Abstract
There has been widespread and growing interest in genetic markers suitable for drawing population genetic inferences about past demographic events and to detect the effects of selection. In addition to single nucleotide polymorphisms (SNPs), microsatellites (or short tandem repeats, STRs) have received great attention in the analysis of human population history. In the SNPSTR database (http://www.imperial.ac.uk/theoreticalgenomics/data-software) we catalogue a relatively new type of compound genetic marker called SNPSTR which combines a microsatellite marker (STR) with one or more tightly linked SNPs. Here, the SNP(s) and the microsatellite are less than 250 bp apart so each SNPSTR can be considered a small haplotype with no recombination occurring between the two individual markers. Thus, SNPSTRs have the potential to become a very useful tool in the field of population genetics. The SNPSTR database contains all inferable human SNPSTRs as well as those in mouse, rat, dog and chicken, i.e. all model organisms for which extensive SNP datasets are available.
INTRODUCTION
The pattern of genetic diversity is dependent on past demographic history (e.g. fluctuations in population size, substructure and migration) as well as gene-specific factors such as mutation rate and selection. This pattern is the result of complicated evolutionary processes and the understanding of these processes can be helpful in the fields of medical genomics, pharmacogenomics, functional genomics and human evolutionary biology (1).
Genetic diversity information is obtained using various molecular markers (polymorphic DNA sequences derived from a single locus). Many different kinds of molecular markers have been used over the years but the two used mainly at the moment in the inference and estimation of population parameters are single nucleotide polymorphisms (or SNPs) and microsatellites (or short tandem repeats—STRs).
SNPs are sequence sites where more than one nucleotide is present in the population (typically some frequent cut-off, such as 1%, is applied, above which a polymorphism is considered a SNP). They are very useful in studying human history and SNP data are abundant thanks to the different SNP projects that have been carried out (2–6). In humans the average nucleotide mutation rate is assumed to be ∼2.5 × 10−8; because of this, SNPs are best used in studying human evolutionary history over longer time scales (1). SNPs may also be considered as carrying very little information because of the small number of possible alleles that can occur at each SNP locus.
Microsatellites are composed of variable numbers of repeats of 2–7 bp (e.g. CA). Microsatellite mutation rate has been estimated to be 10−2−10−5 per generation (1) so they can be used to trace relatively recent demographic events. Soon after microsatellites were discovered (7–10), the necessary theory was developed to relate their patterns of variation to population histories. Our understanding of the underlying microsatellite mutation model, however, is still not very clear, mainly because it is not known how realistic some of the assumptions of these models—such as the simple stepwise mutation model (SSM) (11)—are. Nevertheless some remarkable results have been obtained by a combined analysis of microsatellites and linked SNP data, frequently involving the non-recombining part of the Y-chromosome.
A SNPSTR is a relatively new type of compound genetic marker which combines a STR marker with one or more tightly linked SNPs. This combination of co-inherited markers evolving at different rates may offer the possibility of gaining better resolved insights into population genetic processes compared to when these different marker types are used separately. SNPSTRs were first described by Mountain et al. (12), who developed experimental protocols for autosomal SNPSTRs which contain a SNP and a microsatellite 500 bp apart.
Here, the SNP(s) and the microsatellite are less than 250 bp apart so have the advantage that (i) they are not broken up by recombination, (ii) can be typed straightforwardly in a single PCR reaction, and (iii) they contain slowly evolving binary markers (the SNP) as well as the quickly evolving microsatellites. In principle at least, it should therefore be possible to infer the age of the SNP allele (or the most recent common ancestor of all individuals carrying that allele) from the microsatellite data (using a generic model of the microsatellite mutation process). Each SNPSTR acts as a ‘mini Y-chromosome’ and combining many unlinked SNPSTRs will give us a rich data-source to infer past demographic events (or test for deviations from a neutral model).
In the SNPSTR database we catalogue all inferable SNPSTRs for the five model species, where sufficient SNP information exists in both of NCBI and Ensembl databases. These species are human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), dog (Canis familiaris) and chicken (Gallus gallus) (Table 1). We will first describe the pipeline by which these SNPSTRs were obtained, then we will give a brief description of the current contents of the database, and finally we will explain the different features of the web interface constructed to access the database.
DATABASE CONSTRUCTION
To identify SNPSTRs we started with SNPSTR sequence identification and then used the genomic positions of SNPs to identify nearby genes and disease regions, as well as, to obtain additional genetic variation information. The means chosen to extract the sequences was the Ensembl Perl Application Programming Interface (API). A 1001 bp long sequence was retrieved for each SNP that contains the SNP exactly in the middle. These sequences were scanned for microsatellites with Tandem Repeats Finder (TRF) (13) which locates and displays tandem repeats in DNA sequences.
Variation information was obtained for human SNPSTRs in the form of allele counts using the HapMart tool of the HapMap project database website (http://hapmart.hapmap.org). The aim was to use this information not only to find the polymorphism levels of the SNPs in the different populations in terms of heterozygosity, but also to calculate FST values to identify those SNPs that show population-specific polymorphism patterns.
The second source of extra information obtained was the positions of coding genes. These were used to identify which SNPSTRs were in genes (exons and introns) or in intergenic sequences. If genes are more affected by natural selection you would expect those SNPSTRs in or near genes to show (on average) different diversity patterns than SNPSTRs which are not linked to a gene or disease-associated region. Gene and exon coordinates were obtained again using the Ensembl API.
Finally, disease information was obtained to identify those SNPSTRs that were found in disease areas. Mendelian Inheritance in Man (MIM) disease gene coordinates were obtained using the Ensembl API.
DATABASE CONTENTS
Release 1.0 (July 2006) of SNPSTR database contains 1 735 049 SNPSTRs from the five model species. Of these SNPSTRs, 570 057 are in gene regions, most of them intronic (555 013), and only a few exonic (15 044). Finally, 47 837 of human SNPSTRs occur in areas where there are genes related to disease. A more detailed description of the database can be found in Table 1.
Table 1.
Detailed contents of the SNPSTR database (Release 1.0, July 2006)
| Species | Snpstr | Genic | Exonic | Intronic |
|---|---|---|---|---|
| Human | 611 901 | 200 541 | 5167 | 195 374 |
| Mouse | 832 166 | 284 336 | 8304 | 276 032 |
| Rat | 1607 | 952 | 535 | 417 |
| Dog | 257 182 | 74 550 | 681 | 73 869 |
| Chicken | 32 193 | 9678 | 357 | 9321 |
| Total | 1 735 049 | 570 057 | 15 044 | 555 013 |
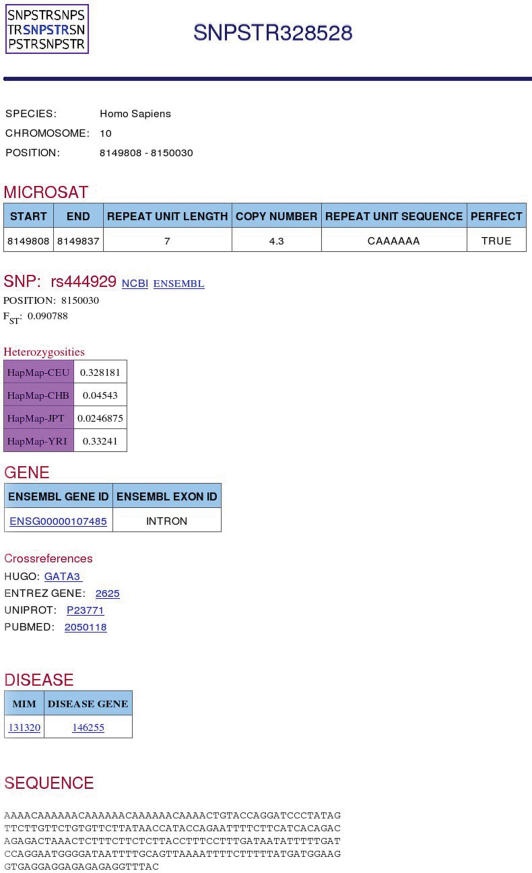
For each SNPSTR the following information is available: SNPSTR database id, species and chromosome where it is found, genomic start and end coordinates (as in Ensembl Built 39), microsatellite information (start and end coordinates, repeat unit length, repeat sequence and copy number, information on whether the microsatellite consists only of perfect repeats or if it contains some non-perfect repeats), SNP information (SNP genomic location and for humans only counts for the four populations, HS and FST values), information on gene when SNPSTR is in gene area (accession numbers from Ensembl, Uniprot, Entrez Gene and HUGO databases as well as Pubmed ID), accession number of nearest OMIM disease where applicable and finally the sequence of the SNPSTR.
The database will be updated when the Ensembl database is updated i.e. approximately every 4 months. This is because the genomic coordinates of the SNPSTRs and the information available for the areas around the SNPSTRs are both based on information from the Ensembl database.
WEB INTERFACE
The SNPSTR database can be accessed through a simple and easy to use CGI/Perl-based web interface at http://www.imperial.ac.uk/theoreticalgenomics/data-software (Figure 1). On-line documentation is provided for each web service. The user can search by accession number, by chromosomal region or by microsatellite repeat sequence. The results can be seen as an html page or can be downloaded as comma-separated or tab-limited files. The user can also download the lists of SNPSTRs classified by chromosome or by microsatellite repeat unit length for each species from the FTP page.
Figure 1.
The front page of the SNPSTR database interface.
Searching by accession number
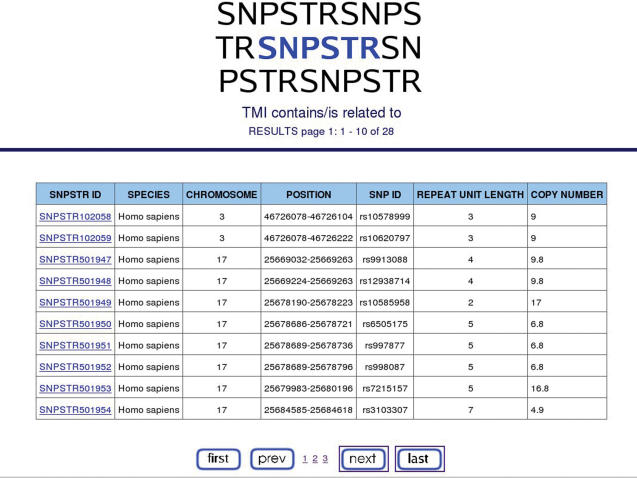
If one knows the SNPSTR id for the SNPSTR of interest, the database can be searched by this id and the entry for this SNPSTR is shown as an html page (Figure 2). However, it is much more likely that the user will want to find if their gene or protein contains any SNPSTRs or if their SNP of interest is part of a SNPSTR. For this reason the database can be searched by SNP ‘rs’ identifier (as according to NCBI and Ensembl databases), gene or protein ID (Ensembl gene id, HUGO gene name or HGNC gene id, EntrezGene gene id, Uniprot protein id), Pubmed ID, MIM gene id or MIM disease ID. In this case, a table with all SNPSTRs is produced with some basic information on each SNPSTR (Figure 3). The user can then click on any SNPSTR number to be taken to the SNPSTR entry html page (Figure 2).
Figure 2.
An example SNPSTR entry.
Figure 3.
Genes may contain more than one SNPSTR.
Searching by region
An alternative way of searching the database is to search by chromosomal region. By choosing a species from the dropdown menu and submitting the chromosome number, start and end base pairs of the region the user wants to search, a table with all the SNPSTRs in the area is obtained similar to the one seen in Figure 3. As above, the user can then click on any SNPSTR number to be taken to the SNPSTR entry html page (Figure 2).
Searching by repeat unit sequence
Finally, one can search by repeat unit sequence by inputting the sequence in the text box and choosing the species of interest. Since the output of this kind of query is likely to be a massive list of SNPSTRs, the user is advised to download the data. If however one chooses to view the data as an html, a table with the basic SNPSTR information is produced. As above, the user can then click on any SNPSTR number to be taken to the SNPSTR entry html page (Figure 2).
Using the ftp site
The user can just download all SNPSTRs as a tab-limited or comma separated file from the ftp site (http://www.imperial.ac.uk/theoreticalgenomics/data-software). SNPSTRs are classified according to chromosome or microsatellite repeat unit length for each species. All files are of the same format (more information on the format can be found in the website).
CONCLUSIONS AND FUTURE WORK
The SNPSTR database is a database of a new type of marker, the compound genetic marker called SNPSTR. All SNPSTRs from five model species (human, mouse, rat, dog and chicken) were extracted using an automated pipeline. It was of particular importance that the database in extensively cross-referenced so each SNPSTR is linked to one or more identifiers from NCBI, Ensembl, HUGO, Uniprot, Pubmed, Entrez Gene and OMIM databases when available. These species were chosen because extensive SNP datasets have been produced by SNP consortia. With the availability of such datasets from other species and the extension of the current datasets (rat and chicken SNP datasets are very limited compared to the human, mouse and dog datasets) the database will expand.
Acknowledgments
We thank Dr Suhail Islam for assistance with setting up the SNPSTR database. I.A. and M.P.H.S. gratefully acknowledge financial support from the Wellcome Trust; M.P.H.S. furthermore acknowledges support through an EMBO Young Investigator Fellowship. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tishkoff S.A., Verrelli B.C. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu. Rev. Genomics. Hum. Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.International Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Rat Genome Sequencing Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 5.International Dog Genome Sequencing Consortium. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 6.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 7.Weber J.L., May P.E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am. J. Hum. Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- 8.Litt M., Luty J.A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 1989;44:397–401. [PMC free article] [PubMed] [Google Scholar]
- 9.Di Rienzo A., Peterson A.C., Garza J.C., Valdes A.M., Slatkin M., Freimer N.B. Mutational processes of simple-sequence repeat loci in human populations. Proc. Natl Acad. Sci. USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowcock A.M., Ruiz-Linares A., Tomfohrde J., Minch E., Kidd J.R., Cavalli-Sforza L.L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 11.Kimura M., Weiss G.H. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mountain J.L., Knight A., Jobin M., Gignoux C., Miller A., Lin A.A., Underhill P.A. SNPSTRs: empirically derived, rapidly typed, autosomal haplotypes for inference of population history and mutational processes. Genome Res. 2002;12:1766–1772. doi: 10.1101/gr.238602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]