Abstract
SNAPPI-DB, a high performance database of Structures, iNterfaces and Alignments of Protein–Protein Interactions, and its associated Java Application Programming Interface (API) is described. SNAPPI-DB contains structural data, down to the level of atom co-ordinates, for each structure in the Protein Data Bank (PDB) together with associated data including SCOP, CATH, Pfam, SWISSPROT, InterPro, GO terms, Protein Quaternary Structures (PQS) and secondary structure information. Domain–domain interactions are stored for multiple domain definitions and are classified by their Superfamily/Family pair and interaction interface. Each set of classified domain–domain interactions has an associated multiple structure alignment for each partner. The API facilitates data access via PDB entries, domains and domain–domain interactions. Rapid development, fast database access and the ability to perform advanced queries without the requirement for complex SQL statements are provided via an object oriented database and the Java Data Objects (JDO) API. SNAPPI-DB contains many features which are not available in other databases of structural protein–protein interactions. It has been applied in three studies on the properties of protein–protein interactions and is currently being employed to train a protein–protein interaction predictor and a functional residue predictor. The database, API and manual are available for download at: http://www.compbio.dundee.ac.uk/SNAPPI/downloads.jsp.
INTRODUCTION
Protein–protein interactions are fundamental to understanding biological networks and cellular processes. Accordingly, many experimental (1–3) and computational (4–10) techniques have been developed to probe and predict interacting protein partners. There are several databases of protein interactions which store the information generated from high throughput experimental methods and literature curation, for example, GRID (11), the IntAct Project (12), BIND (13), MINT (14), DIP (15–17) and the HPRD (18). STRING (19) also contains data derived from database and literature mining and high-throughput experimental data, but in addition contains predictions based on genomic context analysis.
These computational and experimental techniques can yield significant information about possible interactions but they do not provide information about the structure of the interfaces at the atomic level. High-resolution X-ray and NMR structures can provide an atomic level of detail and have therefore been utilised for both investigation and prediction of protein–protein interactions. Analyses of interaction sites from 3-D structures have identified a number of properties that distinguish interaction sites from other areas of protein surfaces, including: residue conservation across species; a tendency to be polar, uncharged and hydrophobic; a planar protruding shape and a higher solvent accessible area (20–25). These properties have been exploited to predict interaction surfaces on protein structures (26–28).
Predictions of protein–protein interactions using structural data have been based on the hypothesis that if two proteins are seen to interact in a known 3-D structure, their homologues will interact in a similar fashion (29,30). A multimeric threading method has been used to extend this approach to distantly related homologous and analogous pairs (31). Structural data for interfaces has also been used to create templates that capture the essential features of interactions sites and which are employed to screen protein structures for the presence of interaction sites (32). Methods of protein–protein interaction prediction have been extensively reviewed by Szilagyi et al. (33).
The advantages of structural data have motivated the creation of several databases of protein-protein interactions and interfaces including 3did (database of 3-D interacting domains) (34), PIBASE (structurally defined protein interfaces) (35), SCOPPI (a structural classification of protein–protein interfaces) (36), PSIBase (Protein Structural Interactome map) (37) and PRISM (PRotein Interactions by Structural Matching) (38).
In this paper, a system is presented which provides a foundation for analysis and prediction of structural data with an emphasis on domain–domain interactions. This system consists of SNAPPI-DB, a database of Structures, iNterfaces and Alignments of Protein–Protein Interactions, and its associated Application Programming Interface (API). SNAPPI-DB, a high performance, object oriented database provides consistent, enhanced quality structural data, enriched with additional data such as multiple domain classifications, quaternary structures and domain-domain interactions. The API facilitates rapid development, is extensible, allows easy access to the data and circumvents the need to write complex SQL queries.
The contents and creation of SNAPPI-DB are discussed, followed by an overview of the API. The system is then compared to other databases of protein–protein interactions observed in structural data. Finally, the unique features of the system and its applications are discussed.
CONTENT AND CREATION
SNAPPI-DB is currently a 38 GB database containing 31 136 Protein Data Bank (PDB) structures and associated data including:
Domains classified to different levels of similarity based on the SCOP and CATH hierarchical classification system and Family level for Pfam.
Domain–domain interactions determined for SCOP, CATH and Pfam domains down to the level of which atoms interact.
Domain–domain interactions classified to different levels of similarity based on the SCOP and CATH hierarchical classification system and Family level for Pfam.
Domain–domain interactions classified by their interacting interfaces (orientation in which the domains are interacting).
Multiple structural alignments of domain interactions from each Family/Superfamily pair for each unique orientation for SCOP, CATH and Pfam domain definitions.
Biological units from Protein Quaternary Structures (PQS) (50).
Unique identifiers to link to the MSD data warehouse (51,52).
Interpro (53) regions/domains.
GO (54) terms.
SWISSPROT (55) identifiers and numbering.
MSD as the data source
The macromolecular structure database (MSD) (51,52) developed at the European Bioinformatics Institute (EBI) was chosen as the raw data source for SNAPPI-DB as it contains the complete contents of the PDB (39,40) together with substantial complementary data pertaining to domains, functional sites, protein families and sequences from other databases. These data include SCOP (41–43), CATH (44–46), Pfam (47–49), SWISSPROT (55), InterPro (53), GO terms (54), PQS (50), secondary structure information and detailed ligand properties. In addition, a key feature of the MSD is the improved consistency compared to PDB flat files.
The design of SNAPPI-DB
The MSD is an extremely useful database that has been key to the development of SNAPPI-DB; however, it was found that the speed of the MSD was not sufficient for high performance analysis at the atom level. In addition, the structure of the MSD is not optimised for analysis at the level of domains and does not contain the additional information stored by SNAPPI-DB on domain–domain interactions and structural multiple alignments of domain–domain interactions classified by their interface.
In order to increase performance and to allow complex analysis with a high degree of abstraction the data relevant to SNAPPI-DB were migrated to an object-oriented database developed with Java Data Objects (JDO) technology. JDO is a persistence framework for the Java language that allows the storage, retrieval and querying of objects. SNAPPI-DB employs the FastObjects community edition JDO implementation (http://www.versant.net/index.html). The application of JDO to storing biological has been described by Srdanovic et al. (56).
In essence, JDO provides an automatic mapping between a data-store and Java objects. This approach has many benefits. Firstly, JDO reduces development time as performing complex queries using this technology is easier than accessing a relational database directly via SQL. Secondly, employing JDO removes the complications inherent in mapping objects to a relational database, a difficulty commonly known as the ‘object-relational impedance mismatch’ problem (57). This feature greatly facilitates handling of biological data which often fits the object model. Finally, the JDO specification is intentionally data-store agnostic and so the JDO interface is the same regardless of the database back-end. Possible data-stores include relational databases, object databases and XML files. The choice of data-store will depend upon the user requirements. For example, a relational database is preferable if queries are to be performed by another application. In the case of high performance data mining an object-oriented data-store has many advantages over other data-store mechanisms such as lack of SQL overhead, speed, easy storage of polymorphic entities and direct two way references (56) and hence was choosen for SNAPPI-DB. However, if required the data could be ported to a relational database and the same API used.
Generation of domain–domain Interactions
Multiple domain definitions
Analysis of protein–protein interactions is usually performed at the level of domains rather than proteins since domains are often considered to be the fundamental functional and structural units. Domains can be assigned differently depending on the domain definition. As some domains in one domain classification do not have corresponding domains in another classification, some domain–domain interactions may not be found if only one of the classifications is used. SNAPPI-DB was employed to analyse the increase in the number of non-redundant domain–domain interactions provided by employing both SCOP and CATH domain definitions. It was observed that the use of both CATH and SCOP domain assignments increases the number of non-redundant interacting domains by between 23.6 and 37.3%. Therefore, it is advantageous to employ both sets of domain classifications simultaneously to investigate interactions at the domain level. Accordingly, both SCOP and CATH domain definitions are included in SNAPPI-DB. Pfam (47–49) was also included as this is a widely used sequence based domain definition which may be utilised to link to proteins which do not have a solved structure.
In addition to these three domain definitions, InterPro (53) domains, GO terms (54) and SWISSPROT (55) indexes are all stored.
Probable quaternary structures
The coordinates that appear in PDB files are those of the asymmetric unit (ASU) which is the fraction of the crystallographic unit cell that has no crystallographic symmetry. This may not be the biologically relevant unit of structure, and so may lack some key protein–protein interactions. In addition, some of the interactions seen in the ASU of the crystal may be artefacts of crystallisation and may not be biologically relevant (58,59).
There are two main sources of quaternary states: the state suggested by the authors of the structure (for all structures deposited since 1999) and computer predictions which apply all relevant symmetry operations and then discriminate between crystal packing artefacts and likely functional protein–protein interactions. The true quaternary state of a complex is not always straightforward to determine and errors are made by both the authors of the structure and in silico predictive methods. PQS (50) was chosen as the source of quaternary structure since although the initial assignments of biological units made by PQS are done by a computer program, they are hand-curated for each structure and errors and inconsistencies in the PQS database are corrected and updates made continuously.
Previously, SNAPPI-DB was used to investigate the additional non-redundant domain–domain interaction interfaces which are observed in the PQS predicted biological units in comparison to the ASUs seen in PDB files (60). It was determined that using PQS instead of the PDB increases the number of additional non-redundant interaction interfaces observed in structural data by 34.5% (1455 interfaces). PQS also removes 2981 interactions from the data set which it classifies as crystal packing artefacts. Therefore, the domain–domain interactions used in SNAPPI-DB are those observed in PQS biological units instead of the ASUs. The interactions which are seen in ASUs and not considered valid by PQS are also available to search in the database if required.
Defining an interaction
Interactions between domains are determined based on distance. Atoms are considered to interact if the distance between them was less than the sum of their van der Waals radii (61) +0.5 Å. Two domains are considered to be interacting if there are ≥10 interacting residue pairs between the domains. The threshold of 10 residues was chosen based on inspection of interaction sites and study of relevant literature. However, since an object-oriented design is adopted this behaviour can be over-ridden by users. For example, the distance based measure could be replaced by solvent accessible area.
Clustering of interactions by Family/Superfamily and interface
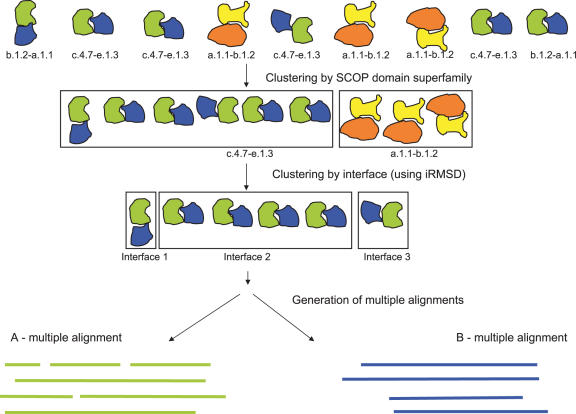
When analysing domain–domain interactions it is often necessary to classify them into pairwise Families/Superfamilies (e.g. in order to deal with redundancy in structural data). The term ‘pairwise Family’ is used to describe the classification of a domain–domain interaction based on the Family classification of each of the interacting domains. Similarly, the term ‘pairwise Superfamily’ is used to describe the classification of a domain–domain interaction based on the Superfamily classification of each of the interacting domains. The database contains domain–domain interactions classified to different levels of similarity (from Class through to Family) based on the SCOP and CATH hierarchical classification system and Family level for Pfam. The first step in Figure 1 shows an example of this process for the SCOP domain classification system to the Superfamily level of similarity.
Figure 1.
The classification of domain–domain interactions. The first step shows clustering by pairwise SCOP domain classification system at the Superfamily level of similarity. Clustering is also performed for the CATH and Pfam classification systems and at all levels of similarity. The second step shows an example of classification of domain–domain interactions by their interface. This classification is determined by the relative orientation of the interacting pair using an implementation of the iRMSD (interaction root-mean square deviation) method described by Aloy et al. (62). The final step shows the generation of a pair of multiple structure alignments, one from each partner of the interaction. These alignments are generated by STAMP (63).
SNAPPI-DB not only classifies interactions by their domain classification but also by the interface with which they are contacting. The second step in Figure 1 shows an example of classification of domain–domain interactions by their interface.
Method of clustering by different interaction interfaces
In order to classify interactions by their interaction interface, the relative orientation of the interacting pair was determined using an implementation of the iRMSD (interaction root–mean–square deviation) method described by Aloy et al. (62). This method determines if two interacting domains are at the same orientation as another pair of interacting domains and thus if they are interacting with the same interfaces. An iRMSD cut-off of <10 Å was applied to distinguish interactions between pairs that have a similar orientation and those that do not.
Structural alignments and positions of interacting residues
Once the interactions are classified by Superfamily/Family and by orientation, structural alignments are generated by STAMP (63). The structural alignments are used to generate a pair of alignments for each set of classified interactions as shown in the final step in Figure 1. SNAPPI-DB contains each of these alignments, the positions of the interacting residues at the surface as well as the transformation matrix required for superposition.
Generation of the data
The downloadable version of SNAPPI-DB contains the data generated after all of these steps have been performed. However, should the user have a local copy of the MSD Oracle relational database they may wish to generate SNAPPI-DB themselves. The system is designed so that this can be done in four easy steps each of which can be customised to allow flexibility.
THE APPLICATION PROGRAMMING INTERFACE
To efficiently deal with the complex and varied nature of the data contained in SNAPPI-DB an easy to use Java API has been developed. The API enables rapid development at a high level of abstraction without any requirement for complex SQL queries. In particular, specific design attention was paid in providing a natural model of protein–protein interaction data and the way bioinformaticians analyse such data. For example, as most analysis is done on the level of domains rather than chains, navigation via domains and domain–domain interactions is as seamless as via PDB entries, and dealing with redundancy in structural data is a core component of the API.
Java 5 is employed since it provides many features that are not available in previous versions of Java such as generics, enhanced ‘for’ loops and autoboxing/unboxing. The same naming convention as the MSD is employed so that users familiar with the MSD can rapidly learn the SNAPPI API. The same unique identifiers are also used so that structures can be mapped back to the MSD.
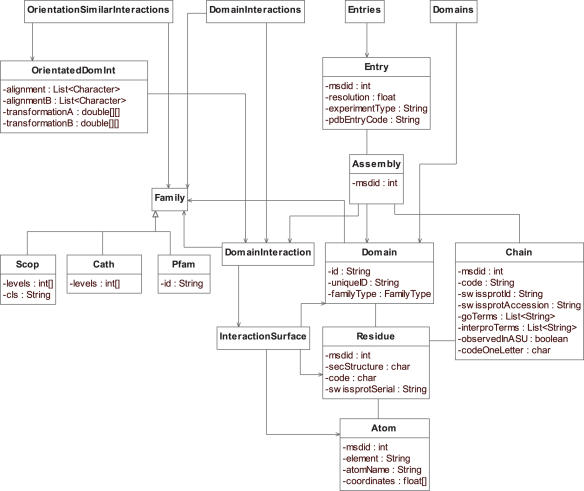
Figure 2 shows a simplified UML diagram of the structure of the API. As the database is object-oriented there is no difference between the relationships of the objects seen in the UML diagram of the API and the way that the objects are stored in the database. Although there are many different ways of navigating through the data in SNAPPI-DB, the database is optimised for searching either from (i) ‘Domains’ class: single domains classified by Family, (ii) ‘DomainInteractions’ class: domain–domain interactions classified by Family pair, (iii) ‘OrientationSimilarInteractions’ class: domain–domain interactions classified by orientation of interaction and (iv) ‘Entries’ class: collection of structures. These four key ways to access the data can be seen at the top of the UML diagram and are summarised below along with pseudo-code.
Figure 2.
Simplified UML diagram of the structure of the SNAPPI-DB API. There are four main points of entry to the data in SNAPPI-DB: (i) ‘Domains’ class: single domains classified by Family, (ii) ‘DomainInteractions’ class: domain–domain interactions classified by Family pair, (iii) ‘OrientationSimilarInteractions’ class: domain–domain interactions classified by orientation of interaction and (iv) ‘Entries’ class: collection of structures.
Navigation via Domains
The ‘Domains’ singleton (sole instance of a class) contains a list of domains classified by their domain Family. Domains can be easily accessed by their domain classification to any level of the domain hierarchy for SCOP and CATH and at the Family level for Pfam. For example, for the SCOP domain definition at the Family level of similarity there is a map which stores the name of the SCOP Family (e.g. a.1.1.1) as the key and a list of all of the domains with this classification as the value. In the pseudo-code in Figure 3, the (Scop.class, 4) is used to denote the Family level of similarity in the SCOP hierarchy. If this analysis was performed at the superfamily level (e.g. a.1.1) then this number would be 3.
Figure 3.
Psuedo-code for navigation via Domains.
One of the key aspects when working with structural data is the problem of redundancy within the data set. The API deals with this explicitly by having the option to select only one domain from a set of domains classified to any level of the domain hierarchy. For example, if the similarity of the domains was set to be the SCOP Family level of similarity then the API would return one domain to represent each Family. If a more stringent level of similarity was set, for example, at the Superfamily level of similarity, then the API would return one domain to represent each Superfamily. Which domain is selected to represent the group is determined by a (user overridable) strategy object passed in. Implementations could, for example, return the highest resolution domain or a randomly selected domain.
Navigation via DomainInteractions
The ‘DomainInteractions’ singleton contains a list of domain–domain interactions grouped by their pairwise domain classification. Each pair of interacting domains can be accessed by their pairwise domain classification to any level of the domain hierarchy for SCOP and CATH and at the Family level for Pfam. For example, for the SCOP domain definition at the Family level of similarity there is a map which stores the name of the pairwise SCOP Family (e.g. a.1.1.1–b.1.2.3) as the key and a list of all of the domain interactions with this classification as the value. In a similar way to Domains, a non-redundant set of domain–domain interactions can easily be generated. Example psuedo-code is shown in Figure 4.
Figure 4.
Psuedo-code for navigation via DomainInteractions.
Navigation via OrientatedDomInts
The ‘OrientationSimilarInteractions’ singleton contains a list of domain–domain interactions classified by their interface orientation. In a similar way to the DomainInteractions above, each domain–domain interaction is classified by their Family pair but in addition to this they are then further classified by the orientation of the interaction giving a collection of lists of domain–domain interactions for each pairwise Family. For example, there is a map with pairwise SCOP Family (e.g. a.1.1.1–b.1.4.7) as the key and a collection of lists of all of the domain–domain interactions with this pairwise Family classification and classified by orientation as the value. Rather than storing DomainInteraction objects in these lists OrientatedDomInt objects are stored. An OrientatedDomInt contains a DomainInteraction and additional information regarding the transform and alignment of the DomainInteraction. Example psuedo-code is shown in Figure 5.
Figure 5.
Psuedo-code for navigation via OrientatedDomInts.
Navigation via Entries
The ‘Entries’ singleton contains a list of PDB Entries. Navigation through each PDB Entry is straightforward as the data are stored in a hierarchal structure as shown in Figure 2. Each Entry contains one or more Assemblies (PQS predicted structures), each Assembly contains one or more Chains. Each Chain contains one or more Residues and each Residue contains one or more Atoms. Each level of the hierarchy also contains other information relevant to the item. For example, each Atom contains the co-ordinate positions of the Atom. The Assemblies also contain domains and domain interactions of SCOP, CATH and Pfam. Example psuedo-code is shown in Figure 6.
Figure 6.
Psuedo-code for navigation via Entries.
UTILITY AND DISCUSSION
Databases of structural domain–domain interactions
When the development of SNAPPI-DB began there were no extensive domain–domain interaction databases based on structural data; however, recently, there have been several databases made available. The fact that so many have been developed shows the timely aspect of investigation of domain–domain interactions using structural data. SNAPPI-DB and its associated API has several features which set it apart from other databases.
SNAPPI-DB contains different forms of derived data, including multiple domain definitions, PQS, GO terms, Interpro, and SWISSPROT and secondary structure information. The database uses the same unique identifiers as the full MSD data warehouse and so extra information required which is contained within the MSD but is not in SNAPPI-DB can easily be obtained. SNAPPI-DB classifies interactions based on the different interfaces with which they interact and provides information about which residues and atoms are in contact. A key advantage of SNAPPI-DB is that each set of classified domain–domain interactions has an associated multiple structural alignment for each partner. These alignments can be used for many tasks such as analysis of conservation patterns for domain–domain interactions or to train protein–protein interaction predictors.
The use of the JDO technology has several advantages (as discussed in depth in the ‘Construction’ Section). One important advantage is that the JDO interface is data store agnostic and so the database could be stored as either a relational database or an object-oriented database. Another key advantage is the high performance nature of SNAPPI-DB. Srdanovic et al. (56) found that the Fast Object Community Edition implementation of JDO had faster performance than the relational database implementation. The authors of the PSIBase system (37) developed a new algorithm (64) for determining interacting domains at the atomic level on the grounds that this task would take months using existing methods. In contrast, determining the interacting domains at the atomic level for 31 136 structures takes ∼3 h on a standard desktop machine (3 GHz PIV, 1 GB RAM) using SNAPPI-DB.
The main advantage of the system comes when the database is used in conjunction with the API. As far as we are aware no other databases come with an associated API and therefore they can not be used at the higher level of abstraction that is provided by this system.
Applicability
SNAPPI-DB has been employed in three different investigations already: an investigation into biological units and their effect upon the properties and prediction of protein–protein interactions (60), a comparison of comparison of SCOP and CATH with respect to domain–domain interactions and investigation into the orientation at which proteins interact. In addition SNAPPI-DB is currently being used to train both a functional residue predictor and a protein–protein interaction predictor. These methods demonstrate the wide applicability of this system in investigations and predictions of protein–protein interactions using structural data.
FUTURE DEVELOPMENTS
It is intended to expand SNAPPI-DB in several ways. At the moment it is not easy to update SNAPPI-DB to take into account new entries being added to the MSD. This problem will be resolved by providing update scripts and by offering multiple versions of SNAPPI-DB and the API for download. As JDO allows storage of the data in any form a relational database form of SNAPPI-DB will also be created so that users that prefer not to access the data via the API can do so. In addition, HMM profiles of the multiple structural alignments will be generated for matching to sequences of putative interacting proteins.
A web interface to SNAPPI-DB, SNAPPI-View, is under development and is currently available to perform simple searches of the database (www.compbio.dundee.co.uk/SNAPPI/search.jsp). SNAPPI-View will be extended to provide functions such as viewing the domain-domain interaction alignments, the structures of the interacting domains and protein interaction networks. As the functionality of the full SNAPPI-View interface is extensive, the web interface will be presented elsewhere.
CONCLUSIONS
In summary, a database of Structures, iNterfaces and Alignments of Protein–Protein Interactions (SNAPPI-DB) and corresponding API has been created. The main features of SNAPPI-DB are:
The API is specifically designed for analysis at the level of domains and domain–domain interactions in addition to PDB entries.
The core data are derived from a consistent and high quality data source, the MSD data warehouse.
The JDO technology provides abstraction from complex SQL queries and allows fast development time.
The object-oriented data store allows high performance and provides a more appropriate model for biological data than does a relational database schema.
SNAPPI-DB uses multiple domain definitions and PQS-generated biological structures.
SNAPPI-DB uses the same unique identifiers as the MSD to facilitate interoperability with the MSD warehouse.
SNAPPI-DB contains many forms of derived data such as SCOP, CATH, Pfam, InterPro, SWISSPROT, GO terms, PQS and secondary structure information.
The domain–domain interfaces are classified at every level of the CATH and SCOP hierarchies and and by interaction interface type.
Multiple structural alignments are provided for domain–domain interactions classified by interface orientation.
AVAILABILITY AND REQUIREMENTS
The SNAPPI package includes the Java 5 API, Ant tasks to generate the compiled code, XML files which contain the details of the objects to be stored, a properties file which stores the file locations and connection details specific to the user, a manual, source code and documentation and of course SNAPPI-DB. The documentation comes in the form of annotated JavaDocs and an in-depth manual. The source code contains many classes of example code.
The SNAPPI-DB package is available for download from www.compbio.dundee.ac.uk/SNAPPI/downloads.jsp. For any help or queries contact emily@compbio.dundee.ac.uk.
SNAPPI-DB and the API are available for both Linux and Windows operating systems and the database will work in parallel for read access. The system will be updated approximately every 6 months, while changes to the derived data such as SCOP and CATH releases will parallel the changes made to the MSD.
The system is distributed under the GPL licence. For alternative non-exclusive licensing options email geoff@compbio.dundee.ac.uk.
Acknowledgments
The authors thank the MSD group at EBI for discussions and information, Dr Jonathan Monk, Mr Eduardo Damato and Dr Jon Barber for network and systems support. Emily Jefferson is supported by a BBSRC (UK Biotechnology and Biological Sciences Research Council) studentship and Thomas Walsh is funded by TEMBLOR, European Community Contract No. QLRI-CT-2001-00015 and Scottish Funding Council, Scottish Bioinformatics Research Network (7030-355-064105-35OF). Funding to pay the Open Access publication charges for this article was provided by BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fields S., Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 3.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Sprinzak E., Margalit H. Correlated sequence-signatures as markers of protein-protein interaction. J. Mol. Biol. 2001;311:681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- 5.Marcotte E.M., Pellegrini M., Ng H.L., Rice D.W., Yeates T.O., Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 6.Ng S.K., Zhang Z., Tan S.H. Integrative approach for computationally inferring protein domain interactions. Bioinformatics. 2003;19:923–929. doi: 10.1093/bioinformatics/btg118. [DOI] [PubMed] [Google Scholar]
- 7.Gomez S.M., Noble W.S., Rzhetsky A. Learning to predict protein–protein interactions from protein sequences. Bioinformatics. 2003;19:1875–1881. doi: 10.1093/bioinformatics/btg352. [DOI] [PubMed] [Google Scholar]
- 8.Marcotte E.M., Xenarios I., Eisenberg D. Mining literature for protein–protein interactions. Bioinformatics. 2001;17:359–363. doi: 10.1093/bioinformatics/17.4.359. [DOI] [PubMed] [Google Scholar]
- 9.Bock J.R., Gough D.A. Predicting protein–protein interactions from primary structure. Bioinformatics. 2001;17:455–460. doi: 10.1093/bioinformatics/17.5.455. [DOI] [PubMed] [Google Scholar]
- 10.Kolesov G., Mewes H.W., Frishman D. Snapper: gene order predicts gene function. Bioinformatics. 2002;18:1017–1019. doi: 10.1093/bioinformatics/18.7.1017. [DOI] [PubMed] [Google Scholar]
- 11.Breitkreutz B.J., Stark C., Tyers M. The grid: the general repository for interaction datasets. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bader G.D., Hogue C.W. Bind—a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–477. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- 14.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 15.Xenarios I., Salwinski L., Duan X.J., Higney P., Kim S.M., Eisenberg D. DIP, the database of interacting proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xenarios I., Rice D.W., Salwinski L., Baron M.K., Marcotte E.M., Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xenarios I., Fernandez E., Salwinski L., Duan X.J., Thompson M.J., Marcotte E.M., Eisenberg D. DIP: the database of interacting proteins: 2001 update. Nucleic Acids Res. 2001;29:239–241. doi: 10.1093/nar/29.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peri S., Navarro J.D., Amanchy R., Kristiansen T.Z., Jonnalagadda C.K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T.K., Gronborg M., et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. String: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones S., Thornton J.M. Principles of protein-protein interactions. Proc. Natl Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teichmann S.A. Principles of protein–protein interactions. Bioinformatics. 2002;18(Suppl. 2):S249. doi: 10.1093/bioinformatics/18.suppl_2.s249. [DOI] [PubMed] [Google Scholar]
- 22.Janin J., Chothia C. The structure of protein–protein recognition sites. J. Biol. Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 23.Nooren I.M., Thornton J.M. Diversity of protein–protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones S., Marin A., Thornton J.M. Protein domain interfaces: characterization and comparison with oligomeric protein interfaces. Protein Eng. 2000;13:77–82. doi: 10.1093/protein/13.2.77. [DOI] [PubMed] [Google Scholar]
- 25.Lo Conte L., Chothia C., Janin J. The atomic structure of protein–protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 26.Jones S., Thornton J.M. Prediction of protein–protein interaction sites using patch analysis. J. Mol. Biol. 1997;272:133–143. doi: 10.1006/jmbi.1997.1233. [DOI] [PubMed] [Google Scholar]
- 27.Landgraf R., Xenarios I., Eisenberg D. Three-dimensional cluster analysis identifies interfaces and functional residue clusters in proteins. J. Mol. Biol. 2001;307:1487–1502. doi: 10.1006/jmbi.2001.4540. [DOI] [PubMed] [Google Scholar]
- 28.Koike A., Takagi T. Prediction of protein-protein interaction sites using support vector machines. Protein Eng. Des. Sel. 2004;17:165–173. doi: 10.1093/protein/gzh020. [DOI] [PubMed] [Google Scholar]
- 29.Aloy P., Russell R.B. Interrogating protein interaction networks through structural biology. Proc. Natl Acad. Sci. USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aloy P., Russell R.B. Interprets: protein interaction prediction through tertiary structure. Bioinformatics. 2003;19:161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 31.Lu L., Lu H., Skolnick J. Multiprospector: an algorithm for the prediction of protein-protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 32.Aytuna A.S., Gursoy A., Keskin O. Prediction of protein-protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics. 2005;21:2850–2855. doi: 10.1093/bioinformatics/bti443. [DOI] [PubMed] [Google Scholar]
- 33.Szilagyi A., Grimm V., Arakaki A., Skolnick J. Prediction of physical protein-protein interactions. Phys. Biol. 2005;2:S1–S16. doi: 10.1088/1478-3975/2/2/S01. [DOI] [PubMed] [Google Scholar]
- 34.Stein A., Russell R., Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis F.P., Sali A. Pibase: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 36.Winter C., Henschel A., Kim W.K., Schroeder M. SCOPPI: a structural classification of protein–protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong S., Yoon G., Jang I., Bolser D., Dafas P., Schroeder M., Choi H., Cho Y., Han K., Lee S., et al. Psibase: a database of protein structural interactome map (psimap) Bioinformatics. 2005;21:2541–2543. doi: 10.1093/bioinformatics/bti366. [DOI] [PubMed] [Google Scholar]
- 38.Ogmen U., Keskin O., Aytuna A.S., Nussinov R., Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westbrook J., Feng Z., Jain S., Bhat T.N., Thanki N., Ravichandran V., Gilliland G.L., Bluhm W., Weissig H., Greer D.S., et al. The protein data bank: unifying the archive. Nucleic Acids Res. 2002;30:245–248. doi: 10.1093/nar/30.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murzin A., Brenner S., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 42.Lo Conte L., Brenner S., Hubbard T., Chothia C., Murzin A. SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res. 2002;30:264–267. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreeva A., Howorth D., Brenner S., Hubbard T., Chothia C., Murzin A. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orengo C., Michie A., Jones S., Jones D., Swindells M., Thornton J. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 45.Pearl F., Lee D., Bray J., Sillitoe I., Todd A., Harrison A., Thornton J., Orengo C. Assigning genomic sequences to CATH. Nucleic Acids Res. 2000;28:277–282. doi: 10.1093/nar/28.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearl F., Todd A., Sillitoe I., Dibley M., Redfern O., Lewis T., Bennett C., Marsden R., Grant A., Lee D., et al. The CATH domain structure database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonnhammer E.L., Eddy S.R., Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 48.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henrick K., Thornton J.M. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 51.Golovin A., Oldfield T.J., Tate J.G., Velankar S., Barton G.J., Boutselakis H., Dimitropoulos D., Fillon J., Hussain A., Ionides J.M., et al. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2004;32:D211–D216. doi: 10.1093/nar/gkh078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velankar S., McNeil P., Mittard-Runte V., Suarez A., Barrell D., Apweiler R., Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. Interpro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boeckmann B., Bairoch A., Apweiler R., Blatter M.-C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srdanovic M., Schenk U., Schwieger M., Campagne F. Critical evaluation of the JDO API for the persistence and portability requirements of complex biological databases. BMC Bioinformatics. 2005;6:5. doi: 10.1186/1471-2105-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ambler S. Agile Database Techniques. John Wiley & Sons; 2003. [Google Scholar]
- 58.Carugo O., Argos P. Protein–protein crystal-packing contacts. Protein Sci. 1997;6:2261–2263. doi: 10.1002/pro.5560061021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ponstingl H., Henrick K., Thornton J.M. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 60.Jefferson E.R., Walsh T.P., Barton G.J. Biological units and their effect upon the properties and prediction of protein–protein interactions. J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2006.09.042. http://dx.doi.org/10.1016/j.jmb.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 61.Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105:1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 62.Aloy P., Ceulemans H., Stark A., Russell R. The relationship between sequence and domain-domain interaction divergence in proteins. J. Mol. Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Russell R.B., Barton G.J. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 64.Dafas P., Bolser D., Gomoluch J., Park J., Schroeder M. Using convex hulls to extract interaction interfaces from known structures. Bioinformatics. 2004;20:1486–1490. doi: 10.1093/bioinformatics/bth106. [DOI] [PubMed] [Google Scholar]