Abstract
Functional glycomics, the scientific attempt to identify and assign functions to all glycan molecules synthesized by an organism, is an emerging field of science. In recent years, several databases have been started, all aiming to support deciphering the biological function of carbohydrates. However, diverse encoding and storage schemes are in use amongst these databases, significantly hampering the interchange of data. The mutual online access between the Bacterial Carbohydrate Structure DataBase (BCSDB) and the GLYCOSCIENCES.de portal, as a first reported attempt of a structure-based direct interconnection of two glyco-related databases is described. In this approach, users have to learn only one interface, will always have access to the latest data of both services, and will have the results of both searches presented in a consistent way. The establishment of this connection helped to find shortcomings and inconsistencies in the database design and functionality related to underlying data concepts and structural representations. For the maintenance of the databases, duplication of work can be easily avoided, and will hopefully lead to a better worldwide acceptance of both services within the community of glycoscienists. BCSDB is available at http://www.glyco.ac.ru/bcsdb/ and the GLYCOSCIENCES.de portal at http://www.glycosciences.de/
INTRODUCTION
Functional glycomics is an emerging field of science, aiming to create a cell-by-cell catalogue of glycosyltransferase expression and detected glycan structures in relation to health and diseases (1–3). Analysis of glycans has proven difficult in the past due to their structural complexity. However, modern analytical methods such as mass spectrometry and NMR have afforded the ability to elucidate most structural details at the concentration levels required for glycomics (4,5). Several national and international initiatives aiming to decipher the biological function of carbohydrates have emerged for the recent years (6–8). In a similar fashion to the finished Human Genome Project—which determined the sequences of the chemical base pairs that make up a human DNA—most of these glycomics projects intend to make their data freely accessible under an open access philosophy. Unfortunately, the exchange of data between different glyco-related databases is seriously hampered by the dearth of generally accepted digital exchange formats and standardized structural and biological descriptions (9). Similar to the genomics and proteomics field, a description of glycan structures would be an appropriate way to establish an efficient connection of glyco-related information resources. However, glycan sequences cannot be described by a simple linear one-letter code as each pair of monosaccharides can be linked in several ways and branched structures can be formed. The GLYCOSCIENCES.de portal (7) demonstrates that data originating from various resources can be efficiently integrated using a linear notation for unique description of carbohydrate sequences (LINUCS) (10). The extended alphanumeric IUPAC description and glycosidic linking information are applied to build up a hierarchy of the various branches starting from the reducing end of the oligosaccharide chain, which is then converted into a linear representation. However, other larger projects use different ways to encode glycan structures. The commercially available GlycoSuiteDB (11) uses the so-called condensed form of the IUPAC description to create a linear representation, where four rules are applied to obtain a unique linear code. The ‘Glycan Database’ of the US Consortium for Functional Glycomics (6) uses the so-called Linear Code™ (12), using a one or two character-based representation of saccharide units and linkages. The ordering of glycan branches is established using a special lookup table where the hierarchy of monosaccharide structures is defined. The KEGG Carbohydrate Matcher (KCaM) (8,13) uses a connection table based graph representation to encode carbohydrate structures, where monosaccharides are represented by nodes and glycosidic bonds as edges.
GLYCOSCIENCES.de, GlycoSuite, CFG Glycan Database and KEGG-Glycan concentrate on glycan structures found in mammalian species. In contrast, the mission of the Russian Bacterial Carbohydrate Structure DataBase (BCSDB) [(14), for URLs see Appendix] is to provide all published glycan structures found in bacteria. Since the monosaccharide namespace as well as the type of linkages found in bacterial polysaccharides differ considerably from those found in mammals, BCSDB uses an internal representation of glycans, which diverges from those used to describe structures found in mammals.
Looking at various existing carbohydrate databases accessible through the Internet, it is obvious that diverse ways to encode and store complex carbohydrates are in use. They all seem to work satisfactorily for the purpose they have been designed for. However, users who would like to access all publicly available glyco-related data spread over many databases have not only to cope with varying graphical and non-graphical interfaces to input glycan structures, but also must be aware that the definition of building blocks and topologies may be different. Each database has developed its own set of rules to solve some problematic encoding situations such as treatment of monovalent substituents, phosphates, sulphates, repeat units, unknown linkages and other uncertain structural features of glycan structures.
It is, of course, an attractive vision [expressed during the Joint Meeting of the Japanese and American Consortia for Glycomics (15)] to have a single user interface which will provide access to all relevant world-wide distributed resources without any technical and administrative barrier. A prerequisite for an efficient exchange of data is the agreement to a generally accepted exchange format as well as a common application programming interface. Consequently, several proposals for an XML-based description of glycan structures have already been published (16,17). To avoid any further confusion about XML descriptions of glycans, the seven larger initiatives in this field [CFG, BCSDB, GLYCOSCIENCES.de, EUROCarbDB, KEGG, HGPI and CCRC (for abbreviations see Appendix)] agreed to further develop the XML description for the encoding of glycan structures on the basis of the already existing GLYcan Data Exchange (GLYDE) (17). The progress discussion is open to all interested scientists and takes place at the forum pages of the EUROCarbDB project.
Concerning the technical realization of the online connection between existing databases, it seems that the Simple Object Access Protocol (SOAP) is now the broadly accepted procedure for automated communication between web-applications. Being designed to communicate via the Internet, it is well suited to be also used for the exchange of glycan-related data between distributed computers. Taken together, it seems like the field has matured to the point where it is feasible to establish an online connection of distributed databases, at least between the larger of the established projects.
BACTERIAL CARBOHYDRATE STRUCTURE DATABASE
The Bacterial Carbohydrate Structure DataBase (BCSDB) [(14), for URLs see Appendix] is a database containing data on natural carbohydrates with known structure. In addition to the structure and bibliography, each record in the BCSDB contains the abstract of the publication, data on the carbohydrate source, methods of structure elucidation, information on the availability of spectral data and assignment of NMR spectra when available, data on conformation, biological activity, chemical and enzymatic synthesis, biosynthesis, genetics and other related data. The search criteria can be fragment(s) of the structure; fragment(s) of the NMR spectrum; and indexed tags, including microorganism, bibliography and keywords.
Currently, the BCSDB contains ∼8200 records on bacterial carbohydrates, including the corresponding part of CarbBank (18) (∼3500 records on structures reported before 1995). This coverage is approaching the total number of bacterial carbohydrate structures ever reported. Data from both literature and CarbBank have been carefully checked for consistency before the upload, and corrected when necessary. The BCSDB interface includes the web-based user part, web-based administrator part and programming gateways for the automated data interchange. The BCSDB is available on the Internet for free usage and validated user data submission.
GLYCOSCIENCES.de
The GLYCOSCIENCES.de portal (7) is an attempt to link glycan-related data originating from various resources through a unique structural description. The LINUCS (LInear Notation for Unique description of Carbohydrate Sequences) (10) notation is used to uniquely encode fully characterized glycans. Currently, the GLYCOSCIENCES portal provides access to ∼24 000 different entries with nearly 14 000 different carbohydrate moieties. These structures are sourced from a number of sources, including the former CarbBank and SugaBase-project (19), automatic extraction from the Protein Data Base (PDB) (20), and the curation of new entries altogether. The structure-oriented approach to the database allows the data related to a single glycan, but originating from various sources (e.g. experimental NMR spectra, theoretically calculated fragment ions for mass spectra interpretation or experimental or simulated 3D structures) to be easily linked and accessed using a single database query. According to the varying needs of specific research questions, the GLYCOSCIENCES portal provides several structure-oriented options to recall glycan-related data. Substructure searches are the most frequently used way to look for glycan structures. The retrieval of glycans matching an exact structure is the most traditional way to access a database. The motif search enables to retrieve all entries, which possess substructures having names such as LewisX, blood group H antigen or GM3. All glycan-related scientific data of the GLYCOSCIENCES.de portal are freely accessible via the Internet following the open access philosophy: ‘free availability and unrestricted use’.
WEB-SERVICES
The SOAP-based web-services are available on the websites of the two projects and are documented in the form of WSDL (for URLs see Appendix) descriptions that provide the possibility of platform-independent formalization of server-side features. WSDL files can be easily integrated into the existing code by using features from various SOAP libraries which allow the transparent work with the SOAP interface under Perl, PHP, Java, etc. Additionally sample PHP clients are available.
DATA TRANSFER FORMATS
GLYcan Data Exchange (GLYDE) version 1.2 (17) was chosen as the structure exchange format. It supports almost all known peculiarities of carbohydrate structures, such as uncertainities in configuration and ring sizes, various combinations of repeating and non-repeating parts, non-carbohydrate linkers, cyclic structures, etc.. GLYDE uses a tree-based approach to structure description. Within this approach the tree root is the reducing and or the rightmost residue in the repeating unit, while all the substituents are the ‘children’ of the residue they are attached to. Configurations, ring-size and other related information is stored as attributes of the residue. The syntax of GLYDE is XML.
To transfer the bibliographic information two approaches are used: the raw data (as array of strings corresponding to authors, title terms, journal name, etc.) or PubMed XML. BCSDB supports both formats, while GLYCOSCIENCES.de currently supports only the former. The former format is simpler in realization but the latter provides more standardization. PubMed XML encodes the bibliographic information using the strictly defined set of rules. More information is available at the NCBI PubMed XML tagged data homepage.
A well-known identifier for an organism is a TaxID provided by NCBI taxonomy database. Both databases provide the search mechanism that uses NCBI TaxID to identify the microorganism. However, the ranking of TaxID is limited to species; thus, no possibility to cross-search for particular strains/serogroups is provided. As this detailed ranking is significant mainly for bacteria, the capability to perform deep species searching is only supported on the BCSDB side of the connection. TaxIDs are stored in the GLYCOSCIENCES.de database together with structures, while BCSDB generates TaxIDs based on genus and species name, making use of an NCBI web service.
EXAMPLES
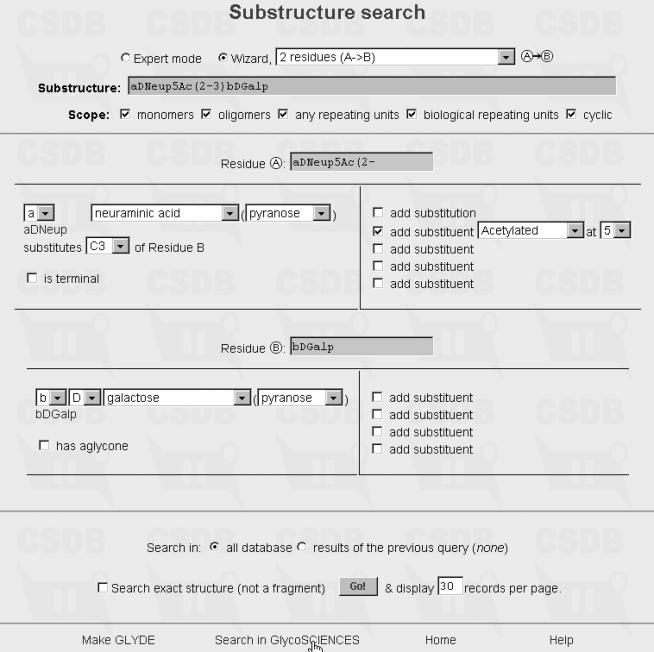
Three examples are given to demonstrate the established interconnection of both data collections. Example 1, using the bibliographic search of GLYCOSCIENCES.de, shows all references found in both resources for author ‘Brade’ in year 2002. GLYCOSCIENCES.de has included only two papers, where NMR spectra are reported. BCSDB lists another five papers where the structures of bacterial polysaccharides are described. Example 2 depicts a substructure search containing a specified disaccharide fragment [α-d-Neup5NAc-(2-3)-β-d-Galp] in GLYCOSCIENCES.de. The structure input option implemented in BCSDB (see Example 2a) is used. The data associated with two entries containing the disaccharide fragment are shown in Example 2b. Example 3 demonstrates a substructure search in BCSDB using GLYCOSCIENCES.de to input the trisaccharide fragment α-d-Galp-(1-3)-α-d-Manp-(1-4)-α-l-Rhap.
Example 1.
Retrieval request of references of author ‘Brade’ in year 2002 in any journal. Used was the GLYCOSCIENCES.de advanced bibliographic search (only results are shown). References from BCSDB contain one structure each.
Example 2a.
Substructure Search for a-d-Neup5NAc-(2-3)-b-d-Galp in Glycosciences.de using the BCSDB Input wizard.
Example 2b.
Result querying GLYCOSCIENCES for all structures containing a specified disaccharide fragment α-d-Neup5Ac-(2→3)β-d-Galp. The data associated with two entries are shown.
Example 3.
Querying BCSDB for all structures containing the specified trisaccharide fragment α-d-Galp-(1-3)-α-d-Manp-(1-4)-α-L-Rhap. The GLYCOSCIENCES.de substructure input spreadsheet is used. The data associated with BCSDB entry 10147 are additionally shown.
CONCLUSIONS
The capability of web services to make distributed scientific data accessible is clearly demonstrated. To our knowledge, the implemented mutual online access between BCSDB and GLYCOSCIENCES.de is the first reported attempt of a structure-based interconnection of two glyco-related databases. For users the advantages are obvious: they can use and have to learn only one interface, always have access to the latest data from both services, and the results of both searches are presented in a consistent way. For the database design and its functionality the establishment of a connection helped to find shortcomings and inconsistencies in both underlying data concepts and structural representations. For the maintenance of the databases, duplication of work can be easily avoided. It can be expected that more frequent use of both services will improve the quality of data. This will hopefully lead to a better worldwide acceptance of both services within the community of glycoscientists.
Since the exchange of data is accomplished through standard, well-documented XML-based descriptions and SOAP protocols; other interested providers of glyco-related databases may easily be linked so that a larger network could grow. It can be envisaged that online connection of thematically related scientific data collections will have a bright future, and not only in the area of glycosciences. One of the main bottlenecks is currently that broadly accepted standard XML exchange formats are often not yet available. It will definitively be a time-consuming task to come to agreements about such standard descriptions within the various communities. With GLYDE 1.2 an XML-based encoding scheme of glycan structures exists, which is sufficiently flexible to link the vast majority of structures contained in BCSDB and GLYCOSCIENCES.de. However, GLYDE 1.2 has some shortcomings regarding uncertainties in terminal residues and other fuzzy encodings, which will become more important for glycomics projects. The current focus of discussion is to base a more flexible encoding on the concept of a connection table approach, instead of a tree-like structure as used in GLYDE 1.2. Recently (September 2006, NIH Meeting ‘Frontier in Glycomics’), the seven larger projects already mentioned above have agreed to support GLYDE-CT as the main database format for the exchange of glycan structures. The hope is of course that only one format will be used by everyone. A less favourable situation would be that several exchange format exit and parsers must be available for each database.
Acknowledgments
P.T. thanks the DKFZ for the fellowship supporting his stay in Heidelberg. The development of GLYCOSCIENCES.de at the DKFZ was supported by a Research Grant from the German Research Foundation (DFG BIB 46 HDdkz 01-01) within the digital library program. The development of the BCSDB was supported by the International Science and Technology Center (project 1197p), the Russian Foundation for Basic Research (project 05-07-90099) and Russian President Grant Commetee (project MK-2005.1700.4). Funding to pay the Open Access publication charges for this article was provided by DFG.
Conflict of interest statement. None declared.
APPENDIX
Web addresses of tools discussed in the paper are as follows:
Apache Axis: http://ws.apache.org/axis
BCSDB: http://www.glyco.ac.ru/bcsdb/
BCSDB: http://www.glyco.ac.ru/bcsdb/help/bcsdb.wsdl
CCRC (Complex Carbohydrate Research Center). http://www.ccrc.uga.edu/
Codehaus Xfire: http://xfire.codehaus.org
Consortium For Functional Glycomics: http://www.functionalglycomics.org/
EUROCarbDB: http://www.eurocarbdb.org
EUROCarbDB forum: http://www.dkfz.de/spec/EuroCarbDB_forum/
GLYCOSCIENCES: http://www.glycosciences.de
GLYCOSCIENCES web-services: http://www.glycosciences.de/soap/soapservice?wsdl
Glyde 1.2: http://www.glyco.ac.ru/bcsdb/help/glyde12.dtd
KEGG-Glycam: http://www.genome.jp/ligand/kcam/
HGPI (Human Disease Pglcomics/Proteome Initiative): http://www.hgpi.jp/
Hibernate: http://www.hibernate.org
Monosaccharoide DB: http://www.dkfz.de/spec/monosaccharide-db/
NCBI electronic utilities: http://www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html
NCBI PubMed XML tagged data: http://www.ncbi.nlm.nih.gov/entrez/query/static/publisher.html
NCBI Taxonomy: http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/
SOAP Lite: http://www.soaplite.com/
Web Service Data Language (WSDL): http://www.w3.org/TR/wsdl/
REFERENCES
- 1.Lowe J., Marth J. A genetic approach to Mammalian glycan function. Annu. Rev. Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 2.Raman R., Raguram S., Venkataraman G., Paulson J.C., Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nature Methods. 2005;1:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 3.von der Lieth C.W., Bohne-Lang A., Lohmann K.K., Frank M. Bioinformatics for glycomics: status, methods, requirements and perspectives. Brief Bioinformatics. 2004;5:164–178. doi: 10.1093/bib/5.2.164. [DOI] [PubMed] [Google Scholar]
- 4.Harvey D. Proteomic analysis of glycosylation: structural determination of N- and O-linked glycans by mass spectrometry. Expert Rev. Proteomics. 2005;2:87–101. doi: 10.1586/14789450.2.1.87. [DOI] [PubMed] [Google Scholar]
- 5.Guerardel Y., Chang L., Maes E., Huang C., Khoo K. Glycomic survey mapping of zebrafish identifies unique sialylation pattern. Glycobiology. 2006;16:244–257. doi: 10.1093/glycob/cwj062. [DOI] [PubMed] [Google Scholar]
- 6.Raman R., Venkataraman M., Ramakrishnan S., Lang W., Raguram S., Sasisekharan R. Advancing Glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- 7.Lutteke T., Bohne-Lang A., Loss A., Goetz T., Frank M., von der Lieth C.W. GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto K., Goto S., Kawano S., Aoki-Kinoshita K., Ueda N., Hamajima M., Kawasaki T., Kanehisa M. KEGG as a glycome informatics resource. Glycobiology. 2006;16:63R–70R. doi: 10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- 9.von der Lieth C.W. An endorsement to create open databases for analytical data of complex carbohydrates. J. Carbohydr. Chem. 2004;23:277–297. [Google Scholar]
- 10.Bohne-Lang A., Lang E., Forster T., von der Lieth C.W. LINUCS: linear notation for unique description of carbohydrate sequences. Carbohydr. Res. 2001;336:1–11. doi: 10.1016/s0008-6215(01)00230-0. [DOI] [PubMed] [Google Scholar]
- 11.Cooper C., Joshi H.J., Harrison M., Wilkins M., Packer N. GlycoSuiteDB: a curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 2003;31:511–513. doi: 10.1093/nar/gkg099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banin E., Neuberger Y., Altshuler Y., Halevi A., Inbar O., Dotan N., Avinoam D. A novel linear code® nomenclature for complex carbohydrates. Trends Glycosci. Glycotechnol. 2002;14:127–137. [Google Scholar]
- 13.Aoki K., Yamaguchi A., Ueda N., Akutsu T., Mamitsuka H., Goto S., Kanehisa M. KCaM (KEGG Carbohydrate Matcher): a software tool for analyzing the structures of carbohydrate sugar chains. Nucleic Acids Res. 2004;32:W267–W272. doi: 10.1093/nar/gkh473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toukach F.V., Knirel Y.A. New database of bacterial carbohydrate structures. proceedings of the XVIII International Symposium on Glycoconjugates; Florence, Italy. 2005. pp. 216–217. [Google Scholar]
- 15.Glycomics, Consortium for Functional. 2004. Joint meeting of the Japanese and American consortia for glycomics. [Google Scholar]
- 16.Kikuchi N., Kameyama A., Nakaya S., Ito H., Sato T., Shikanai T., Takahashi Y., Narimatsu H. The carbohydrate sequence markup language (CabosML): an XML description of carbohydrate structures. Bioinformatics. 2005;21:1717–1718. doi: 10.1093/bioinformatics/bti152. [DOI] [PubMed] [Google Scholar]
- 17.Sahoo S., Thomas C., Sheth A., Henson C., York W. GLYDE—an expressive XML standard for the representation of glycan structure. Carbohydr. Res. 2005;340:2802–2807. doi: 10.1016/j.carres.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 18.Doubet S., Bock K., Smith D., Darvill A., Albersheim P. The complex carbohydrate structure database. Trends Biochem. Sci. 1989;14:475–477. doi: 10.1016/0968-0004(89)90175-8. [DOI] [PubMed] [Google Scholar]
- 19.van Kuik J.A, Hard K., Vliegenthart J.F. 1H NMR database computer program for the analysis of the primary structure of complex carbohydrates. Carbohydr. Res. 1992;235:53–68. doi: 10.1016/0008-6215(92)80078-f. [DOI] [PubMed] [Google Scholar]
- 20.Lutteke T., Frank M., von der Lieth C.W. Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr. Res. 2004;339:1015–1020. doi: 10.1016/j.carres.2003.09.038. [DOI] [PubMed] [Google Scholar]