Abstract
The TBestDB database contains ∼370 000 clustered expressed sequence tag (EST) sequences from 49 organisms, covering a taxonomically broad range of poorly studied, mainly unicellular eukaryotes, and includes experimental information, consensus sequences, gene annotations and metabolic pathway predictions. Most of these ESTs have been generated by the Protist EST Program, a collaboration among six Canadian research groups. EST sequences are read from trace files up to a minimum quality cut-off, vector and linker sequence is masked, and the ESTs are clustered using phrap. The resulting consensus sequences are automatically annotated by using the AutoFACT program. The datasets are automatically checked for clustering errors due to chimerism and potential cross-contamination between organisms, and suspect data are flagged in or removed from the database. Access to data deposited in TBestDB by individual users can be restricted to those users for a limited period. With this first report on TBestDB, we open the database to the research community for free processing, annotation, interspecies comparisons and GenBank submission of EST data generated in individual laboratories. For instructions on submission to TBestDB, contact tbestdb@bch.umontreal.ca. The database can be queried at http://tbestdb.bcm.umontreal.ca/.
INTRODUCTION
Much of the evolutionary diversity and biochemical versatility of the domain Eukarya is contained outside the kingdoms of animals, plants and fungi, in a highly diverse assemblage of poorly studied, mostly unicellular eukaryotes commonly referred to as protists (1–3), many of which are biologically relevant in the fields of human health and agriculture. As the early eukaryotic world must have been exclusively unicellular, protists are the key to understanding the origin and evolution of multicellular eukaryotes. As we know today, close unicellular relatives of the multicellular animals, fungi and land plants are, respectively, choanoflagellates plus Ichthyosporea (4,5), nucleariids [(6–9); E.Steenkamp, S.Baldauf and B.F.Lang, unpublished data], and charophyte algae (10,11). Unfortunately, very few protist genome projects are underway and protist nuclear genomics data are often limited to one or a few standard genes. An effective way of alleviating this shortcoming is to generate expressed sequence tags (ESTs) from cDNA libraries. This technique is fast and cost-effective, and provides a robust approximation of the expressed genetic component of a given organism.
The Protist EST Program (PEP) was a large-scale genomics collaboration among six Canadian research groups with the objective of characterizing the expressed portion of the nuclear genome of a large number of different protist species. Most other protist EST and genome projects and their associated databases focus on pathogenic organisms, e.g. ApiEST-DB [protozoans in the phylum Apicomplexa] (12), CryptoDB [Cryptosporidium] (13), Full-Malaria [Plasmodium species] (14), PlasmoDB [Plasmodium falciparum] (15), TcruziDB [Trypanosoma cruzi] (16), ToxoDB [Toxoplasma gondii] (17) and the protist data contained in GeneDB [17 protist data collections, mostly Trypanosoma and Plasmodium species] (18). The few exceptions such as the Diatom EST Database [Phaeodactylum tricornutum and Thalassiosira pseudonana] (19), dictyBase [Dictyostelium discoideum] (20) and the Porphyra yezoensis EST index (21) tend to have a very specialized focus. PEP, in contrast, aimed to survey a taxonomically broad collection of protists and other poorly studied eukaryotic groups (Table 1). During the PEP project, a total of ∼550 000 ESTs were generated, of which ∼450 000 passed quality cut-offs and 370 000 of these sequences, from 49 organisms, have been made publicly available in the TBestDB database as of July 1, 2006. Approximately 80 000 ESTs from 19 other datasets, including PEP-related and externally generated data, are still under analysis and will be released into the public domain over the next few months. Researchers are invited to submit their data to TBestDB for free processing and annotation, with private access to the results provided for a limited time.
Table 1.
Publicly available sequence content of TBestDB (July 1, 2006)
| Organism name | No. of ESTs | No. of clusters |
|---|---|---|
| Acanthamoeba castellanii | 13 814 | 5262 |
| Acetabularia acetabulum | 3464 | 2573 |
| Allomyces macrogynus | 5073 | 2149 |
| Amoebidium parasiticum | 3623 | 1557 |
| Antonospora (Nosema) locustae | 2376 | 700 |
| Astasia longa | 2730 | 1718 |
| Bigelowiella natans | 3462 | 2318 |
| Blastocystis hominis | 12 759 | 3330 |
| Capsaspora owczarzaki | 8863 | 2516 |
| Chlamydomonas incerta | 5124 | 1388 |
| Cyanophora paradoxa[Durnford group] | 9867 | 2448 |
| Cyanophora paradoxa[Loeffelhardt group] | 4673 | 1478 |
| Diplonema papillatum | 4791 | 3664 |
| Euglena gracilis[Durnford group] | 17 236 | 8651 |
| Glaucocystis nostochinearum | 8745 | 2831 |
| Hartmannella vermiformis | 9505 | 4986 |
| Helicosporidium sp. | 1188 | 701 |
| Heterocapsa triquetra | 6804 | 2038 |
| Histiona aroides | 4009 | 1763 |
| Hyperamoeba dachnya | 2756 | 1762 |
| Isochrysis galbana CCMP 1323 | 12 205 | 6095 |
| Jakoba bahamensis | 4323 | 2286 |
| Jakoba libera | 5452 | 2565 |
| Karlodinium micrum | 16 544 | 11 903 |
| Malawimonas californiana | 4437 | 2314 |
| Malawimonas jakobiformis | 9798 | 4505 |
| Mastigamoeba balamuthi | 19 182 | 4438 |
| Mesostigma viride | 5615 | 1771 |
| Micromonas sp. | 3662 | 2004 |
| Monosiga ovata | 6433 | 2677 |
| Nephroselmis olivacea | 126 | 115 |
| Oxytricha trifallax | 2272 | 1230 |
| Pavlova lutheri | 7590 | 3383 |
| Physarum polycephalum | 9684 | 3078 |
| Polysphondylium pallidum | 4445 | 1247 |
| Polytomella parva | 5062 | 2151 |
| Prototheca wickerhamii | 5641 | 1542 |
| Reclinomonas americana | 17 644 | 6797 |
| Rhizopus oryzae | 12 570 | 5105 |
| Saitoella complicate | 3840 | 1008 |
| Sawyeria marinlandensis | 9300 | 3520 |
| Scenedesmus obliquus | 6615 | 2666 |
| Seculamonas ecuadoriensis | 5256 | 2217 |
| Sphaeroforma arctica | 8006 | 2763 |
| Spizellomyces punctatus | 5365 | 2079 |
| Streblomastix strix | 4475 | 2595 |
| Taphrina deformans | 3919 | 1435 |
| Tetrahymena thermophila | 31 548 | 9050 |
| Trimastix pyriformis | 9615 | 2686 |
| Total | 371 484 | 149 058 |
DATA CONTENT
Information in TBestDB that is publicly accessible at the time of writing is compiled in Table 1. Data include individual EST sequences, consensus sequences and clustering information, conceptual translations, functional annotations drawn from three different sources, as well as metabolic pathway predictions. In addition, the database contains experimental information on cDNA libraries and information on data quality and project status.
EST PROCESSING PIPELINE
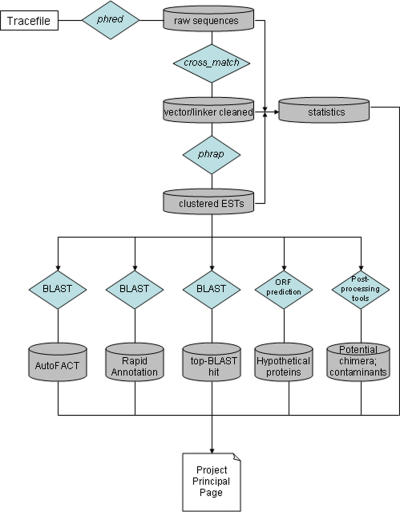
The EST processing pipeline includes three primary steps (Figure 1), starting from the download of sequence submitted by the PEP member laboratories. Annotation is then followed by post-processing steps to detect potential contamination and chimerism.
Figure 1.
EST processing pipeline. EST tracefiles are accepted in .scf or .abi format via a dedicated sftp server. Any EST for which phred cannot read more than 60 nt of high-quality sequence is discarded. The default value for quality is 99% certainty of identification of each residue (ABI sequence technology), but this value has been set to slightly lower thresholds in certain instances where justified by the effective quality. The parameters used for cross_match have been adjusted slightly from the defaults—the minscore value has been changed from 20 to 17, to allow for slightly more relaxed matches, as this was found to give the best identification and masking of short linker sequences. At this point any EST sequence containing fewer than 60 unmasked residues is removed from further consideration. AutoFACT combines the most informative of the top 10 BLAST hits from the European Ribosomal Database (BLASTN), UniRef90 (BLASTX), KEGG (BLASTX), COG (BLASTX), Pfam (RPS-BLAST), and NCBI's nr (BLASTX) and est_others (TBLASTX) databases. Default parameters bitscore >40 and E-value <1 × 10−4 were used. Rapid Annotation is performed using BLASTX against a specialized set of sequences (see Annotation in text) with an E-value cut-off of 1× 10−4. Top-BLAST-hit annotations are from TBLASTX hits to NCBI's nr database using an E-value cut-off of 1 × 10−4. ORF prediction is performed by translating the consensus sequence in all frames, identifying stop codons and marking any potential ORF longer than 20 residues.
Sequence clustering
EST data are accepted as tracefiles in .scf or .abi format. Incoming tracefiles are processed using the phred/phrap package (22), which reads each tracefile, converts it into a sequence file with associated quality assessments for each residue, removes both vector and linker sequences and finally assembles the ESTs into clusters to generate consensus sequences. It should be noted that there is an observed difficulty with phrap in clustering datasets beyond a certain number of readings (starting between 5000 and 10 000 in our experience, depending on the individual dataset), manifesting as a failure to generate some small number, usually <5%, of expected clusters. We have addressed this difficulty by recursively running phrap on the set of unclustered sequences until no new clustering is found.
Statistical breakdown
Once clustering is completed, various statistics are calculated to facilitate the management of ongoing EST projects. Sequence quality is assessed by monitoring maximal and average reading length after quality clipping, and clone insert sizes, before and after vector clipping, are evaluated globally and by library. The overall progress of a project can be assessed on the basis of the distribution and growth of cluster size, and the evolution of redundancy of individual or multiple libraries for a given organism can be monitored, allowing rapid decisions to be made about the most productive directions for further sequencing.
Annotation
TBestDB conducts three kinds of annotation procedures for consensus sequences derived from clustered ESTs. (i) AutoFACT (23) provides the most sophisticated annotations. Using local BLAST comparisons (24), AutoFACT gathers classification information following a hierarchical system, from a collection of seven specialized databases (Table 2). As not all descriptions from top BLAST hits contain biologically meaningful information, AutoFACT adopts an ‘uninformative rule’ to identify the highest scoring BLAST hit that provides a meaningful annotation, generating ∼50% more functionally informative annotations than a top-BLAST-hit approach. Annotations provided by AutoFACT are of high quality, but the process of generating them is time-consuming due to the need for multiple BLAST searches. (ii) The Rapid Annotation procedure was designed to allow quick initial surveys of incoming data. Here, annotations are assigned by searching for sequence similarity to deduced nucleus-encoded proteomes from selected organisms (Arabidopsis thaliana, Ustilago maydis, Neurospora crassa, Homo sapiens, Rickettsia prowazeki and Magnetospirillum magnetotacticum) and deduced mitochondrion-encoded proteins of Reclinomonas americana—all of which have been comprehensively reannotated using AutoFACT—and with collections of representative large and small subunit ribosomal RNAs. Using this procedure, information about ubiquitous proteins and contamination of cDNA libraries with mitochondrial or rRNA sequences is made available to TBestDB users as each new EST dataset is processed. With this system a set of 5000 clusters can be annotated in ∼2 h, which allows for newly submitted data, typically containing 500–1000 EST sequences, to be clustered with existing data from the same organism and the entire dataset to be reannotated within one working day. (iii) Finally, to detect similarities with as-yet-unrecognized hypothetical proteins in published DNA sequences, TBLASTX is run against a local copy of NCBI's non-redundant database and the top hit is shown. The time requirement for this step is quite high, ∼10 min per sequence on our 16-CPU cluster.
Table 2.
Databases searched and classification information assigned by AutoFACT
| Database | Classification Information | Reference |
|---|---|---|
| European Ribosomal Database | Large subunit (LSU) ribosomal RNAs | (34) |
| Small subunit (SSU) ribosomal RNAs | ||
| Gene Ontology terms | (35,36) | |
| UniProt's UniRef 90 | Enzyme Commission numbers | |
| Locus names | ||
| Clusters of Orthologous Groups (COG) | Functional categories | (37,38) |
| Metabolic pathways | (39) | |
| Kyoto Encyclopedia of Genes and Genomes (KEGG) | Enzyme Commission numbers | |
| Locus names | ||
| Protein Families Database (Pfam) | Protein domains | (40) |
| NCBI's non-redundant database (nr) | N/A | (40) |
| NCBI's est_others database |
In addition to the above-mentioned automatic annotations, expert manual annotations are available in some cases, typically provided by the submitter of the sequences. Should all the analyses fail to identify the function of a consensus sequence, it is annotated as of ‘unknown function’. The above annotation procedures are rerun regularly, and in consequence automatically assigned names may change as the reference databases are updated. For this reason any reference to data in TBestDB should use TBestDB's internal cluster IDs in addition to the annotations provided.
Metabolic pathway prediction
AutoFACT annotations are used to build a Pathway Genome Database (25) for each individual organism. On this basis, annotated sequences can be mapped to metabolic pathways available in MetaCyc (26). This allows users to determine which components of a given pathway are present in, or still missing from, the sequenced part of an EST library and, ultimately, to assess the biological versatility of the organisms studied.
POST-PROCESSING
Contamination management
In large sequencing projects, some level of contamination between datasets or from external sources is unavoidable in practice. Sources of contamination include food organisms (bacteria on which many of the organisms documented in TBestDB are grown), symbionts, and human error during culturing, cloning and sequencing. In TBestDB we have implemented an automated system for the identification of potential cross-project contamination, in order to mitigate this problem as far as possible.
Each consensus sequence in TBestDB (query cluster) is searched against the consensus sequences for every other organism in the database (retrieved clusters) using BLASTN. Potential contaminants are identified at a threshold of ≥97% sequence identity over at least 50 nt. rRNA sequences and well-known highly conserved proteins such as actin and ubiquitin, which are also retrieved by these criteria, are explicitly excluded from consideration as contaminants. We automatically remove from the database any query cluster that is found to match a retrieved cluster containing at least three times as many ESTs, as this criterion has proven a reliable identifier of contaminating data. Less clear-cut cases of potential contaminants are flagged, and the source laboratory is asked to examine the flagged sequences to determine whether they should remain in TBestDB.
All of the ESTs belonging to contaminating clusters are moved into a separate database table, where they are used in further rounds of contamination checking. This procedure is necessary so that the curation of different organisms at different times can identify possible common sources of contamination, such as errors introduced by commercial library services shared by several users.
Identification of chimerism
Submitted datasets occasionally include chimeric ESTs (i.e. ESTs containing sequence from two distinct cDNAs), which causes problems during clustering. The identification of such ESTs is not straightforward, but we have implemented automatic tests that identify the bulk of such artifactual sequences.
The simplest test is a search for misplaced poly(A) tracts in the EST sequence. A correctly assembled consensus sequence for a complete cDNA should have a single 3′-terminal poly(A) region. In practice, at least 10 A or T residues (depending on the direction of sequencing) are sufficient to identify the 3′ end of a transcript. Any sequence containing an apparent poly(A) or reverse-complemented poly(A) tail at both ends, or an internal poly(A) or poly(T) tract, is flagged as potentially chimeric.
Chimerism in EST sequences without poly(A) tails is harder to detect. Our current practice is to identify these ESTs by the effects they have on the clustering process. Sections of chimeric ESTs from different origins are expected to match with different sets of sequences. Therefore, clusters containing chimerism should consist of two distinct ‘blocks’ of ESTs usually linked by only a single sequence where the fusion occurs. (This situation is also occasionally encountered when one of the ESTs in a large cluster contains an unexcised intron.) This pattern can be automatically identified by counting the number of ESTs at every position along the cluster and looking for abrupt changes in that number over a short distance. Obviously, this pattern can only be identified in clusters with sufficient coverage—in our experience, clusters containing 10 or more ESTs. In all cases, clusters identified as potentially chimeric are flagged in the database and the decision whether or not to remove chimeric ESTs is left to the submitter of the data.
DATA ACCESS AND PRESENTATION
When users log in to TBestDB they are presented with a list of organisms currently available in the database. Each organism name on the main page links to the organism's principal data page. Access permissions for each organism are determined by the provider of the data; such permissions may allow data to remain private for up to six months so that those who generate a dataset have time to analyse it before it becomes public. An organism's principal data page contains basic library and reading information and links to pages compiling experimental information and the various statistics detailed above. To maintain data currency, most statistics are calculated dynamically upon access. This page also shows all annotated clusters, with the option to order clusters in several ways and to search the various annotation fields for clusters of interest. The cluster ID links to a page containing detailed information related to that cluster, including download functionality for DNA and deduced protein sequences (Figure 2).
Figure 2.
Cluster information page. The head of the cluster information page contains the cluster consensus sequence, links to the ESTs assembled within the cluster and all annotation information. The lower half of the page contains an image illustrating the structure of the cluster. The positions of each EST are indicated. ESTs originating from different libraries are shown in different colours. The read direction of each EST is shown with an arrowhead when that information is available and ESTs that have been internally reverse-complemented by phrap in the process of cluster assembly are indicated in outline. A multiple alignment is then shown depicting the ESTs and clustered consensus sequence in the same pattern (the right-hand portion of the sequence alignment is truncated in order to improve readability of the figure).
The TBestDB main page also links to a set of Pathway Genome DataBases (25) that have been built for each organism for which annotated data are available in TBestDB. Via the pathway viewer (25) integrated With the help of TBestDB, users can inspect specific pathways, enzymatic reactions or compounds of interest, as well as visualize which enzymes and pathways are present within the organism under study or shared with other organisms.
Finally, it is straightforward to perform BLAST searches against all or selected data included in TBestDB to which a user has access. The corresponding query sequences can be uploaded or copy-pasted into a window, and BLAST search functionality is achieved via a link to the web-based sequence analysis workbench AnaBench (27), developed in-house.
IMPLEMENTATION
The TBestDB database is implemented in PostgreSQL 7.4.1 with a web interface written in PHP v4.3.8. The graphics on the cluster pages are generated using the GD module, version 2.0.25. The pipeline is constructed using Perl (5.8.0) scripts to manage the data, call the programs from the phred suite and insert the results into the database. BLAST searches for sequence annotation by AutoFact and TBLASTX searches are run on a separate 16-CPU cluster. All other procedures are executed on PCs with two 2.4 GHz or 2.8 GHz Intel Xeon CPUs.
DISCUSSION
The clustering process implemented in TBestDB features a high level of discrimination, capable of distinguishing closely related homologs. Data from the amoebozoan protist Acanthamoeba castellanii provide relevant examples. Clusters ACL00004208 (containing 32 ESTs) and ACL00004800 (42 ESTs) represent two variants of ribosomal protein S3A, differing only at 3 nt positions within the coding region. Similarly, five variant actin sequences are correctly distinguished in this organism (clusters ACL00003090, ACL00003089, ACL00004196, ACL00004782 and ACL00004755). Of the 1125 nt positions encoding 375 amino acids in actin, only 52 are heterogeneous in these five sequences and all except one of the substitutions are silent. The clustering process is also able to discriminate among clusters that are identical within the coding region but differ within the 3′-terminal untranslated region, either because the different clusters represent distinct alleles or because of variation in the location of the polyadenylation site in transcripts of the same gene.
In cases where consensus EST cluster sequences have counterparts in partial A.castellanii genomic data (28), the match between EST and genomic sequence is almost always 100%, so that the comparison allows ready recognition of introns. For example, ACL00000330 (53 ESTs) encodes a complete ORF for ribosomal protein S3, and comparison with genomic sequence finds an exact match and precisely identifies two GT … AG spliceosomal introns in the latter sequence.
Notably, the datasets collected in TBestDB allow analyses to be conducted on a number of different scales. On the one hand, these data have provided unprecedented insights into the biology of specific protists, which have not been analysed previously at the molecular level either in substantial depth or substantial breadth. For example, the question of residual plastid functions in the non-photosynthetic green algae Prototheca wickerhamii and Helicosporidium sp. has successfully been addressed by surveying nucleus-encoded plastid-targeted proteins (29,30). On a broader scale, the capacity to carry out analyses across a consistently populated and annotated set of taxonomically diverse data allows for rigorous exploration of fundamental biological questions. These questions include the origin of photosynthesis among eukaryotes (31), the extent of lateral gene transfer within various eukaryotic lineages (32) and the basal resolution of the eukaryotic tree (33).
At a more practical level, another valuable feature of TBestDB is that control of access to data is adaptable to meet the needs of individual users. User accounts can be defined to have access to any possible subset of the data within TBestDB. This feature allows users to restrict access to their data for a specified (but limited) period of time prior to release.
In summary, TBestDB provides a powerful and flexible resource for clustering, annotation and distribution of EST data, a combination of features facilitating in-depth analyses of the genetic and biochemical complexity of individual eukaryotic species, systematic comparisons among taxa and global phylogenetic analyses of eukaryotes.
Outlook
We are currently engaged in adding functionality to TBestDB to allow for expert manual curation of specific subsets of the data, initially by the providers of the data in question. In the future, we intend to incorporate additional data from public sources into TBestDB, including EST data from representatives of highly sampled eukaryotes such as vertebrate animals, vascular plants and fungi.
Acknowledgments
The authors would like to thank Sebastien Letort for development of graphics, Sandrine Fraissard for work on detection of chimerism, Maria Yu and Sabrina Rodriguez for their contributions to the development of the TBestDB interface, and Allan Sun and David To for systems administration. Work in the authors' laboratories is supported by operating and equipment funds from Genome Canada, Génome Québec, Genome Atlantic, the Atlantic Canada Opportunities Agency (Atlantic Innovation Fund) and the Canadian Institutes of Health Research (CIHR). The Program in Evolutionary Biology of the Canadian Institute for Advanced Research (CIAR) is acknowledged for interaction, travel and salary support to G.B., B.F.L. and M.W.G. M.W.G. and B.F.L. are also grateful to the Canada Research Chairs Program and Canadian Foundation for Innovation (CFI) for salary and equipment support. We also acknowledge access to the bioinformatics cluster Goldorak of the Bioinformatics Network of Quebec (BioneQ), which is funded by Genome Québec and housed at the Université de Montréal. Funding to pay the Open Access publication charges for this article was provided by the Canadian Institutes for Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Patterson D., Sogin M. Eukaryote origins and protistan diversity. In: Hartman H., Matsuno K., editors. The Origin and Evolution of the Cell. Singapore: World Scientific; 1992. pp. 13–46. [Google Scholar]
- 2.Gray M.W., Burger G., Lang B.F. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 3.Gray M.W., Lang B.F., Burger G. Mitochondria of protists. Annu. Rev. Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- 4.Wainright P.O., Hinkle G., Sogin M.L., Stickel S.K. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 5.Lang B.F., O'Kelly C., Nerad T., Gray M.W., Burger G. The closest unicellular relatives of animals. Curr. Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 6.Leigh J., Seif E., Rodriguez N., Jacob Y., Lang B.F. Fungal evolution meets fungal genomics. In: Arora D., editor. Handbook of Fungal Biotechnology. 2nd edn. New York: Marcel Dekker Inc.; 2003. pp. 145–161. [Google Scholar]
- 7.Barr D.S. An outline for the reclassification of the Chytridiales, and for a new order, the Spizellomycetales. Can. J. Biochem. 1980;58:2380–2394. [Google Scholar]
- 8.Bullerwell C.E., Forget L., Lang B.F. Evolution of monoblepharidalean fungi based on complete mitochondrial genome sequences. Nucleic Acids Res. 2003;31:1614–1623. doi: 10.1093/nar/gkg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James T.Y., Porter D., Leander C.A., Vilgalys R., Longcore J.E. Molecular phylogenetics of the Chytridiomycota supports the utility of ultrastructural data in chytrid systematics. Can. J. Bot. 2000;78:226–350. [Google Scholar]
- 10.Karol K.G., McCourt R.M., Cimino M.T., Delwiche C.F. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- 11.Qiu Y.L., Palmer J.D. Phylogeny of early land plants: insights from genes and genomes. Trends Plant Sci. 1999;4:26–30. doi: 10.1016/s1360-1385(98)01361-2. [DOI] [PubMed] [Google Scholar]
- 12.Li L., Crabtree J., Fisher S., Pinney D., Stoeckert C.J., Jr, Sibley L.D., Roos D.S. ApiEST-DB: analyzing clustered EST data of the apicomplexan parasites. Nucleic Acids Res. 2004;32:D326–D328. doi: 10.1093/nar/gkh112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heiges M., Wang H., Robinson E., Aurrecoechoa C., Gao X., Kaluskar N., Rhodes P., Wang S., He C.Z., Su Y., et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34:419–422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe J., Suzuki Y., Sasaki M., Sugano S. Full-malaria 2004: an enlarged database for comparative studies of full-length cDNAs of malaria parasites, Plasmodium species. Nucleic Acids Res. 2004;32:D334–D338. doi: 10.1093/nar/gkh115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahl A., Brunk B., Crabtree J., Fraunholz M.J., Gajria B., Grant G.R., Ginsburg H., Gupta D., Kissinger J.C., Labo P., et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguero F., Zheng W., Weatherly D.B., Mendes P., Kissinger J.C. TcruziDB: an integrated post-genomics community resource for Trypanosoma cruzi. Nucleic Acids Res. 2006;34:428–431. doi: 10.1093/nar/gkj108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kissinger J.C., Gajria B., Li L., Paulsen I.T., Roos D.S. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31:234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertz-Fowler C., Peacock C.S., Wood V., Aslett M., Kerhhornou A., Mooney P., Tivey A., Berriman M., Hall N., Rutherford K., et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maheswari U., Montsant A., Goll J., Krishnasamy S., Rajyashri K.R., Patell V.M., Bowler C. The Diatom EST Database. Nucleic Acids Res. 2005;33:D344–D347. doi: 10.1093/nar/gki121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chisholm R.L., Gaudet P., Just E.M., Pilcher K.E., Merchant S.N., Kibbe W.A. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34:423–427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikaido I., Asamizu E., Nakajima M., Nakamura Y., Saga N., Tabata S. Generation of 10,154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res. 2000;7:223–227. doi: 10.1093/dnares/7.3.223. [DOI] [PubMed] [Google Scholar]
- 22.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 23.Koski L.B., Gray M.W., Lang B.F., Burger G. AutoFACT: An automatic functional annotation and classification tool. BMC Bioinformatics. 2005;6:151. doi: 10.1186/1471-2105-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Karp P.D., Paley S., Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 26.Karp P.D., Riley M., Paley S.M., Pellegrini-Toole A. The MetaCyc database. Nucleic Acids Res. 2002;30:59–61. doi: 10.1093/nar/30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badidi E., De Sousa C., Lang B.F., Burger G. AnaBench: a Web/CORBA-based workbench for biomolecular sequence analysis and annotation. BMC Bioinformatics. 2003;4:63. doi: 10.1186/1471-2105-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson I.J., Watkins R.F., Samuelson J., Spencer D.F., Majoros W.H., Gray M.W., Loftus B.J. Gene discovery in the Acanthamoeba castellanii genome. Protist. 2005;156:203–214. doi: 10.1016/j.protis.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 29.de Koning A.P., Keeling P.J. Nucleus-encoded genes for plastid-targeted proteins in Helicosporidium: functional diversity of a cryptic plastid in a parasitic alga. Eukaryot. Cell. 2004;3:1198–1205. doi: 10.1128/EC.3.5.1198-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borza T., Popescu C.E., Lee R.W. Multiple metabolic roles for the nonphotosynthetic plastid of the green alga Prototheca wickerhamii. Eukaryot. Cell. 2005;4:253–261. doi: 10.1128/EC.4.2.253-261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Ezpeleta N., Brinkmann H., Burey S.C., Roure B., Burger G., Löffelhardt W., Bohnert H.J., Philippe H., Lang B.F. Monophyly of primary photosynthetic eukaryotes: green plants, red algae and glaucophytes. Curr. Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 32.Watkins R.F., Gray M.W. The frequency of eubacterium-to-eukaryote lateral gene transfers sows significant cross-taxa variation within Amoebozoa. J. Mol. Evol. 2006 doi: 10.1007/s00239-006-0031-0. in press. [DOI] [PubMed] [Google Scholar]
- 33.Keeling P.J., Burger G., Durnford D.G., Lang B.F., Lee R.W., Pearlman R.W., Roger A.J., Gray M.W. Eukaryotic genome diversity and the tree of eukaryotes. Trends Ecol. Evol. 2005 doi: 10.1016/j.tree.2005.09.005. in press. [DOI] [PubMed] [Google Scholar]
- 34.Wuyts J., Perriere G., Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., Apweiler R. The Gene Ontology Annotation (GOA) database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 38.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]