Abstract
The Human Metabolome Database (HMDB) is currently the most complete and comprehensive curated collection of human metabolite and human metabolism data in the world. It contains records for more than 2180 endogenous metabolites with information gathered from thousands of books, journal articles and electronic databases. In addition to its comprehensive literature-derived data, the HMDB also contains an extensive collection of experimental metabolite concentration data compiled from hundreds of mass spectra (MS) and Nuclear Magnetic resonance (NMR) metabolomic analyses performed on urine, blood and cerebrospinal fluid samples. This is further supplemented with thousands of NMR and MS spectra collected on purified, reference metabolites. Each metabolite entry in the HMDB contains an average of 90 separate data fields including a comprehensive compound description, names and synonyms, structural information, physico-chemical data, reference NMR and MS spectra, biofluid concentrations, disease associations, pathway information, enzyme data, gene sequence data, SNP and mutation data as well as extensive links to images, references and other public databases. Extensive searching, relational querying and data browsing tools are also provided. The HMDB is designed to address the broad needs of biochemists, clinical chemists, physicians, medical geneticists, nutritionists and members of the metabolomics community. The HMDB is available at: www.hmdb.ca
INTRODUCTION
Metabolomics is a newly emerging field of ‘omics’ research concerned with the high-throughput identification, quantification and characterization of the small molecule metabolites in the metabolome (1). The metabolome can be defined as the complete complement of all small molecule (<1500 Da) metabolites found in a specific cell, organ or organism. It is a close counterpart to the genome, the transcriptome and the proteome. Together these four ‘omes’ constitute the building blocks of systems biology. Thanks to the Human Genome Project most of the human genome, transcriptome and proteome are now known and the data are electronically available. Unfortunately, the same cannot be said of the human metabolome. To remedy this situation, the Human Metabolome Project (HMP) was launched in 2004 as part of an effort to identify and quantify all detectable metabolites (>1 μM) in the human body. In addition to experimentally identifying and quantifying hundreds of metabolites in different body fluids, this multi-year project was also formally tasked with backfilling and validating the information on all previously identified metabolites and providing this information as a freely available electronic database called the Human Metabolome Database (HMDB).
The foundation to all metabolomics research lies in the work done by hundreds of metabolic biochemists and clinical chemists over the past 70 years. The pathways, reactions and reactants that were identified by these scientists are now available in many excellent on-line metabolic pathway databases such as KEGG (2), BioCyc (3) and Reactome (4). More recently, information about the genes and diseases that are associated with perturbations to these pathways has come on-line through such outstanding resources as OMMBID (5) and OMIM (6). However, the information contained in these databases does not meet the unique data requirements for metabolomics researchers, especially those involved in human metabolomics. This is because human metabolomics is often concerned with rapidly identifying dozens of metabolites at a time and then using these metabolites or combinations of metabolites as disease and/or phenotypic biomarkers. As a result, metabolomics researchers need databases that can be searched using Nuclear Magnetic Resonance (NMR) spectra, mass spectra (MS), chemical structures or chemical formulas—as opposed to sequences or sequence names (7). Likewise metabolomics researchers routinely need to search for metabolite concentrations, properties, locations or metabolite-disease associations. Therefore, metabolomics databases require information not only about compounds and reactions but also data about compound concentrations, biofluid or tissue locations, subcellular locations, physical properties, known disease associations, nomenclature, descriptions, enzyme data, mutation data and characteristic MS or NMR spectra. These data need to be readily available, experimentally validated, fully referenced, easily searched, readily interpreted and they need to cover as much of the human metabolome as possible. In other words, metabolomics researchers need a metabolic equivalent to GenBank or SwissProt. To address these needs, and to serve as a potential model for other metabolomic resources, we have developed the HMDB.
DATABASE DESCRIPTION
Fundamentally the HMDB is a multi-purpose bioinformatics–cheminformatics–medical informatics database with a strong focus on quantitative, analytic or molecular-scale information about metabolites, their associated enzymes or transporters and their disease-related properties. In many respects the HMDB combines the data-rich molecular biology content normally found in curated sequence databases such as SwissProt and UniProt (8) with the equally rich data found in KEGG (about metabolism) and OMMBID (about clinical conditions). It also brings in a large body of independently collected experimental data, including NMR spectra, MS spectra, solubility data and validated metabolite concentrations, to compliment this literature-derived data.
The diversity of data types, the quantity of experimental data and the required breadth of domain knowledge made the assembly of the HMDB both difficult and timeconsuming. To compile, confirm and validate this comprehensive collection of data, more than two dozen textbooks, several thousand journal articles, nearly 30 different electronic databases, and at least 20 in-house or web-based programs were individually searched, accessed, compared, written or run over the course of the previous two years. In addition, more than 2100 confirmatory NMR and MS spectra were collected, 160 experimental solubility determinations were made, 75 organic syntheses were completed and hundreds of high-performance liquid chromatography (HPLC) separations were performed. The team of HMDB contributors and annotators included three organic chemists, six NMR spectroscopists, five mass spectroscopists, two separation specialists, three physicians and 14 bioinformaticians with dual training in computing science and molecular biology/chemistry.
The HMDB currently contains more than 2180 human metabolite entries that are linked to more than 27 700 different synonyms. These metabolites are further connected to some 115 non-redundant pathways, 2080 distinct enzymes, 110 000 SNPs as well as 862 metabolic diseases (genetic and acquired). More than 400 compounds are also linked to experimentally acquired ‘reference’ 1H and 13C NMR and MS/MS spectra. Concentration data (normal and abnormal values) for plasma, urine, CSF and/or other biofluids are also provided for a total of 883 compounds. The entire database, including text, sequence, structure and image data occupies nearly 18 GB of data—most of which can be freely downloaded.
The HMDB is fully searchable with many built-in tools for viewing, sorting and extracting metabolites, biofluid concentrations, enzymes, genes, NMR or MS spectra and disease information. Detailed instructions on where to locate and how to use these browsing/search tools are provided on the HMDB homepage. As with any web-enabled database, the HMDB supports standard text queries (through the text search box located near the top of each page). It also offers general database browsing using the ‘Browse’ and ‘Biofluids’ buttons located in the HMDB menu bar. To facilitate data browsing, the HMDB is divided into synoptic summary tables which, in turn, are linked to more detailed ‘MetaboCards’—in analogy to the very successful DrugCards concept found in DrugBank (9). All of the HMDB's summary tables can be rapidly browsed, sorted or reformatted in a manner similar to the way PubMed (10) abstracts may be viewed. Clicking on the MetaboCard button found in the leftmost column of any given HMDB summary table opens a webpage describing the compound of interest in much greater detail. Each MetaboCard entry contains more than 90 data fields (Table 1) with half of the information being devoted to chemical or physico-chemical data and the other half devoted to biological or biomedical data (disease, biofluid concentration, enzyme, gene, SNP or metabolic pathway information). In addition to providing comprehensive numeric, sequence and textual data, each MetaboCard also contains hyperlinks to many other databases (KEGG, BioCyc, PubChem, ChEBI, PubMed, PDB, SwissProt, GenBank, OMIM and dbSNP), abstracts, digital images and applets for viewing molecular structures (Figure 1).
Table 1.
Summary of the data fields or data types found in each MetaboCard
| Metabolite and medical information | Protein/enzyme information |
|---|---|
| Common name | Enzyme/protein name |
| Description | Enzyme/protein synonyms |
| Synonyms/IUPAC name | Enzyme/protein sequence |
| Chemical structure | Protein number of residues |
| Chemical taxonomy | Protein molecular weight |
| Molecular weight (mono and ave) | Protein pI |
| SMILES (isomeric and canonical) | Protein gene ontology |
| KEGG/PubChem/OMIM/MetaGene links | Protein general function |
| CAS number | Protein specific function |
| InChi identifier | Protein pathways |
| Melting point | Protein reactions |
| Water solubility (predicted and expt) | Protein Pfam domains |
| State (solid, liquid, gas) | Protein signal sites |
| pKa or pI | Protein transmembrane regions |
| LogP or hydrophobicity | Protein metabolic importance |
| MOL/SDF/PDF text files | Protein/enzyme EC link |
| MOL/PDB image files | GenBank, SwissProt, PDB ID |
| NMR spectra (predicted, calculated) | Protein structure data |
| Location (cell, biofluid, tissue) | Protein cellular location |
| Concentration (urine, plasma, CSF) | Gene sequence |
| Associated disorders | GenBank ID |
| Abnormal concentration (urine, plasma, CSF) | Chromosome location |
| Metabolic pathways (KEGG, SimCell) | Chromosome locus |
| Metabolizing enzymes | Protein/enzyme SNPs/mutations |
| Metabolizing ENZYMES | Protein/enzyme references |
A more complete listing is provided under the HMDB ‘Misc’ hyperlink on the HMDB menu bar.
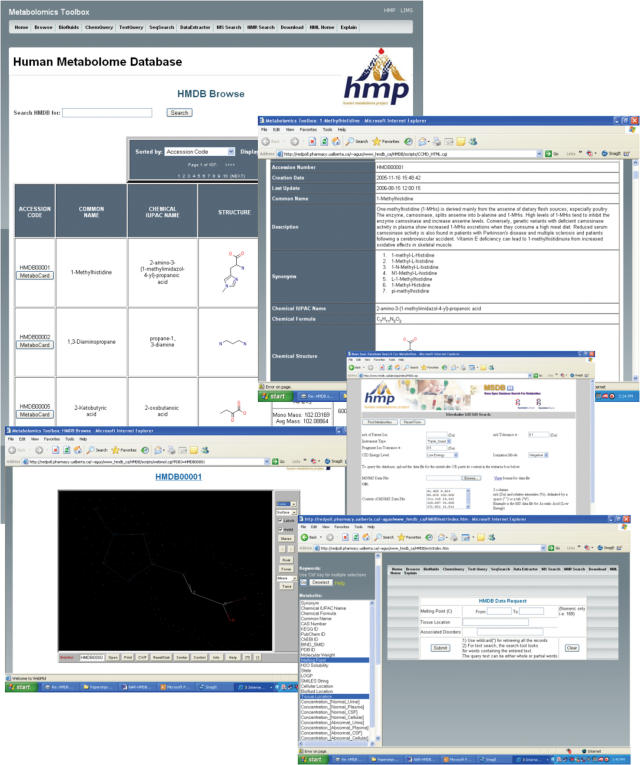
Figure 1.
A screenshot montage of the Human Metabolome Database (HMDB) showing several of HMDB's search and data display tools describing the metabolite 1-Methylhistidine. Not all fields are shown.
A key feature that distinguishes the HMDB from other metabolic resources is its extensive support for higher level database searching and selecting functions. In addition to the data viewing and sorting features already described, the HMDB also offers a chemical structure search utility, a local BLAST search (11) that supports both single and multiple sequence queries, a boolean text search based on GLIMPSE (12), a relational data extraction tool, an MS spectral matching tool and an NMR spectral search tool (for identifying compounds via MS or NMR data from other metabolomic studies).
The HMDB's structure similarity search tool (ChemQuery) is the equivalent to BLAST for chemical structures. Users may sketch [through Advanced Chemistry Development's (ACD) freely available ChemSketch applet] or paste a SMILES string (13) of a query compound into the ChemQuery window. Submitting the query launches a structure similarity search tool that looks for common substructures from the query compound that match the HMDB's metabolite database. High scoring hits are presented in a tabular format with hyperlinks to the corresponding MetaboCards (which in turn links to the protein target). The ChemQuery tool allows users to quickly determine whether their compound of interest is a known metabolite or chemically related to a known metabolite. In addition to these structure similarity searches, the ChemQuery utility also supports compound searches on the basis of chemical formula and molecular weight ranges.
The BLAST search (SeqSearch) allows users to search through the HMDB via sequence similarity as opposed to chemical similarity. A given gene or protein sequence may be searched against the HMDB's sequence database of metabolically important enzymes and transporters by pasting the FASTA formatted sequence (or sequences) into the SeqSearch query box and pressing the ‘submit’ button. A significant hit reveals, through the associated MetaboCard hyperlink, the name(s) or chemical structure(s) of metabolites that may act on that query protein. With SeqSearch metabolite-protein interactions from recently sequenced mammals (chimp, rat, mouse, dog, cat, etc.) may be mapped to these organisms via the human data in the HMDB.
The HMDB's data extraction utility (Data Extractor) employs a simple relational database system that allows users to select one or more data fields and to search for ranges, occurrences or partial occurrences of words or numbers. The Data Extractor uses clickable web forms so that users may intuitively construct SQL-like queries. The data extraction tool allows users to easily construct complex queries as ‘find all diseases where the concentration of homogentisic acid in urine is >1 mM’.
The NMR and MS search utilities allow users to upload spectra (for the MS search) or peak lists (for the NMR search) and to search for matching compounds from the HMDB's collection of MS and NMR spectra. The HMDB contains approximately 3800 predicted 1H and 13C NMR spectra for 1900 compounds. The predicted 1H and 13C NMR spectra were generated using the ACD/HNMR and ACD/CNMR software from Advanced Chemistry Development Inc. Validated Mol files for each compound were used as input for each prediction. In addition, the HMDB contains 930 experimentally collected 1H and 13C NMR spectra for 400 pure compounds (most collected in water at pH 7.0, 10 mM for 1H, 50 mM for 13C). It also contains 1200 MS/MS (Triple-Quad) spectra at three different collision energies for nearly 400 pure compounds. An average of 50 new NMR and MS spectra are being added each month. The HMDB's spectral search utilities allow both pure compounds and mixtures of compounds to be identified from their MS or NMR spectra via peak matching algorithms that were developed in-house. The NMR spectral matching algorithm uses a simple peak matching rule with pre-defined chemical shift tolerances. Query spectra are scored on the number of peak matches to the database spectra. The MS/MS spectral matching algorithm uses a peak matching and spectral scoring concept similar to one previously published by our group (14). The complete set of annotated spectral images (NMR and MS, both experimental and predicted) are retrievable as zip files through the ‘Download’ button located at the top of the HMDB menu.
The link ‘HML Home’ in the HMDB menu bar refers to the Human Metabolite Library. This is a repository of all purchased, synthesized and isolated metabolites that have been acquired by the HMP team. Small quantities of individual compounds or larger collections of metabolites may be purchased (at cost) or freely acquired for collaborative research (via material transfer agreements) through the HML website and its web ordering forms. These compounds may be used as reference or quantitation standards by metabolomics researchers, or the collections may be used for drug screening, crystal screening and enzyme function assays.
Perhaps the most relevant features of the HMDB from the perspective of a medical geneticist or a clinical chemist are its rich content and extensive linkage to metabolic diseases, to normal and abnormal metabolite concentration ranges (in many different biofluids), to mutation/SNP data and to the genes, enzymes, reactions and pathways associated with many diseases of interest. Currently, the HMDB contains 115 metabolic pathway diagrams or metabolic maps. While this number may seem small, the total number of known human pathways in the KEGG database is just 190, with 72 of these being protein-only pathways (i.e. no metabolites). Nevertheless, this total is expected to increase as there are a growing number of novel gene-metabolite regulation pathways being identified via nutrigenomic research—many of which will be included in the HMDB. There are also a number of important drug and xenobiotic metabolism pathways (not in KEGG or Reactome) that will be added over the coming months.
A particularly recent addition to the HMDB is a series of SimCell (15) metabolic wiring diagrams, SimCell models in SBML (Systems Biology Markup Language) and SimCell simulations of nearly 30 well-characterized metabolic pathways. SimCell is a metabolic simulation software package that allows complex metabolic pathways to be modeled at a cellular level and for ‘real-time’ movies of the enzymatic processes to be generated and graphed. The availability of these pre-assembled metabolic models should allow users to simply download the SimCell wiring diagram and conduct ‘in silico’ gene knock-out experiments or test hypotheses ‘in silico’ concerning the possible causes of a suspected genetic disorder.
QUALITY ASSURANCE, COMPLETENESS AND CURATION
The human metabolome is not as well defined as the human genome. The constitution of the metabolome depends on how a metabolite is defined, including its molecular weight cutoff, origin and concentration limit. If every small molecule in the body (food additive, plant extract, drug, drug derivative, toxin, cleaning agent or environmental contaminant) were to be included—from any source at any concentration level—the number of compounds might exceed 100 000. To make the database both relevant and reasonable, the criteria for inclusion in the HMDB is as follows: the compound must weigh <1500 Da, it should be found at concentrations greater than 1 μM (either in normal or in diseased conditions) in one or more biofluids/tissues and it should be of biological origin (either generated by human cells or endogenous gut microflora). Some exceptions are made, including low abundance but biomedically important metabolites (hormones, disease-associated metabolites, essential nutrients and signaling molecules), certain very common drugs (acetaminophen, nicotine) and some ubiquitous food additives (cellobiose, vitamins). Separate databases are being developed by the HMP consortium to include drugs (DrugBank), food additives and toxins.
In annotating the HMDB, every effort is made to ensure that the database is as complete, correct and current as possible. Metabolites are first identified by literature surveys (PubMed, OMIM, OMMBID, text books), data mining (KEGG, Metlin, BioCyc) or experimental methods. If the compound passes the HMDB inclusion criteria, information about the metabolite is entered or prepared by one member of the curation team and separately validated by second member of the curation team. Additional spot checks are routinely performed on each entry by senior members of the curation group, including two PhD-level biochemists. The annotation effort is also aided by several software packages including text mining tools, chemical parameter calculators and protein annotation tools originally developed for DrugBank (9) but modified for the HMDB. These tools collate and display text (and images) from multiple sources allowing the curators to compare, assess, enter and correct metabolite or enzyme/gene information. The source data and software tools used in the annotation process are described in more detail under the ‘Explain’ button located in the HMDB menu bar.
All data is entered into a centralized laboratory information management system (LIMS) allowing all changes and edits to the HMDB to be monitored, dated and automatically transferred. Consistency checks (molecular weight matches chemical formula, state consistent with melting point, no negative molecular weights, name formatting is correct, etc.) are performed every night using an automated checking/correcting script. A second text tracking system has been implemented to monitor and display up-to-date statistics on the number of metabolites, enzymes, non-redundant sequences and other HMDB statistics. This information is displayed on the ‘Download’ page. Proteins that act on, transport or bind to the metabolites in the HMDB are identified and confirmed using multiple sources (PubMed, KEGG, BioCyc, textbooks) as are all metabolite structures, concentrations and pathways (KEGG, PubChem, journals, textbooks). Every effort is being made to validate reported concentration data using independent experimental methods.
DATABASE IMPLEMENTATION
The HMDB is essentially a web-friendly front-end to a sophisticated MySQL relational database (version 4.1.7). Both are maintained on a Linux server equipped with a 2 GHz CPU processor and 1 GB of RAM. Perl scripts are run every night to read selected portions of the MySQL database and to write out the data into GLIMPSE-indexed raw text and XML formats The raw text is dynamically rendered into HTML (with images and hyperlinks) using a series of custom Perl scripts. The MySQL database is part of a generalized metabolomic LIMS system called MetaboLIMS, that allows data about metabolites, pathways, spectra and concentrations to be entered manually. It is a Java application that uses standard Apache web server technologies such as Javaserver Pages (JSP) and Java servlets to manage web-based data input and data queries. MetaboLIMS is also designed to use an imbedded text mining tool called BioSpider (16) to automatically annotate new metabolite entries (descriptions, physical properties and biological data). MetaboLIMS and BioSpider, both of which are available on request from the authors, can be readily used to prepare organism-specific metabolome databases similar to the HMDB.
CONCLUSION
In summary, the HMDB is a comprehensive, web-accessible metabolomics database that brings together quantitative chemical, physical, clinical and biological data about thousands of endogenous human metabolites. We believe the HMDB is unique, not only in the type of data it provides but also in the level of integration and depth of coverage it achieves. The HMDB is also a work in progress. Just like GenBank during the 1990s, the HMDB is going through a rapid phase of growth with constant additions and corrections being made and the release of different database ‘builds’ every two months. Approximately, 100 new compounds are being added every month. These updates will continue throughout most of 2007 after which the HMDB will move from a strictly curated database model to a public-deposition model. Overall, it is hoped that the HMDB will serve as a useful model for future metabolomics databases and that it will serve members of the metabolomics research community as well as educators, students, clinicians and the general public.
Acknowledgments
The authors wish to thank the Canadian Foundation for Innovation (CFI), the Alberta Ingenuity Centre for Machine Learning (AICML) and Genome Alberta, a division of Genome Canada for financial support. Funding to pay the Open Access publication charges for this article was provided by Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.German J.B., Hammock B.D., Watkins S.M. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics. 2005;1:3–9. doi: 10.1007/s11306-005-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krummenacker M., Paley S., Mueller L., Yan T., Karp P.D. Querying and computing with BioCyc databases. Bioinformatics. 2005;21:3454–3455. doi: 10.1093/bioinformatics/bti546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scriver C.R., et al., editors. Online Metabolic and Molecular Basis of Inherited Disease (OMMBID) New York: McGraw-Hill; 2006. http://genetics.accessmedicine.com/). [Google Scholar]
- 7.Smith C.A., O'Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R., Custodio D.E., Abagyan R., Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 8.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manber U., Smith M., Gopal B. WebGlimpse—Combining Browsing and Searching, Usenix 1997 Annual Technical Conference; Anaheim, CA. 1997. pp. 195–206. [Google Scholar]
- 13.Weininger D. SMILES 1. Introduction and Encoding Rules. J. Chem. Inf. Comput. Sci. 1988;28:31–38. [Google Scholar]
- 14.Dworzanski J.P., Snyder A.P., Chen R., Zhang H., Wishart D.S., Li L. Identification of bacteria using tandem mass spectrometry combined with a proteome database and statistical scoring. Anal. Chem. 2004;76:2355–2366. doi: 10.1021/ac0349781. [DOI] [PubMed] [Google Scholar]
- 15.Wishart D.S., Yang R., Arndt D., Tang P., Cruz J. Dynamic cellular automata: an alternative approach to cellular simulation. In Silico Biol. 2005;5:139–161. [PubMed] [Google Scholar]
- 16.Knox C., Shrivastava S., Stothard P., Eisner R., Wishart D.S. BioSpider: A web server for automating metabolome annotations. Pac. Symp. Biocomput. 2007 (in press). [PubMed] [Google Scholar]