genetics research, it is one of the most highly researched organisms in existence. The maize genome has an especially high level of DNA sequence polymorphism and extended regions of non-homology between inbred lines (1,2), hence the diversity represented by the maize gene pool is unparalleled in both a phenotypic and molecular sense. This provides a unique vehicle to explore questions in evolution, domestication, development, trait expression, functional allelic diversity and the interrelated processes that shape such events and their outcomes.
The application of new technologies and bioinformatic tools coupled with thorough phenotypic evaluation for useful traits and molecular characterization of diverse maize germplasm offers the potential for significant discovery via translational genomics. The goal of turning the identification and evaluation of functional and evolutionarily important allelic variation into a comprehensive genomics activity is dependent on being able to associate diverse information in a seamless manner. Maize Genetics and Genomics Database (MaizeGDB) is the community resource charged with developing informatic solutions for storing, displaying and linking comprehensive maize data, so that they are made easily accessible to researchers worldwide. Described here are updates and improvements to MaizeGDB that have been made over the course of the past 2 years.
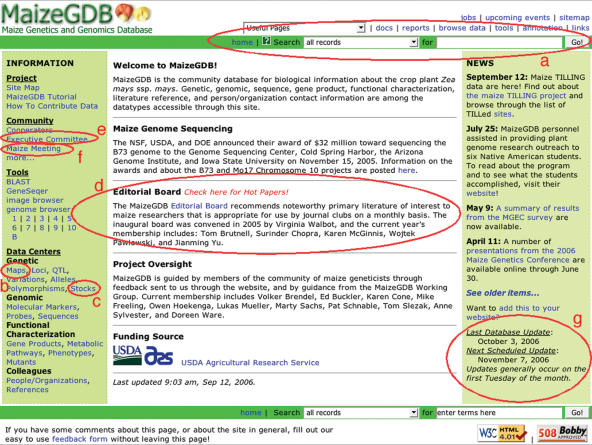
Throughout this paper direct links are listed parenthetically to enable direct access to relevant data. Those data also could be accessed via search and browse mechanisms available on the MaizeGDB front page. For example, to find a map that is listed parenthetically, one could search either ‘all records’ or ‘maps’ using the search bar at the top (and bottom) of any MaizeGDB page (Figure 1a) using the map name. That same data also could be accessed from the front page by navigating to the Maps Data Center using the link shown in Figure 1b. On the Maps Data Center Search page, the Simple or Advanced Search mechanisms could be used to search maps by name, type, or using other limiters. Other data types also can be accessed from the front page using similar tactics.
Figure 1.
Important information, data centers, tools and news items are accessible from the MaizeGDB home page. Data and bulletin boards as well as links to noteworthy or high profile projects are accessible directly on the front page. All MaizeGDB pages have the same header, which is loaded with functionality enabling, e.g. searches of all data from any page (a) and access to tools including the Community Curation Tools. The ‘Maps’ Data Center (b) can limit results by contained loci, chromosome, source or mapping panel and also allows direct access to unique map types including the Recombination Nodule maps through its ‘Map Reports and Tools’ section. Likewise, the ‘Stocks’ Data Center (c) enables queries by focus linkage group, genotypic variation, karyotypic variation and other limitors. Bulletin boards that keep researchers connected with the community include the Editorial Board (d), the Maize Genetics Executive Committee pages (e), and the Annual Maize Genetics Conference, (which is referred to simply as ‘The Maize Meeting’) site (f). Dates of database updates are available directly on the front page (g), as are important major efforts of interest to all maize researchers like the Maize Genome Sequencing project.
NEW DATA TYPES
Since the initial announcement in 2004 that MaizeGDB was up and running (3), the database has expanded to include new data types, such as TILLING data [(4); http://www.maizegdb.org/cgi-bin/displaylocusresults.cgi?term=mtp*], cytological maps with associated images (e.g. http://www.maizegdb.org/cgi-bin/displaymaprecord.cgi?id=892372), and the maize Recombination Nodule maps [(5,6); http://www.maizegdb.org/RNmaps.php]. All new data types are made available alongside related information and are accessible through mechanisms that seamlessly integrate with the site's existing functionalities.
Recombination Nodule maps are uncommon: at present, they are only available for maize, mouse, and tomato (5,7,8). As many researchers are unfamiliar with this unique data type and to demonstrate the utility of the newly available maize Recombination Nodule maps, an example of how they can be used to speed up experiments that utilize the maize translocation stocks follows.
A researcher wants to determine whether the gene product of her new mutant gof1, which maps to 3L within 1 cM of tub6, acts cell autonomously using a mosaic analysis. Since there are no suitable cell autonomous markers known to be proximal to gof1, she decides to use an A-A translocation to bring gof1+ distal to a cell autonomous marker on another chromosome. To do this, she needs to find a translocation stock with a breakpoint on 3L proximal to her gene of interest, and a breakpoint on another chromosome that is distal to a gene that can be used as a cell autonomous marker. To find a list of available translocations, she uses the link on the front page of MaizeGDB to get to the Stocks Data Center (Figure 1c), scrolls down the page (past the green ‘Advanced Stock Query’ box) to Maize Genetics Cooperation Stock Center Resources (9), and clicks the link for the ‘Stock Center Catalog.’ She clicks the link for ‘Reciprocal Translocation (comprehensive list)’ to arrive at http://www.maizegdb.org/cgi-bin/stockcatalog.cgi?id=3, and decides to try 1049B T1-3(5242) (3L.65; 1L.90). In order to determine the approximate positions of these breakpoints relative to genetically mapped loci, she goes to the Maps Data Center (Figure 1b) and navigates to the Recombination Nodule maps. From here, she clicks the link toward the bottom of the page to go to the Morgan2McClintock Translator [(10); http://www.lawrencelab.org/Morgan2McClintock/]. Here, she chooses chromosome 3, pastes the entire ‘Genetic 2005 3’ map from MaizeGDB (accessible at http://www.maizegdb.org/cgi-bin/displaymaprecord.cgi?id=940882) into the text box and hits the button marked ‘Calculate!’. The output file shows that 3L.65 lies about 5 cM proximal to tub6. This means that the breakpoint is proximal to her gene of interest! Using the Translator again for chromosome 1, she finds the position of 1L.90 relative to the ‘Genetic 2005 1’ map (accessible at http://www.maizegdb.org/cgi-bin/displaymaprecord.cgi?id=940880). The output table shows that lw1, a suitable cell autonomous marker (11), lies between cent1 and the breakpoint. Using the stock for T1-3(5242), she can set up a stock heterozygous for gof1, lw1, and the translocation where gof1+ and lw1+ reside on the translocation, and the recessive mutant alleles reside on the normal (non-translocation) chromosomes. Without access to the maize Recombination Nodule map data and the Morgan2McClintock Translator, cytological and genetic maps cannot be integrated directly. Setting up such an experiment would have required many additional tests using various translocation stocks.
DATA UPDATES
All types of data (references, loci, maps, etc.) are updated regularly as time and human resources allow. Listed here are a few of the major recent updates to content.
MaizeGDB's sequence set is made up of all public maize sequences including expressed sequence tag (EST), cDNA, genome survey sequence (GSS), sequence tagged site (STS), HTC and genomic DNA sequences from GenBank (12) as well as the Uniprot (13) protein sequence set. This dataset is updated monthly utilizing a custom pipeline set up by workers at PlantGDB (14). Whereas in the past only the Z.mays ssp. mays sequence set was included in the update, a recent change was made to the sequence set update pipeline to include sequences for all subspecies of Z.mays. Also stored at MaizeGDB is contig membership information for the PlantGDB GSS and PUT (putative unique transcript) contigs as well as the TIGR TC (tentative consensus) EST contig set [12 October 2005 release; (15)].
The IBM2 FPC0507 maps (http://www.maizegdb.org/cgi-bin/displaymapresults.cgi?term=fpc0507) represent the anchored BAC finger print contigs which are currently being used to guide the B73 Maize Genome Sequencing Project. These maps were derived using anchor information for 414 contigs as assigned in the July 2005 manually edited FPC product [(16); http://www.genome.Arizona.edu/fpc/maize]. Represented on the map are loci where at least 2 BACs in the contig were empirically associated with a molecular marker. Positions for loci were derived using a hybrid coordinate comprised of an integer representing the nearest genetic anchor point followed by a decimal and the FPC consensus band (CB) coordinate. Loci are associated with the defining BACs, markers, and marker sequences, and are linked to the current contig representation at Arizona. This map adds over 25 000 new loci to MaizeGDB, most of which are associated with overgo probes designed to detect full-length cDNAs (17). This map serves to integrate the genetic and physical maps and improves high resolution mapping capabilities for researchers working to localize a trait or phenotype of interest to a small region of the genome, thus facilitating, e.g. candidate gene discovery by chromosome walking.
Initiated to assist in anchoring BAC contigs and continued to support maize genetics research, the IBM Neighbors product approximates the genetic map orders of all loci mapped to better than 5 cM and most recently includes the loci which have only been placed onto anchored FPC contigs. Like the anchored BAC FPC contigs, the IBM Neighbors representation utilizes the IBM2 map as the framework. Each version is maintained, and the primary map source for a locus coordinate is displayed. The most recent version (http://www.maizegdb.org/cgi-bin/displaymapresults.cgi?term=ibm2*2005*neighbors) includes well-ordered mutants based on the Genetic 2005 compilation (18), which incorporated the UMC 98 maps (19).
Over 5300 newly mapped loci, based on sequenced probes where most were derived from a cDNA, were provided to MaizeGDB from mapping projects that are using high-resolution intermated recombinant inbred panels of stocks. These came from: the Schnable lab (3391 insertion–deletion loci on 2 map versions, IBM IDP +MMP versions 4 and 5; http://maize-mapping.plantgenomics.iastate.edu), Matthieu Falque [1680 RFLP loci on IBM GNP and GNP LHRF maps; (20)], and the Community Mapping Service (289 loci; described below).
NEW FUNCTIONALITIES
Along with additions of new data, MaizeGDB personnel remain committed to maintaining and improving upon the interface to the database. New map displays have ‘sequence view’ (which shows the sequences associated with each locus on the map) and ‘primer view’, (which shows the primers and probes for each locus on the map) options, which are accessible toward the top of all default map views (e.g. http://www.maizegdb.org/cgi-bin/displaymaprecord.cgi?id=143431). Search algorithms have been refined to allow researchers to simply enter a term in the search field toward the top of any page at MaizeGDB (see Figure 1a) and quickly find relevant results, and summary information is now available on search results pages so that the most relevant records can easily be identified. For example, if a researcher were to search all records with the string ‘r1’, over 170 loci would be found. To help with locating the locus for an exact match, i.e. the locus r1 colored1, the exact match is shown at the top of the list of results. In addition, though the appearance of many data displays has not changed, the underlying code has been rewritten to improve load times and to optimize computational efficiency.
In an effort to improve access to diverse maize data, the MaizeGDB interface has been modified to include an abundance of linkages to other databases including the Plant Ontology Consortium [(21); http://www.plantontology.org], Gramene [(22); http://www.gramene.org] and the Maize Sequencing Project's genome browser. Data displays provide abundant context-sensitive linkages to records in other databases, enabling users to visit, e.g. a gene record at MaizeGDB and with just a click quickly find related sequences, annotated maps and similar sequences at other websites.
Recently the Community Curation Tools (accessible through the ‘tools’ link at the top of any page at MaizeGDB; Figure 1) were updated to enable the entry of QTL data. Insights gained from experience with QTL data entry into the legacy MaizeDB (23) were leveraged in planning the functionality of this module, and linking of trait and map location to germplasm containing the superior allele is enforced. New automatic nomenclature features ensure consistency and minimize data entry effort. General functionality of the QTL module is consistent with that of the other Community Curation Tools described previously (24).
COMMUNITY SUPPORT ACTIVITIES
MaizeGDB hosts an Editorial Board (Figure 1d) whose members communicate with MaizeGDB personnel monthly to report their selections of current and historic literature germane to maize research. The list of Editorial Board selections is appropriate to guide journal clubs or for use by individuals keen to remain abreast of the advances relevant to maize research. It can be accessed at http://www.maizegdb.org/editorial_board.php.
MaizeGDB personnel support the Maize Genetics Executive Committee (MGEC; Figure 1e) by providing a venue for them to contact and interact with the community of maize researchers. MaizeGDB personnel create and administer customized community surveys and handle and monitor elections for the MGEC, (which involves the creation of methods for anonymous balloting and key-based restrictions to preclude stuffing the ballot box). These services help the MGEC to understand the needs of and communicate clearly with the community of maize researchers. Note also that author M. L. Schaeffer is a member of the MGEC.
The Annual Maize Genetics Conference (see Figure 1f) is growing by leaps and bounds, with a nearly 20% increase in attendance within the last 3 years and a near doubling in the number of abstracts submitted. Workers at MaizeGDB created a set of tools to accept abstract submissions and to manage review of the abstracts by Conference Steering Committee members. MaizeGDB personnel also maintain the mailing list for the Conference Steering Committee, and authors T. E. Seigfried and M. L. Schaeffer serve on the conference steering committee in an ex officio capacity and also assemble and print the conference program.
MaizeGDB hosts the Maize Newsletter (MNL), and MaizeGDB Curator M. L. Schaeffer is a co-editor of that publication. The main MNL site is at MaizeGDB (http://www.maizegdb.org/mnl.php), and new volumes are staged at the University of Missouri-Columbia (http://www.agron.missouri.edu). Contributions from collaborators are posted as received, and, with minor editing, redacted for printing once a year. The MNL also includes annual reports from the Maize Genetics Cooperation–Stock Center, the MaizeGDB, and any new map syntheses developed by MaizeGDB and collaborators. Funding for the redaction, printing, and mailing are from an endowment established by contributions from collaborators.
CIMDE is the community mapping service originally provided by personnel working on the Maize Mapping Project [(25); http://www.maizemap.org/bioinformatics.htm]. Author M. L. Schaeffer currently manages CIMDE. Map positions are determined using 580 framework loci and MapMaker software (26), and are returned within 2 weeks of submission of raw map scores. When those data become public, they are incorporated into the Community IBM Map (cIBM) along with related documentation, such as the contributor, nucleotide sequence accessions, encoded proteins and literature citations. The most recent update (cIBM2005; http://www.maizegdb.org/cgi-bin/displaymapresults.cgi?term=cIBM*2005) includes the published maps of the Genoplante Consortium (20) and data submitted for inclusion in the public IBM Neighbors map (described above). The data sources are attributed, and sequence accessions related to mapped loci as well as sequence details, such as primers required to reproduce the mappings, are actively solicited and annotated.
DATABASE AND AVAILABILITY
Standard operating procedures, accessibility and machine architecture are reviewed in detail elsewhere (24). The following is a brief description of how the project databases are administered and explains availability and methods of access. The MaizeGDB schema is accessible online at http://www.maizegdb.org/MaizeGDBSchema.pdf. Presently there are three copies of the database and interface, which exist on three identical servers. The interface on each server interacts with data on the local copy of the database, thereby allowing the maintenance of a production environment (i.e. the copy accessed through http://www.maizegdb.org), a curation or staging environment, and an isolated testing and development environment. The development environment functions as a playground where data manipulation and interface development are tested. The curation database stores the most current data. As data are entered into the curation database (by researchers using the Community Curation Tools and by professional curators), they are initially listed as non-public and can only be viewed by MaizeGDB staff members. Once the data are reviewed, a curation level tag is changed so that the new records will become publicly accessible. Updates to production are carried out by replacing the existing production copy of the database with a duplicate of the curation database and the latest sequence files from PlantGDB. This update generally occurs on the first Tuesday of each month (see Figure 1g). The curation database is backed up on a daily basis and is available for download (http://goblin1.zool.iastate.edu/~oracle/) for those who have Oracle RDBMS installed locally. Requests to gain read-only SQL access to the database should be directed by email to mgdb@iastate.edu. Data housed at MaizeGDB are in the public domain, hence they are freely available for use without a license.
FUTURE PLANS
The genome of maize inbred line B73 is being sequenced, and the creation and public availability of the official site (called the Maize Genome Browser; http://www.maizesequence.org) is in the offing. The Maize Genome Browser has embedded links to MaizeGDB throughout, and context-sensitive links from MaizeGDB into the Maize Genome Browser are planned. By creating links to the Maize Genome Browser, MaizeGDB can connect researchers with up-to-date views of the maize genome as it is sequenced without bearing the responsibility of supporting an independent genome browser for maize. To learn more about the Maize Genome Sequencing Consortium's plans and to find updates on their progress, visit http://www.maizegdb.org/sequencing_project.php.
In most cases, a model organism's official gene models are housed at the model organism database (MOD). As the maize genome is being sequenced and it is anticipated that MaizeGDB (the MOD for maize) will be charged with storing and making available the official gene models, plans are in the works to put together an infrastructure for supporting the storage of this new data type and to create a version control system to allow for the storage of each major gene model release.
Not all datatypes are currently integrated with sequence data. Breeders find it difficult to locate all genomic and phenotypic data for plant germplasm collections because breeding and sequence data are housed in separate, disconnected databases. Although MaizeGDB stores maize data related to genetics and genomics, most historical, geographic origin, characterization and evaluation data associated with the National Plant Germplasm System's Plant Genetic Resources collections are housed in the Germplasm Resource Information Network (GRIN; http://www.ars-grin.gov/npgs/). Work to integrate MaizeGDB with GRIN is a high-priority item for development in the coming year so that breeders are enabled to more easily associate genetic and genomic data with traditional crop improvement information.
Acknowledgments
The authors thank the MaizeGDB Working Group (E. Buckler, K. Cone, M. Freeling, O. Hoekenga, L. Mueller, P. Schnable, T. Slezak, A. Sylvester, D. Ware, and especially M. Sachs and V. Brendel) for their help and guidance. The contributions of Editorial Board (2006 membership includes T. Brutnell, S. Chopra, K. McGinnis, W. Pawlowski, M. Timmermans and J. Yu) are most appreciated. The authors also thank H. Sanchez-Villeda and M. McMullen for contributing a naming and tracking tool for loci defined by FPC; S. Schroeder, H. Sanchez-Villeda and others for maintaining the software and Web services associated with the CIMDE mapping services; S. Baran for coding the Community Curation Tools and G. Davis, M. McMullen, and E. Coe for advice on mappings (though the authors take full credit for any errors). The authors thank V. Brendel's PlantGDB Group (M. Wilkerson, Q. F. Dong and A. Fu) for creating and maintaining the maize sequence pipeline. Funding to pay the Open Access publication charges for this article was provided by USDA-ARS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brunner S., Fengler K., Morgante M., Tingey S., Rafalski A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell. 2005;17:343–360. doi: 10.1105/tpc.104.025627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu H., Dooner H. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl Acad. Sci. USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence C., Dong Q., Polacco M., Seigfried T., Brendel V. MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res. 2004;32:D393–D397. doi: 10.1093/nar/gkh011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Till B., Reynolds S., Weil C., Springer N., Burtner C., Young K., Bowers E., Codomo C., Enns L., Odden A., et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004;4:12. doi: 10.1186/1471-2229-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson L., Doyle G., Brigham B., Carter J., Hooker K., Lai A., Rice M., Stack S. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics. 2003;165:849–865. doi: 10.1093/genetics/165.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson L., Salameh N., Bass H., Harper L., Cande W., Weber G., Stack S. Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics. 2004;166:1923–1933. doi: 10.1534/genetics.166.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Froenicke L., Anderson L., Wieinberg J., Ashley T. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 2002;71:1353–1368. doi: 10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherman J., Stack S. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon. esculentum) Genetics. 1995;141:683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholl R., Sachs M., Ware D. Maintaining collections of mutants for plant functional genomics. In: Grotewold E., editor. Plant Function Genomics. Vol. 236. Totowa, NJ: Humana Press; 2003. pp. 311–326. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence C., Seigfried T., Bass H., Anderson L. Predicting chromosomal locations of genetically mapped loci in maize using the Morgan2McClintock Translator. Genetics. 2006;172:2007–2009. doi: 10.1534/genetics.105.054155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hake S., Freeling M. Analysis of genetic mosaics shows that the extra epidermal cell divisions in Knotted mutant maize plants are induced by adjacent mesophyll cells. Nature. 1986;320:621–623. [Google Scholar]
- 12.Wheeler D.L., Church D.M., Federhen S., Lash A.E., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Sequeira E., Tatusova T.A., et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apweiler R., Bairoch A., Wu C., Barker W., Boeckmann B., Serenella F., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2005;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong Q., Lawrence C., Schlueter S., Wilkerson M., Kurtz S., Lushbough C., Brendel V. Comparative plant genomics resources at PlantGDB. Plant Physiol. 2005;139:610–618. doi: 10.1104/pp.104.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan A., Pertea G., Cheung F., Lee D., Zheng L., Whitelaw C., Pontaroli A., SanMiguel P., Yuan Y., Bennetzen J., et al. The TIGR Maize Database. Nucleic Acids Res. 2006;34:D771–D776. doi: 10.1093/nar/gkj072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pampanwar V., Engler F., Hatfield J., Blundy S., Gupta G., Soderlund C. FPC web tools for rice, maize, and distribution. Plant Physiol. 2005;138:116–126. doi: 10.1104/pp.104.056291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardiner J., Schroeder S., Polacco M., Sanchez-Villeda H., Fang Z., Morgante M., Landewe T., Fengler K., Useche F., Hanafey M., et al. Anchoring 9371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 2004;134:1317–1326. doi: 10.1104/pp.103.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coe E., Schaeffer M. Genetic, physical, maps and database resources for maize. Maydica. 2005;50:285–303. [Google Scholar]
- 19.Polacco M., Sandhez-Villeda H., Coe E. A consensus genetic map, intermated B73 x Mo17 (IBM) neighbors. Maize Genet. Coop. News Lett. 2003;77:137–179. [Google Scholar]
- 20.Falque M., Decousset L., Dervins D., Jacob A., Joets J., Martinant J., Raffoux X., Ribiere N., Ridel C., Samson D., et al. Linkage mapping of 1454 new maize candidate gene loci. Genetics. 2005;170:1957–1966. doi: 10.1534/genetics.104.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaiswal P., Avraham S., Ilic K., Kellogg E., McCouch S.R., Pujar A., Reiser L., Rhee S., Sachs M., Schaeffer M., et al. Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp. Funct. Genomics. 2005;6:388–406. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaiswal P., Ni J., Yap I., Ware D., Spooner W., Youens-Clark K., Ren L., Liang C., Zhao W., Ratnapu K., et al. Gramene: a bird's eye view of cereal genomes. Nucleic Acids Res. 2006;34:D717–D723. doi: 10.1093/nar/gkj154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne P., Berlyn M., Coe E., Davis G., Polacco M., Hancock D., Letovsky S. Reporting and accessing QTL information in USDA's Maize Genome Database. J. Agric. Genomics. 1995;1:1–11. [Google Scholar]
- 24.Lawrence C., Seigfried T., Brendel V. The maize genetics and genomics database. The community resource for access to diverse maize data. Plant Physiol. 2005;139:610–618. doi: 10.1104/pp.104.059196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Villeda H., Schroeder S., Polacco M., McMullen M., Havermann S., Davis G., Vroh-Bi I., Cone K., Sharopova N., Yim Y., et al. Development of an integrated laboratory information management system for the maize mapping project. Bioinformatics. 2003;19:2022–2030. doi: 10.1093/bioinformatics/btg274. [DOI] [PubMed] [Google Scholar]
- 26.Lander E., Green P., Abrahamson J., Barlow A., Daly M., Lincoln S., Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]