Abstract
Although recent studies have revealed that the majority of human genes are subject to regulation of alternative promoters, the biological relevance of this phenomenon remains unclear. We have also demonstrated that roughly half of the human RefSeq genes examined contain putative alternative promoters (PAPs). Here we report large-scale comparative studies of PAPs between human and mouse counterpart genes. Detailed sequence comparison of the 17,245 putative promoter regions (PPRs) in 5463 PAP-containing human genes revealed that PPRs in only a minor fraction of genes (807 genes) showed clear evolutionary conservation as one or more pairs. Also, we found that there were substantial qualitative differences between conserved and non-conserved PPRs, with the latter class being AT-rich PPRs of relative minor usage, enriched in repetitive elements and sometimes producing transcripts that encode small or no proteins. Systematic luciferase assays of these PPRs revealed that both classes of PPRs did have promoter activity, but that their strength ranges were significantly different. Furthermore, we demonstrate that these characteristic features of the non-conserved PPRs are shared with the PPRs of previously discovered putative non-protein coding transcripts. Taken together, our data suggest that there are two distinct classes of promoters in humans, with the latter class of promoters emerging frequently during evolution.
With the completion of the human and mouse genome sequencing projects (Waterston et al. 2002; International Human Genome Sequencing Consortium 2004) as well as the large-scale compilation of full-length cDNA information (Zhang et al. 2000; Okazaki et al. 2002; Strausberg et al. 2002; Imanishi et al. 2004; Ota et al. 2004), it has gradually become clear that the genome systems in higher mammals are far more complex than previously thought. Now, the once-dominant static view that a single locus corresponds to only one transcript and one protein has been shown to be of very limited validity. Rather, it is more common for a single locus to produce several transcript variants. In about half of human genes, on average, four different transcripts are produced by alternative splicing and as a consequence translated into proteins of divergent biological functions (Modrek and Lee 2002; Imanishi et al. 2004). Similarly, recent studies also demonstrated that diversification via transcriptional regulation is no less common in human genes (Landry et al. 2003). By use of alternative promoters (APs), which consist of different modules of transcriptional regulatory elements, diversified transcriptional regulation is enabled within a single locus (Landry et al. 2003; Carninci et al. 2005; Cheng et al. 2005; Kim et al. 2005; Kimura et al. 2006).
The functional diversification of a single gene enabled by the use of alternative splices (ASs) and APs is thought to be the molecular basis whereby the human genome is able to establish highly complex systems, such as the brain and immune systems (King and Wilson 1975), in spite of the fact that the total number of human genes, which is estimated at 20,000–25,000 (International Human Genome Sequencing Consortium 2004), is not so different from those of yeast, fly, and worm (Goffeau et al. 1996; C. elegans Sequencing Consortium 1998). Even compared to other fellow mammals, such as mice, dogs, and cows, humans have some strikingly different physiological and anatomical and metabolic characteristics, although the basic gene sets are highly comparable. Indeed, several papers have appeared, suggesting that species-specific AS and APs are responsible for certain types of species-specific organismal characteristics, regarding signal transduction, growth factor responses, neuronal connections, drug metabolism, and so on (Grandien et al. 1997; Luzi et al. 2000; Tautz 2000; Dermitzakis and Clark 2002; Su and Gladyshev 2004; Pan et al. 2005; Wu 2005).
To address questions currently of interest in genome, evolutionary and pharmaceutical sciences, large-scale attempts to discover and characterize ASs/APs in human genes have been started. We have also been identifying and characterizing the transcriptional start sites (TSSs) and the adjacent putative promoter regions (PPRs) using the data of our 1.8 million human full-length cDNAs. These cDNAs were collected from cDNA libraries constructed by a cap-targeting method, oligo-capping (Suzuki and Sugano 2003; Yamashita et al. 2006). We have recently reported that the use of APs is very common in human genes. Among 15,262 human protein-coding genes examined, putative alternative promoters (PAPs; PAP is defined as a promoter [PPR] group that consists of multiple individual promoters [PPRs]) were observed in 7674 (52%) (Kimura et al. 2006). In this data set, 1803 PAPs showing clear tissue-biased usages were included. All of the retrieved results and related raw data have been made publicly and freely available without any restrictions from our database, DBTSS (Yamashita et al. 2006; http://dbtss.hgc.jp).
In spite of potential importance of widespread PAPs in humans, it is not still clear why there are so many PAPs. In the present study, in order to understand what biological relevance those PAPs have and how they have been shaped during evolution, we carried out a large-scale comparative study of PAPs between human and mouse putative counterpart genes. For this purpose, we first prepared the TSS and PPR data for mice based on mouse full-length cDNA sequences collected from the mouse full-length cDNA project (Okazaki et al. 2002). Similar to our findings in humans, widespread presence of PPRs was also observed in mice. However, intriguingly, sequence comparisons of the individual PPR members revealed that only a minor population of the human PAP relationships was evolutionarily conserved. Further detailed computational inspection followed by experimental characterization led us to conclude that there are two evolutionary tracks of promoters with distinct characteristics from each other. Here we report our large-scale comparative studies of PAPs of human and mouse genes.
Results
Identification of widespread presence of PAPs in mice and sequence comparison between human and mouse PPRs
For the comparative study of PAPs (groups of PPRs), we collected the TSS information and retrieved the adjacent PPRs for mice using the same procedure as previously described for humans (Kimura et al. 2006). The 5′-end information of 580,204 mouse full-length cDNAs was clustered so that the individual PPRs were separated from each other by >500 bp in a given PAP. As a result, a data set of 19,023 PPRs in 13,704 mouse protein-coding genes (so-called RefSeq Genes; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene) was prepared (for further details, see Methods and Supplemental Fig. 1). As shown in Table 1, in this data set, PAPs (genes containing multiple PPRs) were identified in 3816 genes (28% of the total 13,704 genes examined), showing that the presence of PAPs is widespread in mice as well as in humans. Although the number and frequency of PAPs in mice (3816 genes; 28%) is smaller than that in humans (7674 genes; 52%), this should reflect the difference in the redundancy of the cDNA data between humans and mice.
Table 1.
PAPs identified in human and mice
The table shows the number of loci containing indicated number of PPRs. The average numbers of included TSS positions and total cDNA clones are also shown.
The retrieved mouse PPRs were subjected to comparative studies of PAPs in humans and mice. For our 7674 human PAP-containing genes, TSS information for mouse counterpart genes (according to Homologene; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene ) was found for 5463 genes. In total, the 5463 gene pairs included 17,245 and 8622 PPRs in humans and mice, respectively. As for each of these PPRs, sequence alignments between humans and mice were generated (−500 bp to +0 bp of the TSSs were used; the position of the most frequently used TSS was defined as 0; this range was set to avoid sequence overlap between different PPRs within a particular PAP). All human and mouse PPR pairs belonging to the same mutually best-hit homologous gene were considered. For the sequence alignment, we used LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html). We used this local alignment program because it is relatively robust for gaps and thus was expected to generate precise sequence alignments of promoters, although its application for genome-wide comparison is impossible due to its computational cost (also see the references Suzuki et al. 2004; Yamashita et al. 2006). All of the raw data and sequence comparison of the PPRs at each gene and other related information are publicly available from our database, DBTSS (http://dbtss.hgc.jp).
Lack of evolutionary conservation of a major part of the PPR members of a PAP
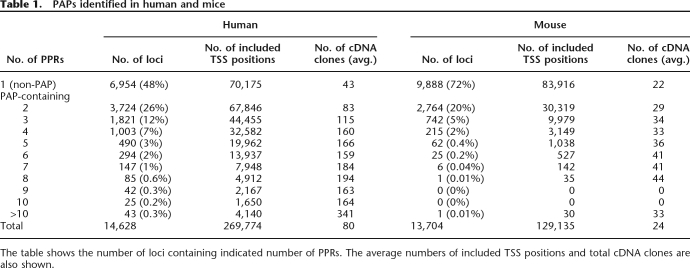
As a result of the sequence comparison, clear conservation was found for 5735 PPRs (33% of the total 17,245 human PPRs examined; Fig. 1A). In 807 human genes (15% of the total 5463 human PAP-containing genes examined), we found two or more “conserved” PPRs, consisting of a “conserved” PAP relationship (Fig. 1B; for typical examples and detailed information, see Supplemental Fig. 1B and Supplemental Table 3). As the analyses of the functional-diversification transcriptional regulation should be rather straightforward in this population, first priority for future analyses should be put on further detailed experimental characterization of the multifaceted use of the promoters. As for clues for those purposes, information regarding GO terms attached to the loci is presented in Supplemental Table 5.
Figure 1.
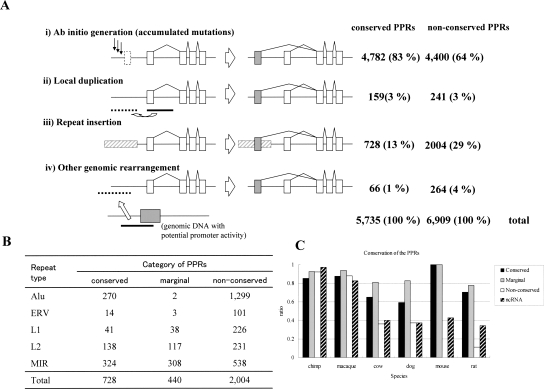
Evolutionary conservation of PAPs. (A) Patterns of evolutionary conservation and the number of PPRs belonging to each of the categories. Lines and arrows show the genomic sequence and mapped positions of the TSSs, respectively. Alignable regions are indicated as regions connected by vertical lines. For details, also see the text. (B) Number of loci in which the indicated number of PPR members are “conserved.” (C) Composition of the PAP in terms of average numbers of “conserved,” “marginal,” and “non-conserved” PPRs. When a PAP is consisted of the indicated number of PPR members (X-axis), how many of the PPRs are “conserved” (solid bar), “marginal” (gray bar), or “non-conserved” (white bar) on average are shown.
However, to our surprise, it was rather rare that the multiple PPR members within a single PAP were conserved altogether. It was far more common that the PPR sequence could be aligned only for one PPR in a PAP, while the remainder of the PPRs could not be aligned at all (Fig. 1A). As shown in Figure 1C, the number of “conserved” PPRs did not increase in proportion to the increase of the number of PPR members in the PAP. Even within a PAP consisting of more than five individual PPRs, the average number of “conserved” PPRs remained nearly one (see the solid bars in Fig. 1C). The increased parts were mostly accounted for PPRs for which no clear conservations were observed (“marginal” or “non-conserved”; see below).
For those PPRs for which no significant alignments could be generated, we analyzed genome–genome BLASTZ alignments in UCSC Genome Browser (http://genome.ucsc.edu/). Among 11,510 human PPRs (67% of the 17,245 PPRs) for which no mouse counterpart PPRs could be found, 4601 PPRs (27% of the 17,245 PPRs) were located within alignable regions, although no mouse TSSs were observed in their proximal regions. There were two possibilities to explain this: (1) cDNA coverage was insufficient; (2) promoter activities were lost in mice in spite of the fact that certain levels of sequence similarity remained. To decide between these two possibilities, we compared the number of TSSs allocated to each of the PPRs. In 1014 cases, insufficient coverage of the cDNAs was unlikely to be accounted for the absence of TSSs. In these cases, the statistical estimation (with the cutoff of P< 0.05) based on the comparison of the number of TSSs between humans and mice indicated that there must be at least one TSS at the corresponding position in mice, too (intuitively, it is understood as a case in which no TSS was observed from a particular mouse genomic region [non-PPR] although there are many human TSSs identified from the corresponding human genomic region [PPR]; for examples and further details, see Supplemental Fig. 1B). It was instead likely that the corresponding genomic regions had come to have the promoter activities only on the human side. Special cases of these observations in which the distances of the TSS clusters (PPRs) are small are also reported as “TSS turnover” by a recent study using CAGE tag analysis (Frith et al. 2006). Although further validation of this notion remains necessary, it is an intriguing possibility that these cases represent snapshots of the birth of promoters, a moment when a particular DNA has just acquired promoter activity.
On the other hand, although we scrutinized the genome–genome alignments as well as the PPR–PPR alignments, we could not find any significant alignments for the remaining 6909 PPRs (40% of the 17,245 PPRs). In these cases, the corresponding genomic sequences together with the corresponding TSSs were completely missing from the mouse side.
According to these observations, we classified the individual PPRs into three groups: “conserved (genome aligned with TSS support),” “marginal (genome aligned without TSS support),” and “non-conserved (genome not aligned),” respectively, as illustrated in Figure 1A. In the following study, we will focus the discussion on the comparison between “conserved” and “non-conserved” PPRs, but the “marginal” PPRs showed features generally similar to “non-conserved” PPRs in each analysis.
Characteristic features of “conserved” and “non-conserved” PPRs
We first examined whether there are qualitative differences between “conserved” and “non-conserved” PPRs. As shown in Figure 2, we found a number of substantial characteristic features differing between them:
Figure 2.
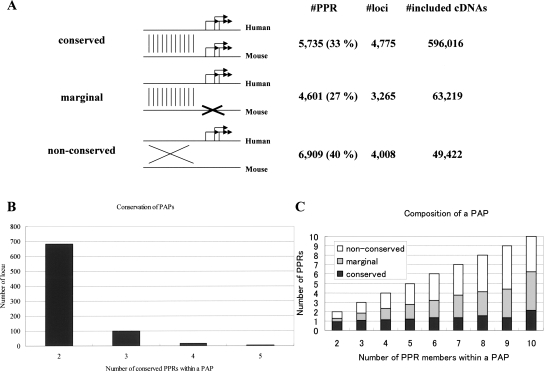
Characteristic features of conserved and “non-conserved” PPRs. (A) Frequencies of CpG islands and TATA boxes and overall G+C content in each category of PPRs. The statistical significances of the differences in the frequencies between “conserved” and “non-conserved” at the indicated positions are all P < 1.0 × −100 (*: t-test; ** and ***: χ2 test). (B) Distribution of the G+C contents. What the lines represent is shown in the inset. (C) Distance covering 75% of the TSSs (X-axis) was examined for each category of the PPRs. Frequency of the PPRs (Y-axis) belonging to each population is shown. (D) Number of PPRs which were used at the indicated relative frequencies (judged from the included cDNA numbers) is shown. (E) Number of PPRs which alter indicated length of the amino acids is shown. (F) Number of PPRs which alter indicated portion (ratio) of the amino acids is shown. (G) Number of PPRs that are located at the indicated position relative to RefSeq is shown. The relative position was designated as RefSequation 5′-end = 0 and RefSequation 3′-end = 1. Note that a minus value indicates a position upstream of the 5′-end of the RefSeq. (H) The number of PPRs which produce transcripts encoding amino acids of the indicated length. Schematic representations of the definitions of the X-axes are shown in the bottom margins for C, D, F, and G.
The “conserved” and “non-conserved” PPRs have sequence features distinct from each other (Fig. 2A). We found that, compared to the “conserved” PPRs, the “non-conserved” PPRs were poor in CpG islands (P < 1 × 10−100: χ2 test; all of the P-values appearing in this section are on the comparison between the “conserved” PPRs and the “non-conserved” PPRs) and were enriched in TATA-like elements (TATA boxes predicted using relaxed parameters; P < 1 × 10−100: χ2 test). Also, the G+C content of the “non-conserved” PPRs was significantly deviated toward A+T compared to the “conserved” PPRs (P < 1 × 10−100: t-test). Indeed, the distribution of the G+C content of the “non-conserved” PPRs resembled that of the average genomic DNA, unlike that for the “conserved” PPRs (Fig. 2B). Also see Supplemental Table 7 for predicted transcription factor binding sites enriched in “conserved” or in “non-conserved” PPRs.
In the “non-conserved” PPRs, the distribution patterns of TSSs showed significantly more fluctuation than those in the “conserved” PPRs (P < 8 × 10−40: Wilcoxon test; Fig. 2C).
The “non-conserved” PPRs were of minor usage (Fig. 2D; P < 1 × 10−100: Wilcoxon test). While 4662 (68%) of the “non-conserved” PPRs were in the population of the PPRs with relative usage of <10%, only 654 (11%) of the “conserved” PPRs belonged to this population. Also, the number of cDNAs corresponding to each of the PPRs indicated that the usage of “non-conserved” PPRs was significantly lower than that of “conserved” PPRs (on average, 104 and seven cDNAs for “conserved” and “non-conserved” PPRs, respectively; P < 1 × 10−100: Wilcoxon test; see Supplemental Fig. 2). Thus, the transcriptional level of the “non-conserved” PPRs seemed to be minor in terms of absolute levels as well as relative levels.
The “non-conserved” PPRs changed the amino acid sequences very drastically (sometimes invoking >300 amino acid changes), while the major part of the “conserved” PPRs caused alterations in small parts of the N-terminal amino acid sequences (Fig. 2E). This tendency became clearer when altered portions (ratios) of amino acid lengths relative to the entire amino acid lengths were evaluated (Fig. 2F; “non-conserved” PPRs sometimes invoking >50% amino acid changes). This is a consequence of the fact that conserved PPRs were located around the 5′-end, mostly associated with the differential use of the non-coding first exons or in small coding alterations, while the non-conserved PPRs were located throughout the regions from the first exons to the last exons in the genes, influencing a major part of the coding regions (Fig. 2G). Interestingly, as a consequence, 2124 (31%) of the “non-conserved” PPRs encoded amino acid (aa) sequences of <100 aa, while only 466 (8%) of the “conserved” PPRs were in this category (Fig. 2H). It appeared likely that the transcripts produced from the “non-conserved” PPRs should frequently function as non-protein-coding transcripts.
Experimental characterization of the “non-conserved” PPRs and their resemblance to the PPRs of “ncRNAs”
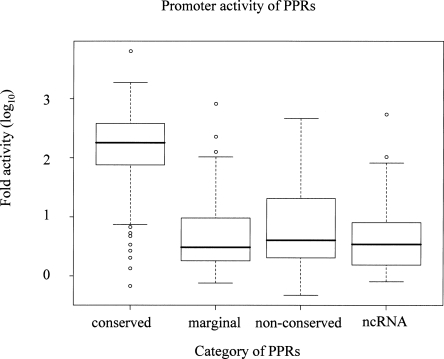
In order to experimentally validate if there are actually so many “non-conserved” PPRs in the human genome and to evaluate the range of the strength of their promoter activities, we performed luciferase reporter gene assays using physically cloned PPR DNA fragments. For this purpose, we constructed an oligo-cap (5′-end) cDNA library of human embryonic kidney (HEK) 293 cells and produced 12,504 5′-end sequences (GenBank accession nos.: BP870448–BP873619; BP244227–BP249739; details of the overview of the promoter activities within HEK293 cells will be published elsewhere). These cDNAs collectively represented 2170 PPRs in the above human PPR data set. By this, we were able to confirm the expression of the genes encoding them as well as the positions of their PPRs in HEK293 cells directly. We set PCR primers (corresponding to −1 kb to +200 bp) for 1100 PPRs. The PPR clones obtained were transiently transfected into HEK293 cells and their promoter activities were measured. We successfully cloned and obtained reproducible promoter activity data for 321 “conserved” PPRs, 56 “marginal,” and 59 “non-conserved” PPRs. As shown in Figure 3, the promoter activities of “conserved” PPRs were significantly higher than those of “non-conserved” PPRs (P < 1 × 10−100: Wilcoxon test). In spite of the clear difference, in both cases, each of the observed promoter activities was significantly higher than the averaged promoter activities of 250 randomly isolated genomic DNA fragments (for details, see Supplemental Table 4).
Figure 3.
Promoter activities of the “conserved” and “non-conserved” PPRs in HE293 cells. Distributions of the observed promoter activities are shown for each category of the PPRs. The activities are shown on a log scale with a base of 10. The average promoter activity found for 250 random genomic fragments was designated as 1 (log 0).
Having determined the range of transcriptional activities of “non-conserved” PPRs, which frequently drive transcripts encoding no or very small proteins, we wished to analyze the PPRs of a class of so-called long non-protein coding transcripts, which were discovered in recent full-length cDNA studies. (Note: We will simply call them “ncRNAs” hereafter; they are sometimes called transcripts of unknown functions [TUFs]; see Mattick and Makunin 2006; Willingham and Gingeras 2006) Among the 768 “putative ncRNAs” which were identified from our previous human large-scale full-length cDNA analyses and annotation project, FLJ (Ota et al. 2004), 49 were clearly confirmed to be transcribed in HEK293 cells by RT-PCR analyses (Supplemental Fig. 3 and Supplemental Table 6). The PPRs of 35 of these 49 “ncRNAs” were successfully cloned and were subjected to luciferase assays (Supplemental Table 4). As shown in Figure 3, we found that these promoter activities were rather in a similar range with those of “non-conserved” PPRs than those of “conserved” PPRs (“conserved”–“ncRNA”: P < 1 × 10−100; “non-conserved”–“ncRNA”: P = 0.3: Wilcoxon test). Sequence analysis also showed these PPRs share the features of “non-conserved” PPRs; ncRNAs are GC-poor (45%; “conserved”–“ncRNA”: P < 4 × 10−11; “non-conserved”–“ncRNA”: P = 0.1: t-test), CpG island-less (26%; “conserved”–“ncRNA”: P < 7 × 10−13; “non-conserved”–“ncRNA”: P = 0.9: χ2 test), enriched in TATA-like elements (63%; “conserved”–“ncRNA”: P < 2 × 10−8; “non-conserved”–“ncRNA”: P = 0.2: χ2 test), and the major part is lacking corresponding regions in the mouse genome according to UCSC BLASTZ alignment (70% were located outside of BLASTZ-alignable regions in mice).
Possible origin of the “non-conserved” PAPs
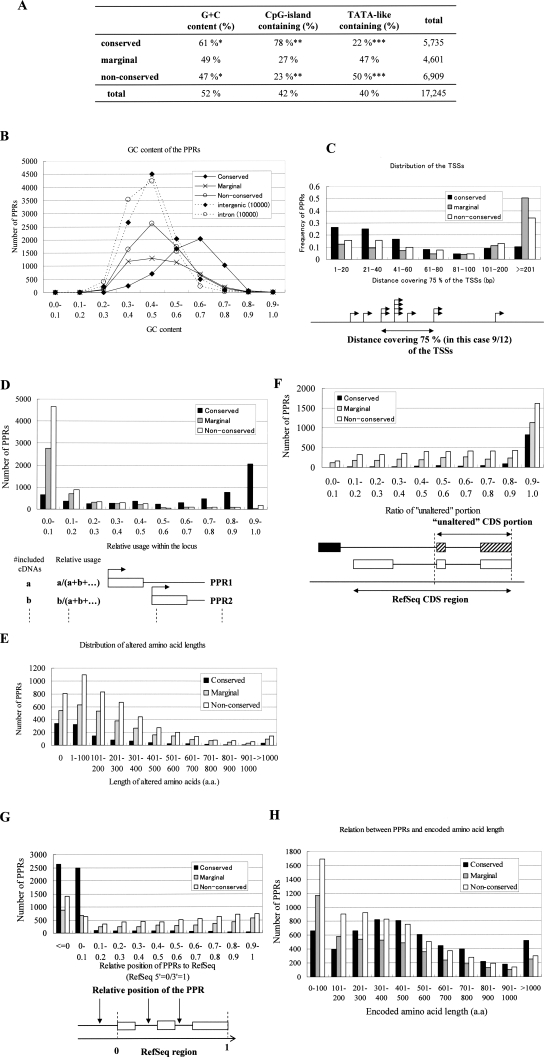
We also examined the possible origin of the “non-conserved” PPRs. It has been proposed that the PPRs could be generated by any of the following mechanisms (Landry et al. 2003): (1) Ab initio generation (accumulated mutations); (2) local duplication; (3) repeat insertion; (4) other genomic rearrangements (Fig. 4A; for definitions, see Methods). We classified the “conserved” and “non-conserved” PPRs identified by the present study into the above categories. The most frequently observed category was “ab initio generation” for both classes. In these cases, there were no corresponding genomic sequences at all in the mouse genome, thus the PPRs seemed to have emerged from average genomic sequences only in humans or have been lost only in mice.
Figure 4.
Possible origin of the conserved and non-conserved PAPs. (A) The numbers of the PPRs which showed the indicated patterns of possible evolutional origin are shown. (B) Repetitive elements identified in each category of the PPRs. (C) The frequency of the human PPRs for which corresponding PPRs or syntenic genomic regions could be identified in the genomes of the indicated species is shown.
We also found that repetitive sequence elements were enriched in the “non-conserved” PPRs (2004; 29%; Fig. 4B) compared to the “conserved” PPRs (728; 13%; P < 1 × 10−100: χ2 test). Especially, the so-called retroelement-type repetitive elements, such as L1 and Alu, mostly accounted for this differential distribution. There are a number of reported examples in which such classes of retroelements were integrated in the vicinity of future TSSs and acquired transcriptional regulatory activities via slight changes in their sequences (Norris et al. 1995; Vansant and Reynolds 1995; Hamdi et al. 2000). Considering that L1 and Alu spread throughout the human genome after the human and mouse lineages separated, integration of those elements could explain the generation of species-specific promoters.
As for the “non-conserved” PPRs, as reference genomic sequences have become available for several other mammals due to recent genome sequencing projects (see the Web site of NHGRI, http://www.genome.gov/), the genomic regions in chimpanzees, macaque monkeys, dogs, and cows were analyzed in a similar way as for the human and mouse comparison; first, the PPRs were tentatively defined as the 5′-end adjacent regions of annotated genes and available ESTs of full-length cDNAs aligned with human PPRs using LALIGN; for those for which no clear alignments were generated, respective genomic sequences in chimpanzees, macaque monkeys, dogs, and cows were further searched according to the BLASTZ alignment in UCSC Genome Browser. We found that the “non-conserved” PPRs were swiftly lost in proportion to the evolutional distances, and no more than 30% of them were identified in dogs, cows, and rats (Fig. 4C). On the other hand, at least 60% of the (human–mouse) “conserved” PPRs were found in other organisms’ genomes. Even considering the incompleteness of the genome sequencing in some of these species, we concluded that a major part of the “non-conserved” PPRs appear to have emerged evolutionarily in a lineage- or species-specific manner and are likely to have evolved very rapidly.
Discussion
Here we have described a large-scale comparative study of PAPs of human and mouse genes. Taking advantage of the collection of the 5′-end information of human and mouse full-length cDNAs, we used the well-defined 5′-end cDNA information for the identification and analyses of the PPR members. This allows a thorough comparison both of the transcriptional activity (PPRs) and sequence similarities and differences in both mouse and human. Both species have widespread PAPs, but the patterns of sequence conservation vary dramatically among the PAPs.
Interestingly, while we found that two or more PPRs were conserved in 807 genes, we unexpectedly observed that such conserved PAP relationships were only a minor fraction. In most cases, only one PPR was conserved within a given PAP, while the rest was non-conserved. It is unlikely that this general lack of conservation resulted from misidentification of the PPRs. It is true that, if PPRs were identified by dubious “full-length” cDNAs, the regions adjacent to their 5′-ends would merely be intronic sequences, and thus would be expected to be non-conserved. However, in the present study, we carefully removed potential erroneous oligo-cap cDNAs from our data set (see Methods; also see Kimura et al. 2006). Actually, in almost all cases, the PAPs were separated by mutually exclusive use of the first exons. Moreover, luciferase assays of representative PPRs showed significant promoter activities in HEK293 cells. For these reasons, we concluded that most of the PAPs identified in the present study are upstream sequences of the true TSSs, from which transcription is actually initiated in vivo, and indeed there are thousands of non-conserved active promoters.
Our finding that a large population of the “non-conserved” PAPs was located well inside of the gene seems in line with the findings obtained from recent ChIP-on-chip analyses. Binding analyses of common transcription factors, including SP1, MYC, TP53, and CREB, revealed that there are comparable numbers of docking sites for them at the internal part of the genes as well as at the 5′-ends (Cawley et al. 2004; Impey et al. 2004). The widespread transcription of these intragenic PPRs is also supported by expression analyses using genome tiling arrays (Kapranov et al. 2002, 2005). Moreover, it has been further demonstrated that at least some of these intragenic transcription events occur in response to extra-cellular signals or in a tissue-/developmental stage-specific manner (Cawley et al. 2004; Impey et al. 2004). Interestingly, it was recently reported that a significant number of genes are involved in repressing uncontrolled transcription from the internal part of the genes by promoting the formation of proper chromosome structures in these regions and that disruption of these genes resulted in abnormal embryonic development, possibly allowing unfavorable transcriptions (Tominaga et al. 2005). These reported evidences together with our data should strongly suggest that there are actually widespread intragenic alternative promoters which play specific biological roles at least in a number of genes.
The presumed biological roles of “non-conserved” PPRs immediately raise the question of how these roles would be realized. One mechanism may be encoding alternative proteins with modified functions or proteins with identical functions expressed in different conditions. In the case of the human SHC1 gene, transcripts derived from the proximal alternative promoter encode a protein lacking the interaction domain for binding with some of the interacting partners, and thereby serve as modulators of signaling pathways (Luzi et al. 2000). Besides, transcriptional regulations which produce significantly different proteins between humans and rodents or other mammals have already been reported, although the number of examples is still limited (Wang and Negishi 2003; Owens et al. 2005). For example, the ACACB gene in humans has two alternative promoters, one of which is well-conserved between humans and rats, whereas the other is not, producing a biologically functional regulatory protein in skeletal muscle only in humans (Oh et al. 2005). Considering our finding that thousands of PAPs are “non-conserved,” while one of the PPR members is almost always “conserved” (Fig. 1C), it might be a general feature in the functional diversification of mammalian genes that the proteins which serve as functional modulators, for example, by being involved in fine-tuning of the control mechanisms of cellular biological processes, have been built-in in an ex post facto manner by dynamic evolutional alterations.
Another mechanism for the “non-conserved” PPRs to realize their functions may be producing ncRNAs (Figs. 2H). A recent stream of reports have shown that ncRNAs serve important regulatory roles (Hornstein and Shomron 2006; Willingham and Gingeras 2006) via various mechanisms, such as by sense–antisense interactions with target transcripts and by directly interacting with proteins, thereby modulating the strength of protein–protein interactions (Mattick and Makunin 2006). A recent estimate indicated that thousands of ncRNAs are transcribed from the human genome, and at least 30% of human genes are subject to regulation by these RNAs (Lewis et al. 2005). Therefore, it may not be surprising that a significant fraction of the widespread “non-conserved” PPRs give rise to ncRNAs. Actually the sequence features and transcriptional activities of “non-conserved” PPRs were shown here to resemble those of the promoter regions of previously identified “ncRNAs.”
However, it was surprising that, regardless of whether they encoded proteins or not, “non-conserved” PPRs which seemed ultimately to be associated with regulatory roles were commonly found to be in a different evolutionary track from canonical PPRs. Although further detailed studies on the nucleotide changes accompanied by consequent functional changes in the promoter activities would be necessary to reveal at which point of the evolutionary stage the “non-conserved” PPRs emerged and which of them are on the way to positive, purifying, or neutral selections, it was significant to observe that the most frequent and dynamic aspects of functional diversification of genes in higher mammals should generally be orchestrated via a core “conserved” promoter playing the main tune in an ensemble of accessory “non-conserved” promoters.
The finding of the widespread presence of “non-conserved” PAPs is somewhat reminiscent of the case of ASs: The major population of ASs identified in both humans and mice was also shown to be evolutionarily non-conserved and is of minor usage. The recently born ASs are regarded as primitive forms, presumably serving as an evolutional reservoir for new transcript variants. Likewise, ab initio emergence of the promoters may take place relatively frequently among the wide variety of genomic sequences. Generally, the so-called consensus sequences for many of the transcription factor binding sites as well as those for splice junctions and other splicing enhancers are short and frequently found throughout any genomes. Basic sequence materials which can potentially consist of a promoter or a splicing junction have been constantly forming during evolution and are abundantly found throughout long mammalian genomes (Rockman and Wray 2002). It is likely that, because of their evolutionally new lineage, “non-conserved” PPRs still preserve traces of universal genomic sequences. It is possible that at least some, even many, of these PRPs are evolutionarily neutral, having no detectable biological role. However, it is as well possible that promoters specialized in transcribing regulatory transcripts may flexibly utilize this evolutionary reservoir of novel promoters to deal with dynamic change of regulatory networks, the nature of which enables organismal adaptation to rapidly change environmental requirements.
Very recently, a paper appeared from another group, also analyzing the properties of PAPs using our data set (Baek et al. 2007). As described by the study of Baek and colleagues, it is also important to analyze characteristics of genes having no alternative promoters (single promoters), although we have focused the present study on the analysis of genes having alternative promoters, which occupy roughly half of the human genes. Indeed, we were amazed at the complex nature of the mammalian transcriptomes, which is being revealed by our and other studies, as well as their flexibility, enabling numerous features unique to each organism. In the present study, we used the 5′-end sequences of putative full-length cDNAs and avoided using recently produced and massively compiled data from tag-based approaches, such as CAGE and 5′-end SAGE data (Hashimoto et al. 2004; Carninci et al. 2006). We took this approach because analyses of the latter would require different data processing and analyses, hindering a uniform interpretation of the data. If those tag data are also taken into consideration, further non-conserved PPRs should be identified from both humans and mice. We speculate so, because we consider it likely that more transcripts of even minor expression levels would be identified from non-conserved genomic regions when transcriptome analyses in humans and mice are further deepened. Also, some of the cases which were categorized as “marginal” in this study would turn out to be actually “non-conserved,” if no TSS appears even with such deep transcriptome data. Rapid progress in both genomic and full-length cDNA sequencing projects in more than a dozen higher mammals, such as primates, cattle, dogs, cats, and organisms at various evolutional stages, should shortly enhance our understanding of the highly diversified transcriptional regulatory mechanisms in further detail. Now, attempts to achieve a comprehensive understanding of the unique aspects of transcriptional networks of genes encoded within the respective genomes should be within the range of practical investigations. With that knowledge, we will at last be able to understand how regulatory blueprints of genomes have been fabricated, eventually resulting in the evolution of the great diversity of life, including humans.
Methods
Mapping and clustering of the 5′-end data
The mouse PPRs data set was generated similarly as the case in the human PPRs. The 5′-end information for 580,204 mouse full-length cDNAs which were obtained from the 119 kinds of cap-trapper full-length cDNA libraries (see Supplemental Table 1) was collected and mapped onto the mouse genomic sequence (mm5; as of UCSC Genome Browser). TSSs were clustered so that the distances of the TSSs from each other were >500 bp. Details of the procedure were described previously (Kimura et al. 2006). For statistics of the data processing, see Supplemental Table 2. For the raw data of individual PPRs, see Supplemental Tables 3–5. For graphical views, visit our database, DBTSS (Yamashita et al. 2006; http://dbtss.hgc.jp). Also, see references Suzuki and Sugano (2003) and Kimura et al. (2006) for a discussion of the possible contamination of erroneously cloned truncated cDNAs.
Sequence alignment
For comparing the sequences of PPRs between human and mouse genes, the putative counterpart gene sets were defined according to the information described in Homologene. For each of the PPRs, the sequence alignments were generated for the retrieved 500-bp sequences using a sequence alignment program, LALIGN. For details for setting the parameters, see the reference Suzuki et al. (2004). For the genome–genome alignment data, the information generated by UCSC Genome Browser using BLASTZ was used.
Procedures used in computational characterizations
The presence of CpG islands was determined according to the standard procedure described previously (Gardiner-Garden and Frommer 1987). The CpG islands covering the TSSs were counted as “CpG island-containing” PPRs. For the search of TATA boxes, MATCH was run for TRANSFAC database version 8.2 (Matys et al. 2006), and the hits from the matrices V$TATA_01 and V$TATA_C with the search conditions of −90 to +23 (plus strand), cutoff value of 0.77, were counted as “TATA-containing” promoters. This range was selected because previous studies demonstrated that these conditions should give the overall optimized accuracies of specificity and selectivity (Tsunoda and Takagi 1999).
In order to evaluate the relative usage of the “non-conserved” against “conserved” PPRs, the numbers of cDNAs belonging to every PPR were counted separately (as representing the individual expression level of the PPR), and the proportion of them relative to the total number of cDNAs belonging to the corresponding locus was calculated (as representing the total expression level). Those thereby-calculated relative usages of the PPRs were compared between the “conserved” and “non-conserved” PPRs.
In order to identify putative protein-coding regions in the transcripts whose TSSs were defined by each of the PPRs, the 5′-end sequences were connected to RefSeq sequences from the position where they overlapped. The possible protein coding regions were determined from the resultant virtual hybrid transcripts, and putative amino acid lengths were calculated. According to the obtained information, the ratios of transcripts from commonly used regions relative to transcripts covering the entire amino acid sequences of RefSeq were also calculated.
The indicated categories of possible evolutional origins of the PPRs were defined as follows: (1) Ab initio generation: the case which could not be defined by 2–4; (2) local duplication: the case in which BLASTN search detected a homologous sequence within the local region; from the terminal exon of the upstream adjacent gene to the 3′-end of the last exon of the gene; (3) repeat insertion: the case in which the repetitive element (as defined by UCSC Genome Browser) was found in the PPR; (4) other genomic rearrangement: the case in which BLASTN search detected a homologous sequence outside of the local region defined in 2.
For analyzing the conservation of the “non-conserved” PPRs in other organisms, the surrounding sequences of the 5′-ends of the cDNAs were retrieved from the chimpanzee, macaque, cow, dog, and rat genomic sequences as of UCSC Genome Browser. The surrounding sequences were defined as those of annotated genes and ESTs, which should be the closest equivalents of the human and mouse data, although the accumulation of data for them was relatively poor. The obtained sequences were analyzed similarly in the cases of humans and mice using LALIGN and by considering the genome alignments registered in UCSC.
For evaluating statistical significance, χ2 test, Wilcoxon test, or t-test were performed, using a statistical analysis software suite, R (http://www.r-project.org/). Which method was used is indicated at the corresponding position.
Luciferase assays
Genomic DNAs corresponding to the PPRs or other categories were amplified by PCR, using the genomic DNA (Strategene) and KOD PCR kit (Toyobo). The PCR conditions were as described in the manufacturers’ instructions and the primer sequences used for each amplification are shown in Supplemental Material. Because the representation of “marginal” and “non-conserved” PPRs had been small in the obtained PPR clone set, possibly reflecting the fact that the promoter activities of the “non-conserved” PPRs are weak and thus are less frequently represented by a limited number of the cDNA sequences, we reset the PCR primers, attempting to clone them intensively, so that the data set contained >50 PPRs in each category.
For amplifications of the random genomic DNAs, the primers having the cloning sites only were used with low annealing temperature. The products were size-fractionated by agarose gel electrophoresis. The recovered fragments were sequenced and the redundancy was removed. In total, 250 genomic DNA were selected from non-promoter regions. Each of the mapped positions of the fragments is shown in Supplemental Material. The amplified genomic DNAs were cloned into the luciferase vector using the Gateway System (Invitrogen). The plasmid DNAs were purified using Qiaprep Ultra (Qiagen) and transfected into HEK293 cells using Fugene6 (Roche) according to the manufacturers’ instructions. The luciferase assays were performed 48 h after the transfections using a dual luciferase kit (Promega). Every assay was performed in triplicate.
Acknowledgments
We thank K. Abe and K. Imamura for technical support. We also thank E. Nakajima for careful reading of the manuscript. This work was supported by grants from the New Energy and Industrial Technology Development Organization (NEDO) project of the Ministry of Economy, Trade, and Industry (METI) of Japan; the Japan Key Technology Center project of METI of JAPAN; and a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. BP870448–BP873619 and BP244227–BP249739.]
Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6030107.
References
- Baek D., Davis C., Ewing B., Gordon D., Green P., Davis C., Ewing B., Gordon D., Green P., Ewing B., Gordon D., Green P., Gordon D., Green P., Green P. Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 2007;17:145–155. doi: 10.1101/gr.5872707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Taylor M.S., Engstrom P.G., Frith M.C., Engstrom P.G., Frith M.C., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E.T., Clark A.G., Clark A.G. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Frith M.C., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Carninci P., Hayashizaki Y., Sandelin A., Hayashizaki Y., Sandelin A., Sandelin A. Evolutionary turnover of mammalian transcription start sites. Genome Res. 2006;16:713–722. doi: 10.1101/gr.5031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Hoheisel J.D., Jacq C., Johnston M., Jacq C., Johnston M., Johnston M., et al. 1996Life with 6000 genes Science 274546 , 563–567. [DOI] [PubMed] [Google Scholar]
- Grandien K., Berkenstam A., Gustafsson J.A., Berkenstam A., Gustafsson J.A., Gustafsson J.A. The estrogen receptor gene: Promoter organization and expression. Int. J. Biochem. Cell Biol. 1997;29:1343–1369. doi: 10.1016/s1357-2725(97)89967-0. [DOI] [PubMed] [Google Scholar]
- Hamdi H.K., Nishio H., Tavis J., Zielinski R., Dugaiczyk A., Nishio H., Tavis J., Zielinski R., Dugaiczyk A., Tavis J., Zielinski R., Dugaiczyk A., Zielinski R., Dugaiczyk A., Dugaiczyk A. Alu-mediated phylogenetic novelties in gene regulation and development. J. Mol. Biol. 2000;299:931–939. doi: 10.1006/jmbi.2000.3795. [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Suzuki Y., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Kasai Y., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Morohoshi K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Yamada T., Sese J., Morishita S., Sugano S., Matsushima K., Sese J., Morishita S., Sugano S., Matsushima K., Morishita S., Sugano S., Matsushima K., Sugano S., Matsushima K., Matsushima K. 5′-End SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Shomron N., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Imanishi T., Itoh T., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Itoh T., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Tamura T., Yamaguchi-Kabata Y., Tanino M., Yamaguchi-Kabata Y., Tanino M., Tanino M., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., McWeeney S., Dunn J.J., Mandel G., Goodman R.H., Dunn J.J., Mandel G., Goodman R.H., Mandel G., Goodman R.H., Goodman R.H. Defining the CREB regulon: A genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Cheng J., Long J., Helt G., Dike S., Gingeras T.R., Long J., Helt G., Dike S., Gingeras T.R., Helt G., Dike S., Gingeras T.R., Dike S., Gingeras T.R., Gingeras T.R. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Sekine M., Tsuritani K., Wakaguri H., Tsuritani K., Wakaguri H., Wakaguri H., et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Wilson A.C., Wilson A.C. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Landry J.R., Mager D.L., Wilhelm B.T., Mager D.L., Wilhelm B.T., Wilhelm B.T. Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Luzi L., Confalonieri S., Di Fiore P.P., Pelicci P.G., Confalonieri S., Di Fiore P.P., Pelicci P.G., Di Fiore P.P., Pelicci P.G., Pelicci P.G. Evolution of Shc functions from nematode to human. Curr. Opin. Genet. Dev. 2000;10:668–674. doi: 10.1016/s0959-437x(00)00146-5. [DOI] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I. V., Makunin I. V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Reuter I., Chekmenev D., Krull M., Hornischer K., Chekmenev D., Krull M., Hornischer K., Krull M., Hornischer K., Hornischer K., et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B., Lee C., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Norris J., Fan D., Aleman C., Marks J.R., Futreal P.A., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P., Fan D., Aleman C., Marks J.R., Futreal P.A., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P., Aleman C., Marks J.R., Futreal P.A., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P., Marks J.R., Futreal P.A., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P., Futreal P.A., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P., Wiseman R.W., Iglehart J.D., Deininger P.L., McDonnell D.P., Iglehart J.D., Deininger P.L., McDonnell D.P., Deininger P.L., McDonnell D.P., McDonnell D.P. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J. Biol. Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- Oh S.Y., Lee M.Y., Kim J.M., Yoon S., Shin S., Park Y.N., Ahn Y.H., Kim K.S., Lee M.Y., Kim J.M., Yoon S., Shin S., Park Y.N., Ahn Y.H., Kim K.S., Kim J.M., Yoon S., Shin S., Park Y.N., Ahn Y.H., Kim K.S., Yoon S., Shin S., Park Y.N., Ahn Y.H., Kim K.S., Shin S., Park Y.N., Ahn Y.H., Kim K.S., Park Y.N., Ahn Y.H., Kim K.S., Ahn Y.H., Kim K.S., Kim K.S. Alternative usages of multiple promoters of the acetyl-CoA carboxylase beta gene are related to differential transcriptional regulation in human and rodent tissues. J. Biol. Chem. 2005;280:5909–5916. doi: 10.1074/jbc.M409037200. [DOI] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Nikaido I., Osato N., Saito R., Suzuki H., Osato N., Saito R., Suzuki H., Saito R., Suzuki H., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Wakamatsu A., Hayashi K., Sato H., Nagai K., Hayashi K., Sato H., Nagai K., Sato H., Nagai K., Nagai K., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Owens I.S., Basu N.K., Banerjee R., Basu N.K., Banerjee R., Banerjee R. UDP-glucuronosyltransferases: Gene structures of UGT1 and UGT2 families. Methods Enzymol. 2005;400:1–22. doi: 10.1016/S0076-6879(05)00001-7. [DOI] [PubMed] [Google Scholar]
- Pan Q., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Bakowski M.A., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Morris Q., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Zhang W., Frey B.J., Hughes T.R., Blencowe B.J., Frey B.J., Hughes T.R., Blencowe B.J., Hughes T.R., Blencowe B.J., Blencowe B.J. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Rockman M.V., Wray G.A., Wray G.A. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., Shenmen C.M., Schuler G.D., Altschul S.F., Schuler G.D., Altschul S.F., Altschul S.F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Gladyshev V.N., Gladyshev V.N. Alternative splicing involving the thioredoxin reductase module in mammals: A glutaredoxin-containing thioredoxin reductase 1. Biochemistry. 2004;43:12177–12188. doi: 10.1021/bi048478t. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Sugano S., Sugano S. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 2003;221:73–91. doi: 10.1385/1-59259-359-3:73. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yamashita R., Shirota M., Sakakibara Y., Chiba J., Mizushima-Sugano J., Nakai K., Sugano S., Yamashita R., Shirota M., Sakakibara Y., Chiba J., Mizushima-Sugano J., Nakai K., Sugano S., Shirota M., Sakakibara Y., Chiba J., Mizushima-Sugano J., Nakai K., Sugano S., Sakakibara Y., Chiba J., Mizushima-Sugano J., Nakai K., Sugano S., Chiba J., Mizushima-Sugano J., Nakai K., Sugano S., Mizushima-Sugano J., Nakai K., Sugano S., Nakai K., Sugano S., Sugano S. Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions. Genome Res. 2004;14:1711–1718. doi: 10.1101/gr.2435604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Evolution of transcriptional regulation. Curr. Opin. Genet. Dev. 2000;10:575–579. doi: 10.1016/s0959-437x(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Tominaga K., Kirtane B., Jackson J.G., Ikeno Y., Ikeda T., Hawks C., Smith J.R., Matzuk M.M., Pereira-Smith O.M., Kirtane B., Jackson J.G., Ikeno Y., Ikeda T., Hawks C., Smith J.R., Matzuk M.M., Pereira-Smith O.M., Jackson J.G., Ikeno Y., Ikeda T., Hawks C., Smith J.R., Matzuk M.M., Pereira-Smith O.M., Ikeno Y., Ikeda T., Hawks C., Smith J.R., Matzuk M.M., Pereira-Smith O.M., Ikeda T., Hawks C., Smith J.R., Matzuk M.M., Pereira-Smith O.M., Hawks C., Smith J.R., Matzuk M.M., Pereira-Smith O.M., Smith J.R., Matzuk M.M., Pereira-Smith O.M., Matzuk M.M., Pereira-Smith O.M., Pereira-Smith O.M. MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 2005;25:2924–2937. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda T., Takagi T., Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- Vansant G., Reynolds W.F., Reynolds W.F. The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element. Proc. Natl. Acad. Sci. 1995;92:8229–8233. doi: 10.1073/pnas.92.18.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Negishi M., Negishi M. Transcriptional regulation of cytochrome p450 2B genes by nuclear receptors. Curr. Drug Metab. 2003;4:515–525. doi: 10.2174/1389200033489262. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Willingham A.T., Gingeras T.R., Gingeras T.R. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Wu Q. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics. 2005;169:2179–2188. doi: 10.1534/genetics.104.037606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita R., Suzuki Y., Wakaguri H., Tsuritani K., Nakai K., Sugano S., Suzuki Y., Wakaguri H., Tsuritani K., Nakai K., Sugano S., Wakaguri H., Tsuritani K., Nakai K., Sugano S., Tsuritani K., Nakai K., Sugano S., Nakai K., Sugano S., Sugano S. DBTSS: DataBase of Human Transcription Start Sites, progress report 2006. Nucleic Acids Res. 2006;34:D86–D89. doi: 10.1093/nar/gkj129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.H., Ye M., Wu X.Y., Ren S.X., Zhao M., Zhao C.J., Fu G., Shen Y., Fan H.Y., Lu G., Ye M., Wu X.Y., Ren S.X., Zhao M., Zhao C.J., Fu G., Shen Y., Fan H.Y., Lu G., Wu X.Y., Ren S.X., Zhao M., Zhao C.J., Fu G., Shen Y., Fan H.Y., Lu G., Ren S.X., Zhao M., Zhao C.J., Fu G., Shen Y., Fan H.Y., Lu G., Zhao M., Zhao C.J., Fu G., Shen Y., Fan H.Y., Lu G., Zhao C.J., Fu G., Shen Y., Fan H.Y., Lu G., Fu G., Shen Y., Fan H.Y., Lu G., Shen Y., Fan H.Y., Lu G., Fan H.Y., Lu G., Lu G., et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]