Abstract
REBASE is a comprehensive database of information about restriction enzymes, DNA methyltransferases and related proteins involved in the biological process of restriction-modification. It contains fully referenced information about recognition and cleavage sites, isoschizomers, neoschizomers, commercial availability, methylation sensitivity, crystal and sequence data. Experimentally characterized homing endonucleases are also included. All newly sequenced genomes are analyzed for the presence of putative restriction systems and these data are included within the REBASE. The contents or REBASE may be browsed from the web (http://rebase.neb.com/rebase/rebase.ftp.html) and selected compilations can be downloaded by ftp (ftp.neb.com). Additionally, monthly updates can be requested via email.
INTRODUCTION
The last description of REBASE in the 2005 NAR Database Issue (1) contained more than 3600 biochemically characterized restriction-modification (RM) systems and included an analysis of ∼200 bacterial and archaeal genomes that had been deposited in the RefSeq Database of GenBank (2). In the interim, the number of sequenced bacterial and archaeal genomes has risen to more than 400 and the total number of RM systems in REBASE is now almost 3800 well-characterized systems and more than 4000 systems, whose existence is predicted on the basis of a bioinformatic analysis of DNA sequences in GenBank. These putative RM genes are named systematically according to recently published nomenclature rules (3) and all have the suffix ‘P’ to indicate their putative status. The REBASE website (http://rebase.neb.com) summarizes all information known about every restriction enzyme and any associated proteins. This includes the source, commercial availability, sequence data, crystal structure information, cleavage sites, recognition sequences, isoschizomers and methylation sensitivity. Within the reference section of REBASE, links are maintained to the full text of all papers whenever that is readily available on the web. Also, there is extensive reciprocal cross-referencing between REBASE and NCBI, including links to GenBank and PubMed and NCBI's LinkOut utility. Links to other major databases such as SWISS-PROT (4), PDB (5) and Pfam (6) are also maintained. There are currently 3805 biochemically or genetically characterized restriction enzymes in REBASE and of the 3698 Type II restriction enzymes, 611 are commercially available, including 262 distinct specificities. As can be seen from Figure 1, the putative restriction and modification enzymes now exceed those for which detailed biochemical characterization is available. This is expected to continue into the future. However, it should be noted that because of the large number of sequenced examples of biochemically characterized restriction systems, the putative recognition sequences and in some cases the cleavage sites of predicted restriction enzymes can be inferred. These inferences are all included within REBASE where they are clearly marked as predictions. Among the 2709 restriction enzyme genes and 4485 DNA methyltransferase genes that can be identified in GenBank, the sequenced microbial genomes now account for 3180 of these genes!
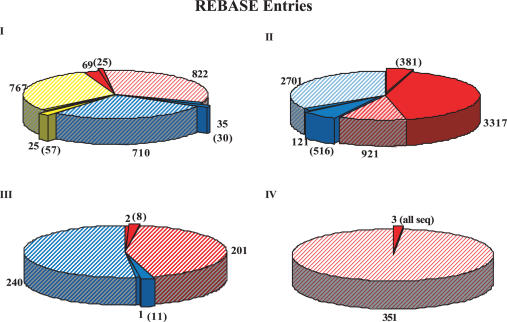
Figure 1.
The distribution in REBASE of the components of the four Types of restriction systems is shown. Red, restriction enzyme genes; blue, methyltransferase genes; and yellow, specificity subunits. Full colors indicate genes whose products have been biochemically characterized, whereas shaded areas represent inferred function based on bioinformatic analysis of DNA sequences. The pop-out slices (with numbers in parentheses) indicate those genes where sequence is available for biochemically characterized enzymes. The adjacent numbers represent those for which only biochemical evidence is available.
The identification of putative RM genes in DNA sequences is achieved by analyzing each sequence for overall sequence similarity to REBASE gene sequences. For DNA methyltransferases, which are the primary indicator of an RM system, the presence, proper order and characteristic spacing of well-conserved motifs is used to suggest likely candidates. The more widely divergent genes that encode the restriction enzymes always reside close to the genes for their cognate methyltransferases, but usually they cannot be recognized directly because they lack any sequence similarity to any other genes in GenBank.
Given the wealth of experimental data, both published and unpublished, REBASE can be a valuable resource during the annotation of bacterial and archaeal genomes. When newly sequenced genes show strong matches to the genes of the Type II restriction systems, their specificity can often be predicted with high accuracy. In particular, because the Type II restriction enzyme genes belong to the category of rapidly evolving genes, strong matches are a particularly good indicator of identical recognition specificity. With the plethora of restriction systems that occur in all sequenced microbial genomes, annotators are encouraged to use the resources of the REBASE database or to contact the REBASE staff if help is needed.
From the REBASE website users can use the programs available from the REBASE Tools icon to analyze DNA sequences for the presence of restriction enzyme recognition sites, show all possible restriction patterns of a given DNA based on the known enzymes, predict the recognition sequences of an unknown restriction enzyme and ‘Blast’ a new sequence against all the sequences in REBASE. Additionally, users can keep current with the most recently discovered enzymes and newest references (NEWS icon), browse a variety of useful compilations, including sequence collections and crystal data (Lists icon), update the enzyme files being used by software packages such as GCG, DNA Strider, EMBOSS (Files icon) and submit new findings for inclusion in REBASE (Submit icon).
The Genomes icon leads to a compilation of data for 385 bacterial and 29 archaeal genomes, illustrated by schematic representations of the whole genomes and the individual RM systems within them, providing a quick overview of the RM system content of sequenced genomes.
Choose the Methylation sensitivity icon for all enzymes with methylation sensitivity data, isoschizomers showing differential sensitivity, effects of overlapping methylation and other useful information involving this complex area of study.
We offer both simple (from the home page) and advanced searching (Search icon), where one can search multiple fields, choose result columns and sort order.
Acknowledgments
Special thanks are due to the many individuals who have so kindly contributed their unpublished results for inclusion in this compilation and to the REBASE users who continue to guide our efforts with their helpful comments. We are especially grateful to Karen Otto for secretarial help. This database is supported by the National Library of Medicine (LM04971) and New England Biolabs, Inc. Funding to pay the Open Access publication charges for this article was provided by New England Biolabs, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE—restriction enzymes and methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson D.A., Karsch-Mizrachi I., Lipmann D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Vitinaite J., Blumenthal R.M., Degtyarev S.K.H., Dryden D.T.F., Duibvig K., et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeckmann B., Bairoch A., Apweiler R., Blatter M.-C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finn R.D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich S.V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]