Abstract
Embryonic stem (ES) cells are pluripotent cell lines with the capacity of self-renewal and the ability to differentiate into specific cell types. We performed the first genome-wide analysis of the mouse ES cell transcriptome using ∼250,000 gene trap sequence tags deposited in public databases. We unveiled >8000 novel transcripts, mostly non-coding, and >1000 novel alternative and often tissue-specific exons of known genes. Experimental verification of the expression of these genes and exons by RT-PCR yielded a 70% validation rate. A novel non-coding transcript within the set studied showed a highly specific pattern of expression by in situ hybridization. Our analysis also shows that the genome presents gene trapping hotspots, which correspond to 383 known and 87 novel genes. These “hypertrapped” genes show minimal overlap with previously published expression profiles of ES cells; however, we prove by real-time PCR that they are highly expressed in this cell type, thus potentially contributing to the phenotype of ES cells. Although gene trapping was initially devised as an insertional mutagenesis technique, our study demonstrates its impact on the discovery of a substantial and unprecedented portion of the transcriptome.
The completion of the sequencing and annotation of the mouse genome (Waterston et al. 2002) suggested that our understanding of the number and function of most mammalian genes would be rapidly accomplished. Recently, however, the FANTOM Consortium has demonstrated quite evidently that the annotation of the genome is far from being completed and that an ever increasing portion of the genome is understood to encode what has been defined in this recent study as transcriptional forests, that is, regions of the genome that present a complex array of sense and anti-sense, coding and non-coding transcripts (Carninci et al. 2005). Despite the striking results obtained in the study, the authors conclude by giving evidence for the incompleteness of the current collection and the need for further elucidation of the transcriptome.
Although embryonic stem (ES) cells are likely to be one of the richest sources of transcriptional diversity, expressing ∼60% of known genes (Zambrowicz et al. 1998), paradoxically there is an evident lack of substantial EST or full-length cDNA sequences derived from these cells. Several small-scale EST-based studies have been performed on several stages of embryo development (Ko et al. 2000) as well as blastocysts (Sasaki et al. 1998). Moreover, several gene expression profiling experiments have been conducted on ES cells, with conflicting results (discussed in Vogel 2003), but only one study has addressed the question of the identification of novel genes expressed in ES cells by generating ∼10,000 ESTs from ES cells, which unearthed 977 novel genes, of which only 377 were not supported by other EST/cDNA evidence (Sharov et al. 2003).
Gene trapping has become the most widely used approach to produce mutations on a large scale in the genome of ES cells. Before the completion of the first draft of the mouse genome, great emphasis had been placed on the value of gene trapping as a gene identification tool (Skarnes 1993), and although it has been shown several times that integration often happens in sites as yet not annotated with gene structures (Wiles et al. 2000; Hansen et al. 2003), no further analysis has been carried out to verify this on a larger scale. Although the identification of sequence tags from gene trapping is similar in nature and quality to EST sequences, their capture depends only in part on transcription levels (since some vectors are able to trap genes that are not expressed in ES cells), while it depends fully on integration of the vector and its splicing with an endogenous gene.
Since the identification of novel genes in the ES cell transcriptome has a more general impact on our understanding of the genome and genes that are encoded within it, we have used ∼250,000 traps from all available public projects to reannotate the mouse genome as well as shed light on gene trapping hotspots in ES cells. We show that the use of a resource that has not been used extensively in the context of genome annotation reveals thousands of novel features of the mouse genome. Our analysis results in the discovery of >8000 novel transcripts and >1000 novel exons within existing RefSeq genes. We provide experimental evidence indicating that at least 70% of our predictions are truly transcribed in ES cells and other tissues, including an example of very specific expression by in situ hybridization. Moreover, we extensively characterize gene trapping hotspots, and prove experimentally that hotspots are mostly associated with genes that are significantly expressed in ES cells. This set of genes shows minimal overlap with previous expression-based assays and therefore provides a new set of genes of potential interest to unravel further the molecular mechanisms of ES cells.
Results
Clustering gene trap sequences in the genome
We collected 249,827 traps from the GSS section of GenBank produced by several public and private gene trapping projects. In 95.2% of the cases, sequence tags have been obtained by 5′- or 3′-RACE-PCR of the fusion transcript between the reporter gene and the endogenous gene (“mRNA” traps). In the remaining cases, sequences were obtained by inverse-PCR, revealing the exact genomic insertion site (“genomic” traps).
Using a stringent in-house automated pipeline (see Methods), we mapped sequence tags to the genome and found a clear location for ∼65% of them (153,807 “mRNA” and 7630 “genomic DNA”), while 26% (65,020 tags) present ambiguous mapping due to poor quality of deposited sequences, and 9% (23,370 tags) present no match in the genome. Approximately 43% of unmapped traps can be explained by the poor quality of the trap sequence (traps with <50 nt of unambiguous sequence), while the remaining (∼5% of all traps) can be attributed to genome coverage issues or to spurious sequences in the data set. Unmapped traps and “genomic” traps were discarded from this analysis, as they cannot be used reliably to identify novel transcripts.
We assembled all remaining traps, showing sequence overlap on the same strand of each chromosome by at least one base pair in clusters (referred to from here on as “trapclusters”). This analysis yielded 31,854 trapclusters, with an average size of ∼300 bp, on average composed of two exons. We found that 58.4% of the trapclusters (17,316) are composed by a single sequence tag. Although so many traps are found in singletons, almost 50% of the traps are found in <5% of the clusters of large size. In other words, traps are either found in very small clusters or in hotspots that contain even hundreds of traps (Supplemental Fig. S1). This distribution reflects the fact that, on the one hand, most trapping events are unique (suggesting that the technique is far from saturation) and, on the other hand, that insertional “hotspots” exist within the genome.
We found that 12,509 trapclusters are spliced on the genome. We therefore used these clusters to check for the presence of canonical splice junctions. Canonical splice sites were found in 10,810 trapclusters (i.e., 86.4%). We also verified for reverse CT-AC junctions (which could have resulted from mis-annotation), but these accounted only for 23 trapclusters. The remaining 1676 include very few known infrequent splice sites (26 GC-AG and 28 AT-AC, as seen in Burset et al. 2000), but mostly they are likely to be due to problems in the transcript–genome alignment, given by poor quality of the trap sequence tags.
In order to assess the ability of sequence tags to detect novel genes, we decided to compare our data set with available collections of transcribed sequences, namely Fantom3, based on full-length cDNAs (Carninci et al. 2005), and Unigene, based on clustering of single pass EST sequences (Schuler 1997). The overlap between the data sets shows that trapclusters present the highest proportion (40%) of unique sequences among the three data sets, suggesting that the ES cell transcriptome might reveal molecular “signatures” different from those described by Fantom3 and Unigene in different tissues and cells (Supplemental Fig. S2).
Next, we compared our data set to the RefSeq data set: the analysis showed that 44% of trapclusters overlapped with RefSeq. Investigating further trapclusters that do not overlap RefSeq but overlap novel genes predicted by Ensembl (a further 9%), cDNAs identified by Fantom3 (a further 7%) or EST clusters contained in Unigene (a further 2%), we still identify 38% of trapclusters that indicate completely novel putative features of the transcriptome (Supplemental Fig. S3). Vice versa, 47% of RefSeq genes (7858 out of 16,635) have been trapped, and a similar proportion of genes is obtained when verifying how many orthologs of known human disease genes have been trapped (∼50%, listed in Supplemental Table S1). The distribution of trapped genes across chromosomes is in accordance with gene density (Supplemental Fig. S4). All the data can be visualized as DAS tracks, using the DAS server at http://das.tigem.it/cgi-bin/dashome/das on the Ensembl 32 version of the mouse genome at http://jul2005.archive.ensembl.org.
Gene traps identify >1000 novel exons within known genes
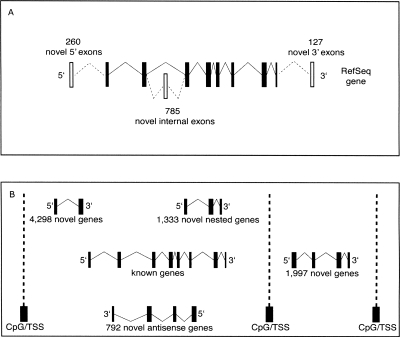
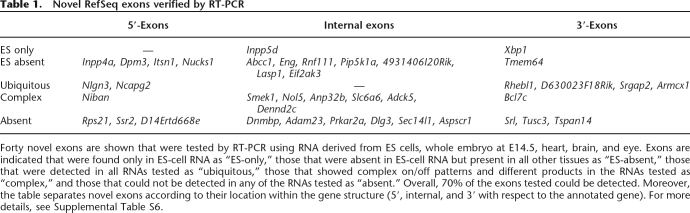
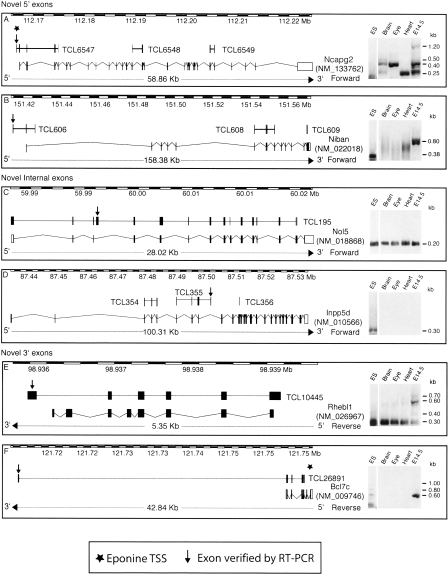
We then investigated trapcluster sequences showing a partial overlap with current RefSeq gene structures that could indicate novel potential exons. This analysis yielded 1172 novel exons identified on 830 RefSeq genes, primarily internal exons (785), as well as 5′-exons (260) and 3′-exons (127) (Fig. 1A). We decided to verify 40 of these candidate exons by RT-PCR by designing a primer on the candidate exon and a primer on the closest exon of the annotated gene and obtained a positive result on ES cell RNA in 40% of the cases. Extending the RT-PCR analysis to RNA samples such as adult brain, eye, heart, and whole embryo at embryonic day 14.5 (E14.5) identified as positive a further 30% yielding an overall rate of positively verified exons of 70% (Table 1). The latter category, labeled as “ES-absent,” was composed of six exons trapped by a poly(A)-type vector (thus possibly not transcribed in ES cells) and six exons trapped by an SAbgeo-type vector, probably expressed below detection levels in ES cells and up-regulated upon differentiation. These data confirm that gene trapping can capture both expressed and not expressed genes, depending on the type of vector used. Some examples of known genes to which our analysis added novel exons are shown in Figure 2.
Figure 1.
Discovery of novel transcriptomic features based on trapclusters. (A) Prediction of 1172 novel exons identified on 830 RefSeq genes, primarily novel internal exons (785), as well as 5′ novel exons (260) and 3′-exons (127). (B) Prediction of 1997 novel genes and 6423 novel transcripts found within known gene loci (of which 1333 are nested and 792 putative anti-sense).
Table 1.
Novel RefSeq exons verified by RT-PCR
Forty novel exons are shown that were tested by RT-PCR using RNA derived from ES cells, whole embryo at E14.5, heart, brain, and eye. Exons are indicated that were found only in ES-cell RNA as “ES-only,” those that were absent in ES-cell RNA but present in all other tissues as “ES-absent,” those that were detected in all RNAs tested as “ubiquitous,” those that showed complex on/off patterns and different products in the RNAs tested as “complex,” and those that could not be detected in any of the RNAs tested as “absent.” Overall, 70% of the exons tested could be detected. Moreover, the table separates novel exons according to their location within the gene structure (5′, internal, and 3′ with respect to the annotated gene). For more details, see Supplemental Table S6.
Figure 2.
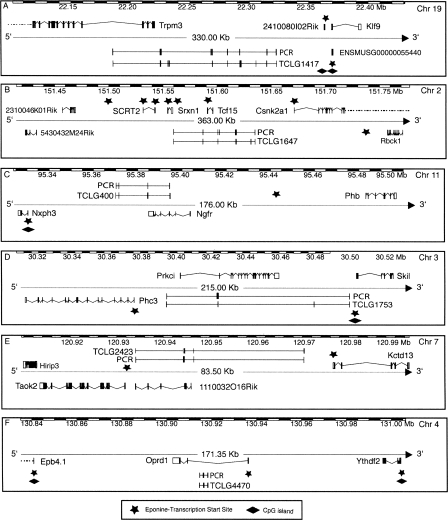
Discovery of novel exons on known RefSeq genes. The figure shows six examples of RefSeq genes to which novel exons (indicated by the arrow) were added using gene trap data, confirmed by RT-PCRs conducted on ES cell RNA as well as four other RNA samples shown in the panel on the right of each diagram. TCL6547 was verified as an alternative 5′-UTR exon of the Ncapg2 gene found to be expressed in all RNA samples tested, showing several splicing variants. The TCL606 cluster also confirms a new 5′-UTR exon (belonging to the Niban gene); however, its expression was only confirmed in ES cells and whole embryo (and not in heart, brain, or eye). The TCL195 cluster represents a novel alternatively spliced “cassette” exon added between exon 3 and exon 4 of the Nol5 gene, which is found to be expressed in all samples tested, always yielding the same PCR product. TCL355 adds an internal exon to the Inpp5d gene, and its sequence terminates at this exon. This cluster was found to be expressed only within ES cells. The TCL10445 cluster adds a 3′-exon to the Rhebl1 gene, and the transcript that includes this exon skips the last two constitutive exons of this gene in all RNA samples tested, while the isoform that includes the exons between is only found in whole embryo RNA. TCL26891 adds a further 3′-exon >30 kb away from the last exon of the Bcl7c gene. This exon is only found expressed in ES-cell RNA and whole embryo RNA.
Gene traps identify >8000 novel transcripts
We decided to inspect further the large set of trapclusters (66%) that did not overlap known genes. Owing to the fragmented nature of trapclusters, most of them were found isolated, not overlapping with other clusters or known genes, making it difficult to assign gene boundaries. In order to reduce this large data set into an approximate potential number of novel genes, therefore, we investigated the presence of CpG islands and transcription start sites predicted by Eponine (Down and Hubbard 2002) around trapclusters. This allowed us to group adjacent (but not overlapping) trapclusters into a set of 8420 novel transcripts divided into 1997 “novel genes” (found regions between CpG islands bare of any annotation) and 6423 “novel transcripts” located within known transcriptional forests. Of the latter, 1333 are “nested,” that is, in the same direction as the known transcript of the locus but fully contained within its introns, while 792 are in opposite direction to the known transcript (i.e., putative anti-sense transcripts) (Fig. 1B).
We verified the overlap of the 8420 novel transcripts with ab initio predictions made by GENSCAN, which showed that 59% (4990 out of 8420) are, indeed, also predicted computationally. In order to assess to what extent these novel transcripts could also represent transcripts as yet not identified within the human genome and other mammalian genomes, we analyzed multispecies alignments underlying our novel transcript data set. This analysis showed that 65% were found in regions alignable to the human genome via an MLAGAN mammalian multispecies alignment. Having obtained a location on the human genome, we were able to inspect the homologous region for presence of known genes (which were found in 61% of the cases, 3309 out of 5462 conserved novel transcripts), as well as for evidence of transcription based on a tiling array data set (Cheng et al. 2005) (65% of the cases, 1107 out of 1697 conserved novel transcripts located in human chromosomes inspected by Cheng et al.). By comparison, performing the same analysis on known RefSeq genes shows that 92% are alignable to the human genome and 80% overlap with the tiling array data set.
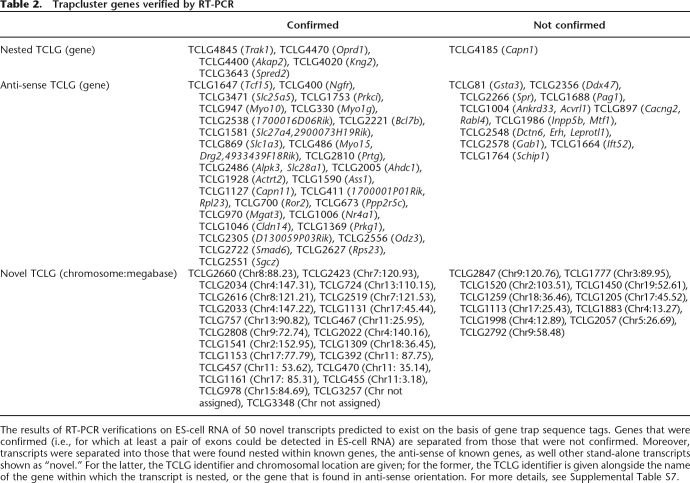
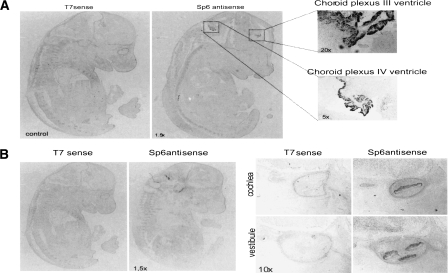
We performed RT-PCR experiments to test the existence of 80 randomly chosen sequences (1%) from the data set of 8420 novel transcripts, as well as the splicing of all the exons contained within them. The results showed that ∼71% of these genes (57/80) are expressed in ES cells (Table 2), and >50% of their exons are also confirmed to be expressed. As a further proof of the significance of our RT-PCR results, we have performed a similar test on a set of negative controls, that is, 10 RT-PCRs performed using 20 existing trap primers assorted randomly, as well as the primers for trap TCLG470 as a positive control, and while the positive control was confirmed, all other primer combinations yielded negative results. These results, when compared to our 70% validation rate for trap cluster genes, indicate that our 70% validation rate is highly significant (P-value = 4.904 × 10−5). Some examples of genes that have been verified are shown in Figure 3. The data obtained computationally (human alignments and overlap with tiling array data) coincide with the wet lab data obtained (71% RT-PCR verified) supporting ∼65%–70% of the transcripts predicted, thus indicating that our data set should contain at least 5500 real novel transcripts. It should be noted that the majority of these sequences appears to be non-coding as ∼13% of the transcripts have an open reading frame longer than 100 amino acids or a significant BLAST hit to the Uniref90 protein database (and only 2.5% have both). We decided to verify further the expression of non-coding transcripts within our data set by performing an in situ hybridization on a mouse embryo at the E14.5 developmental stage of a non-coding transcript found in anti-sense orientation with respect to the Trpm3 gene, TCLG1417, which had shown positive results by RT-PCR as described in Figure 3. This novel gene showed extremely specific expression at the developmental stage tested, with a signal localized only in the cochlea and the choroid plexus (Fig. 4A,B).
Table 2.
Trapcluster genes verified by RT-PCR
The results of RT-PCR verifications on ES-cell RNA of 50 novel transcripts predicted to exist on the basis of gene trap sequence tags. Genes that were confirmed (i.e., for which at least a pair of exons could be detected in ES-cell RNA) are separated from those that were not confirmed. Moreover, transcripts were separated into those that were found nested within known genes, the anti-sense of known genes, as well other stand-alone transcripts shown as “novel.” For the latter, the TCLG identifier and chromosomal location are given; for the former, the TCLG identifier is given alongside the name of the gene within which the transcript is nested, or the gene that is found in anti-sense orientation. For more details, see Supplemental Table S7.
Figure 3.
Discovery of novel genes based on trapclusters. The figure shows six examples of novel multiexon genes predicted using gene trap data verified by RT-PCR on ES-cell RNA as well as CpG island and Eponine transcription start site annotation. TCLG1417 is a transcript without an ORF found in reverse orientation and partial overlap with the Trpm3 gene with seven out of 10 predicted exons confirmed to be transcribed in ES cells (more expression info in Fig. 4). TCLG1647 is also found in opposite orientation to a known gene, Tcf15, but it is actually larger and contains the known gene within its intron. This trapcluster gene was predicted to contain seven exons, but PCR verification resulted in the merge of two proximal exons, the addition of a novel exon that was not present in the gene trap collection, and two exons that could not be linked to this transcript. TCLG400 is also opposite and in partial overlap to a known gene, Ngfr, and all its four exons were confirmed by RT-PCR. Only three out of five exons of TCLG1753 were connected in a single, large transcript that contains the Prkci gene on the opposite strand. TCLG2423 is found opposite to the 1110032O16Rik gene, and all its four exons were confirmed by RT-PCR. TCLG4470 is a compact three-exon transcript found opposite and nested to the Oprd1 gene, confirmed by RT-PCR.
Figure 4.
In situ hybridization of trapcluster gene TCLG1417 on E14.5 mouse embryo. The figure shows the in situ hybridization of trapcluster gene TCLG1417 on a mouse embryo at the E14.5 developmental stage. This gene shows a highly specific signal. (A) The signal detected within the choroids plexus at 1.5×, 5×, and 20× magnifications. (B) The signal within the developing auditory and vestibular pathways, specifically the developing cochlea and vestibule at 1.5× and 10× magnification.
Functional classification of trappable genes
A gene ontology analysis shows that the spectrum of genes that have been trapped in ES cells is quite wide, as reported before (Hansen et al. 2003); however, there is statistically significant enrichment (P < 0.001) for several KEGG pathways involved in the basic metabolism of protein translation and degradation (e.g., the ribosome and the proteasome) and energy metabolism (oxidative phosphorylation and ATP synthesis), as well as nucleic acid metabolism (pyrimidine and purine metabolism as well as aminoacyl-tRNA biosynthesis). A similar analysis performed on Gene Ontology classes revealed >300 classes with significant enrichment, all related to intracellular cell compartments, metabolic and physiological biological processes, and catalytic molecular functions, in particular, classes related to the metabolism of DNA, RNA, and proteins (see Supplemental Table S2 for full details). In contrast, genes that were not trapped presented a significant bias for the neuroactive ligand–receptor interaction pathways (most neural receptors such as GPCRs, GABA receptors, etc., are not trapped), the cytokine–cytokine receptor pathways (including most chemokine ligands and TNF family members), and the complement and coagulation cascades, indicating that membrane and extracellular genes are very unlikely to be trapped, confirming the need for specialized vector design (i.e., secretory trap) to saturate the genome (see Supplemental Table S3).
Hypertrapped genes are expressed at high levels, but not detected by previous expression profiling studies
As discussed earlier, the clustering of traps showed a small set of clusters containing a large portion of traps and most clusters being composed of a few traps. The former are “gene trapping hotspots” that have been observed before (Hansen et al. 2003) but have not been investigated in any further detail. We have verified that these hotspots do not relate to specific genomic regions; thus, the other two factors that could theoretically influence the rate of trapping are the size of the gene locus (the more space for the insertion to occur, the higher the chances of the insertion) and the chromatin accessibility of the region, which is tightly linked with the levels of expression of the genes within it, although we cannot exclude a possible bias determined by the type of vector used. When we calculated the distribution of trapped RefSeq genes versus the gene length, we, indeed, found that the rate of trapping increased with gene length, confirming that the insertion of gene trap vectors is influenced by gene size (Supplemental Fig. S5).
Therefore, we normalized our data set with respect to gene length (for details, see Methods) in order to identify genes that could be trapped at high rates owing to expression levels. This led to the identification of 383 RefSeq genes (from here on referred to as “hypertrapped”), which represent 5% of the frequency distribution but contain 20% of all the gene traps sequenced (30,754 traps, >37% of the traps found in known RefSeq genes) (more details in Supplemental Table S4). A gene ontology analysis revealed biases similar to those shown by the entire list of trapped genes. The only significant difference was that hypertrapped genes are more significantly enriched for ubiquitin-conjugating enzymes.
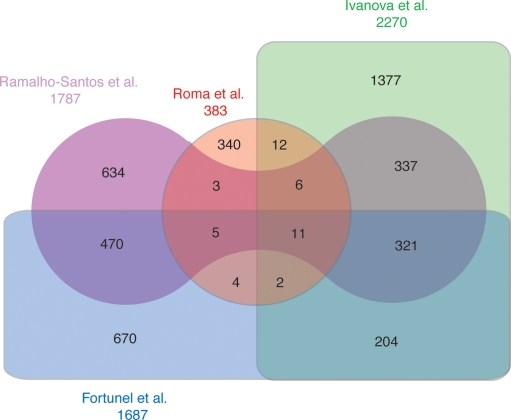
Expression profiling on ES cells was conducted in the past by several groups (Vogel 2003) and presented a set of 332 genes found to be expressed at high levels in ES cells in three different studies. Our set of hypertrapped genes shows minimal overlap with these studies: only 11 genes overlap all four data sets, and 340 out of 383 hypertrapped genes show no overlap with any of the published data sets (Fig. 5). To test whether hypertrapped genes indicate genes with high levels of expression in ES cells, we performed real-time RT-PCR experiments to compare the level of expression of 10 genes from the hypertrapped gene list and, as a control, 10 randomly selected genes that were trapped only once or twice. We compared the level of expression in ES cells of these genes to the Pou5f1 (formerly known as Oct4) gene, a well known marker expressed in pluripotent and germ line cells.
Figure 5.
Overlap of hypertrapped RefSeq genes with published ES-cell genes derived from expression profiling. A four-way Venn diagram showing the overlap between our data set of hypertrapped genes and three previously published data sets of genes highly expressed in ES cells obtained by expression profiling. The diagram shows that although the expression profiles show an overlap of >300 genes, only 11 of those are found also in our data set. Moreover, 340 hypertrapped genes are not overlapping any of the previously published expression-based data sets.
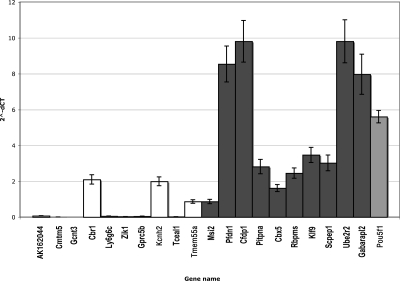
The results indicate that 80% of the hypertrapped genes we tested presented levels of expression that were significantly higher than the control set and comparable to Pou5f1 (Fig. 6). Only one of the hypertrapped genes tested, Scpep1, is present in two of the three previously published data sets. Hypertrapped genes, therefore, constitute a novel set of genes that are likely to be expressed at significant levels in ES cells and might be relevant to unravel further the molecular mechanisms underlying ES cells. There are also several gene trapping hotspots that do not fall in annotated regions of the genome, since among the novel transcripts identified there are also 87 that can be categorized as being “hypertrapped” and warrant further investigation (listed in Supplemental Table S5).
Figure 6.
Real-time RT-PCR verification of level of expression of hypertrapped RefSeq genes. The bar chart shows the levels of expressions of 10 hypertrapped genes (dark gray) and 10 genes trapped one or two times (white), as well as the Pou5f1 gene (light gray), a marker of pluripotent cell lines. Eighty percent of hypertrapped genes are expressed at significantly higher levels than genes trapped at the median rate of one trap per gene.
Discussion
In our study, we exploited the large data set of publicly available sequences derived from gene trapping experiments to investigate whether they allowed us to understand further the ES cell transcriptome, as well as the mouse genome at a broader level. The most striking result of our analysis is the unveiling of thousands of novel transcripts, which indicated that 38% of the trapclusters cannot be mapped to regions of the genome that have already been annotated with gene structures by RefSeq, Ensembl, Fantom, or Unigene.
The proportion of RefSeq genes that have been trapped (∼50%) could appear to differ from the claims made by the Lexicon group (Zambrowicz et al. 2003), which indicated that their gene trap collection covered ∼60% of known mouse genes. However, they selected for this assessment only a sentinel set of 3904 full-length mouse cDNAs having an identified human ortholog, mapped to a specific chromosomal location in the mouse genome, and represented in the RefSeq database.
The novel exons predicted on RefSeq known genes can be attributed to alternative isoforms missing from the current annotation of the gene. The splicing patterns obtained, in particular for 5′- and 3′-exons, were often diverse, indicating a richness in alternative splicing within these regions. The fact that more internal exons than external ones are discovered using gene trapping is in line with the fact that the technique provides sequences from integration events that happen within introns. Our RT-PCR validation indicates that 70% of these are likely to be expressed, and likely to be tissue-specific.
The fact that at least 40% can be detected in ES cells, with a further 30% verified by testing only four more different RNA sources, indicates that it is likely that an even higher proportion of our novel exons would be verified if many more developmental stages and tissues were assayed. These results highlight the fact that genes that have undergone trapping in ES cells might be expressed at very low levels within these cells, but can be found at higher levels in specific tissues and cell types upon differentiation, as seen in the example shown by in situ hybridization of TCLG1417. This also suggests that the reason why gene trapping in ES cells could reveal so many novel genes not found in previous cDNA and EST databases is that they are probably expressed at high levels at specific time points and in cell types that have not been used to produce libraries for EST collection.
Trapclusters were annotated with Gene Ontology and KEGG identifiers, in order to understand differences between the sets of genes that were trapped, not trapped, or hypertrapped. Hypertrapped and trapped categories both contain genes that are related to all basic molecular functions of a cell, such as transcription, translation and degradation of proteins. Hypertrapped genes show a balanced subselection of the same types of genes. The most interesting result was that related to genes that have not been trapped (see Supplemental Table S2). Importantly, entire pathways and gene families (those involving membrane receptors in particular) are clearly not trapped, indicating that it is unlikely that genes within those families and pathways will be trapped using current vector designs. Some of these genes, such as rhodopsin-like receptors and some GPCRs, are known to be mostly single-exon genes, which is probably the main reason why they are not trapped. Interestingly, the set of genes that are not trapped shows a significant bias for genes that are involved in defense mechanisms and response to external stimuli. It would be highly desirable to obtain gene trap sequences from other gene trap vectors that would enable trapping of such genes. Interesting vectors that are able to trap such genes effectively have been presented (Medico et al. 2001; De-Zolt et al. 2006) and perhaps ought to be used on larger-scale studies to enable trapping of genes that are involved in secretory pathways, response to external stimuli, defense mechanisms, and inflammation responses.
As discussed above, hotspots are likely to be caused by both levels of expression of the endogenous gene, as well as large introns, allowing multiple gene trap vector insertions. The bimodal distribution mirrors the fact that ES cells are known to express a large number of genes at basal levels, and a few hundred genes at high levels (for review, see Sharov et al. 2003). We were able to verify that trapping hotspots are, indeed, associated to genes with long introns and moreover reflect genes that are significantly expressed in ES cells, compared to a well-known marker of ES cells, such as Pou5f1.
The list of hypertrapped genes indicates the high levels of transcription, translation, and degradation that are happening constantly within ES cells, since most genes that were found to be hypertrapped were related to transcription, ribosomes, and ubiquitination. Our comparison with published “stemness” genes derived from expression profiling (Vogel 2003) showed a remarkably low overlap, and, in particular, the genes that were found by real-time PCR to be expressed at high levels within our set of hypertrapped genes are not present in the data sets published. It is known that Pou5f1 requires finely tuned levels of expression; thus, this result points to possible limitations of expression profiling and indicates a set of genes that are significantly expressed in ES cells that warrant further investigation.
Predicted novel genes were confirmed by a variety of techniques including RT-PCR, real-time PCR, as well as in situ hybridization, as well as several computational approaches (multispecies alignments, comparison with tiling array data), suggesting that at least 65% of our trapclusters are truly expressed genes in ES cells. It was very encouraging to obtain such a specifically localized signal by in situ hybridization on the TCLG1417 gene, especially considering that it is a novel non-coding gene, and the heated debate on non-coding genes that do not fall in the much studied microRNA category. Its expression specificity would suggest a role within auditory pathways; thus, it would be particularly interesting to pursue it further.
Taken together, our results indicate that gene trapping in ES cells holds a fundamental value for biology at large that transcends the usefulness of gene trapping as a mutagenesis tool. Our results clearly indicate the existence of thousands of novel genes and transcripts that had not been annotated yet. Only when expression arrays include and measure every genic component of the genome, and experiments on these arrays account for all developmental stages and cell types, will we be able, hopefully, to dissect gene networks completely and accurately.
Methods
Bioinformatics analysis of gene trap sequence tags
A total of 249,827 traps were collected from the GSS section of the NCBI GenBank (October 2005), which, in turn, were generated from several gene trap projects: 10,350 BayGenomics, 4879 CMHD, 9736 ES-cells, 1627 FHCRC, 13,031 GGTC, 198,902 Lexicon, 8301 Sanger, 1346 TIGEM, and 1655 Vanderbilt. Repeated elements were identified by using RepeatMasker (http://www.repeatmasker.org) and Repbase Update (http://www.girinst.org) (Jurka et al. 2005). An in-house automated analysis pipeline was developed (1) to map each trap to the mouse genome, (2) to predict the trapped gene and the most likely insertion site based on the structure of the vector used, (3) to cluster traps based on their mapping, and (4) to retrieve the relevant annotation present at the relevant genomic locations.
Each trap was aligned against a repeat masked version of the mouse genome (May 2005 Assembly; http://www.ncbi.nlm.nih.gov/genome/guide/mouse/) using WUBLAST (Altschul et al. 1990) with an E-value cutoff of 10−5. The BLAST output was parsed to extract genomic locations for each query sequence by using BioPerl modules (Stajich et al. 2002), with a cutoff of 96% percentage identity. In order to choose the best alignment, we selected only the best genomic locus for each sequence based on the identity, the length coverage, and the number of exons. Since many genes have multiple copies and, therefore, sequences may have multiple, almost equally good alignments in different genomic locations, we optimized our algorithm in order to distinguish the real trapped gene from recent pseudogenes and to choose all the possible mappings for each sequence in case of duplicated genes.
Moreover, for each trap, we predicted the trapped gene and the putative vector location based on the known vector specifications reported in the literature by using a local version of the mouse Ensembl database (release 32) and the Ensembl API (Curwen et al. 2004).
Of the total 161,437 traps successfully mapped onto the mouse genome, we selected 153,807 clone sequences annotated in GSS as “mRNA.” These sequences were clustered into 31,854 trapclusters based on an overlap of their locations in the genome on the same chromosome strand by at least one base pair.
The Ensembl database was also used as the source to annotate trapclusters. For each putative exon of the trapcluster, we verified if it overlapped an exon of a known RefSeq gene (only curated mRNAs having accession prefix NM and NR were taken into consideration; Pruitt et al. 2005), genes predicted by the Ensembl pipeline, but not present in the RefSeq-curated data set (Birney et al. 2006), cDNAs isolated by the FANTOM3 project (Carninci et al. 2005), and EST clusters collected in the Unigene data set (Schuler 1997). Human orthologs of the trapped genes were also retrieved from Ensembl for genes involved in the development of genetic diseases, as reported in the On-Line Mendelian Inheritance in Man (OMIM) database (Hamosh et al. 2005). All data were stored in a MySQL database.
Identification of splice sites
We tested the presence of misoriented trapclusters by checking for GT-AG (sense) versus CT-AC (anti-sense) splice junctions. Since sequence and alignment quality problems could hide the exact position of the splice junctions, we looked at the presence of both canonical splice donor (GT) and acceptor (AG) within a range of ±5 bases.
Comparison of the trapclusters with Fantom and Unigene data sets
Fantom transcripts were downloaded from the Fantom3 Web site (http://fantom.gsc.riken.go.jp/) and mapped to the mouse genome using our mapping pipeline. Alignment information of the Unigene sequences was retrieved from the Ensembl database through the Ensembl API. Comparison among trapclusters, Fantom, and Unigene was performed based on sharing at least 1 bp on the same chromosome strand using a cutoff for all the sequences of 96% identity with the genome.
Gene ontology analysis
A gene ontology analysis for both trapped and not trapped genes was performed using the DAVID Web tool (Dennis et al. 2003) using a P-value lower than 0.001 (http://david.niaid.nih.gov/david/version2/index.htm).
Identification of hypertrapped RefSeq genes
We identified known RefSeq genes that are hypertrapped using this formula:
where t is the number of traps, e is the number of trapped exons, n is the number of total exons, I is the length in intronic base pairs, and selecting genes showing the top 5% R-values.
Real-time PCR
A 2× PCR supermix from Bio-Rad (iQTM SYBR Green supermix) containing Taq DNA polymerase (iTaqTM polymerase), MgCl2, dNTPs, SYBR Green I, and fluorescein was used. Primers were added to the reaction mix at a final concentration of 400 nM. One microgram of RNA purified from ES cells and DNase I-digested was reverse transcribed as previously described. The cDNA was added at a dilution of 1:3.
Each sample was amplified in triplicate. The real-time quantitative RT-PCR was performed using an iCycler iQ system (Bio-Rad). Cycling conditions were 3 min at 95°C, followed by 40 cycles of 10 sec at 95°C, 30 sec at 60°C, and 45 sec at 72°C. The fluorescence data used for quantitation were collected at the end of each 72°C step, and the threshold cycle (ct) was automatically determined using the accompanying iCycler iQ software by calculating the second derivative of each trace and looking for the point of maximum curvature.
The primers used for each gene are available on request. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as reference gene.
RT-PCR
To perform RT-PCR, total RNA from undifferentiated ES cells (E14Tg2A.4 clone) was extracted using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. One microgram of total RNA, DNase I digested, was reverse-transcribed to cDNA with SuperScript II (Invitrogen) using random hexamers. One-tenth of the cDNA sample was subjected to PCR amplification with specific primers.
Identification of novel genes and transcripts
CpG islands and transcription start sites were obtained from the Ensembl database. CpG islands in Ensembl are predicted by looking for sequences longer than 200 bp with a GC content >50% and an observed-to-expected ratio of CpG dinucleotides above 0.6, while transcription start sites are predicted using Eponine (Down and Hubbard 2002). These locations allowed us to distinguish “trapcluster genes,” that is, trapclusters that were found within two CpG islands/Eponine predictions where no gene had been annotated and are thus likely to be part of a completely new locus, and “trapcluster transcripts” that fell downstream from the CpG islands/Eponine predictions of a known gene locus, although not showing any sequence overlap with it. Only trapcluster genes not overlapping RefSeq genes or Ensembl gene predictions were considered novel genes.
Moreover, for novel genes, we calculated the longest open reading frame using BioPerl scripts (Stajich et al. 2002) and verified whether they had a significant hit in the Uniref90 database (Wu et al. 2006) using BLASTP (Altschul et al. 1990) with an E-value cut-off of 10−5.
We also compared the whole data set with ab initio computational gene predictions generated by GENSCAN. Finally, we used multispecies alignments to verify the presence of our sequences on the human genome and to assess their potential overlap with novel sites of transcription revealed by the genome tiling array data set available at http://transcriptome.affymetrix.com/publication/transcriptome_10chromosomes (Cheng et al. 2005).
The identification of novel hypertrapped genes was performed using this formula: R = t × 1/(log10)I, where t is the number of traps and I is the length in intronic base pairs.
In situ hybridization
The DNA fragments used as probes were obtained by PCR and cloned in the PCRTOPO 2.1 vector containing both T7 and Sp6 promoters. The primers used to amplify the probe are forward, 5′-TGAAAGCCACAGGACAAGAAG-3′; reverse, 5′-CAAGCTT CAAATAGCATGTTT-3′.
The embryos were removed by Caesarean section, according to the institutional guidelines and approved by the Local Committee for “Ethical Experimental Activities on Animals.” Embryos at E14.5 were immersed in 4% paraformaldehyde in PBS (pH 7.4) overnight. Then, the embryos were dehydrated in 10%, 20%, and 30% sucrose and embedded in O.C.T. compound (Tissue Tek). Cryostat sections (16 Tm) were cut and affixed to Superfrost/PLUS slides. In situ hybridization was performed using standard procedure. Photographs were taken using a fluorescence microscope, Zeiss Axioplan 2.
Overlap analysis between published data sets and hypertrapped genes
Published expression profiles of ES cells (discussed in Vogel 2003) were downloaded and compared to our list of hypertrapped genes using Unigene identifiers. The overlaps between published data sets were derived from the comparison made by Fortunel et al. (2003).
Acknowledgments
We thank Marco De Simone, Mario Traditi, and Alessandro Davassi for their technical support; as well as Andrea Ballabio, Remo Sanges, Vincenza Maselli, and Vincenzo Gennarino for their useful suggestions; and Remo Sanges and Chiara Migliore for their assistance. This work was supported by the Fondazione Telethon and the European Union (grant no. 512003).
Footnotes
[Supplemental material is available online at www.genome.org and http://trapcluster.tigem.it/download.php.]
Article published online before print. Article and publication data are at http://www.genome.org/cgi/doi/10.1101/gr.5720807
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Cox T., Cunningham F., Curwen V., Cutts T., Cunningham F., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M., Seledtsov I.A., Solovyev V.V., Seledtsov I.A., Solovyev V.V., Solovyev V.V. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., FANTOM Consortium, RIKEN Genome Exploration Research Group Genome Science Group (Genome Network Project Core Group). et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M., Clamp M., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M., Clamp M., Andrews T.D., Clarke L., Mongin E., Searle S.M., Clamp M., Clarke L., Mongin E., Searle S.M., Clamp M., Mongin E., Searle S.M., Clamp M., Searle S.M., Clamp M., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Yang J., Gao W., Lane H.C., Lempicki R.A., Gao W., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- De-Zolt S., Schnutgen F., Seisenberger C., Hansen J., Hollatz M., Floss T., Ruiz P., Wurst W., von Melchner H., Schnutgen F., Seisenberger C., Hansen J., Hollatz M., Floss T., Ruiz P., Wurst W., von Melchner H., Seisenberger C., Hansen J., Hollatz M., Floss T., Ruiz P., Wurst W., von Melchner H., Hansen J., Hollatz M., Floss T., Ruiz P., Wurst W., von Melchner H., Hollatz M., Floss T., Ruiz P., Wurst W., von Melchner H., Floss T., Ruiz P., Wurst W., von Melchner H., Ruiz P., Wurst W., von Melchner H., Wurst W., von Melchner H., von Melchner H. High-throughput trapping of secretory pathway genes in mouse embryonic stem cells. Nucleic Acids Res. 2006;34:e25. doi: 10.1093/nar/gnj026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J., Hubbard T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunel N.O., Otu H.H., Ng H.H., Chen J., Mu X., Chevassut T., Li X., Joseph M., Bailey C., Hatzfeld J.A., Otu H.H., Ng H.H., Chen J., Mu X., Chevassut T., Li X., Joseph M., Bailey C., Hatzfeld J.A., Ng H.H., Chen J., Mu X., Chevassut T., Li X., Joseph M., Bailey C., Hatzfeld J.A., Chen J., Mu X., Chevassut T., Li X., Joseph M., Bailey C., Hatzfeld J.A., Mu X., Chevassut T., Li X., Joseph M., Bailey C., Hatzfeld J.A., Chevassut T., Li X., Joseph M., Bailey C., Hatzfeld J.A., Li X., Joseph M., Bailey C., Hatzfeld J.A., Joseph M., Bailey C., Hatzfeld J.A., Bailey C., Hatzfeld J.A., Hatzfeld J.A., et al. Comment on “ ‘Stemness’: Transcriptional profiling of embryonic and adult stem cells” and “A stem cell molecular signature.”. Science. 2003;302:393. doi: 10.1126/science.1086384. [DOI] [PubMed] [Google Scholar]
- Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A., Amberger J.S., Bocchini C.A., McKusick V.A., Bocchini C.A., McKusick V.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Floss T., Van Sloun P., Fuchtbauer E.M., Vauti F., Arnold H.H., Schnutgen F., Wurst W., von Melchner H., Ruiz P., Floss T., Van Sloun P., Fuchtbauer E.M., Vauti F., Arnold H.H., Schnutgen F., Wurst W., von Melchner H., Ruiz P., Van Sloun P., Fuchtbauer E.M., Vauti F., Arnold H.H., Schnutgen F., Wurst W., von Melchner H., Ruiz P., Fuchtbauer E.M., Vauti F., Arnold H.H., Schnutgen F., Wurst W., von Melchner H., Ruiz P., Vauti F., Arnold H.H., Schnutgen F., Wurst W., von Melchner H., Ruiz P., Arnold H.H., Schnutgen F., Wurst W., von Melchner H., Ruiz P., Schnutgen F., Wurst W., von Melchner H., Ruiz P., Wurst W., von Melchner H., Ruiz P., von Melchner H., Ruiz P., Ruiz P. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Klonowski P., Kohany O., Walichiewicz J., Kohany O., Walichiewicz J., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Ko M.S., Kitchen J.R., Wang X., Threat T.A., Wang X., Hasegawa A., Sun T., Grahovac M.J., Kargul G.J., Lim M.K., Kitchen J.R., Wang X., Threat T.A., Wang X., Hasegawa A., Sun T., Grahovac M.J., Kargul G.J., Lim M.K., Wang X., Threat T.A., Wang X., Hasegawa A., Sun T., Grahovac M.J., Kargul G.J., Lim M.K., Threat T.A., Wang X., Hasegawa A., Sun T., Grahovac M.J., Kargul G.J., Lim M.K., Wang X., Hasegawa A., Sun T., Grahovac M.J., Kargul G.J., Lim M.K., Hasegawa A., Sun T., Grahovac M.J., Kargul G.J., Lim M.K., Sun T., Grahovac M.J., Kargul G.J., Lim M.K., Grahovac M.J., Kargul G.J., Lim M.K., Kargul G.J., Lim M.K., Lim M.K., et al. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development. 2000;127:1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- Medico E., Gambarotta G., Gentile A., Comoglio P.M., Soriano P., Gambarotta G., Gentile A., Comoglio P.M., Soriano P., Gentile A., Comoglio P.M., Soriano P., Comoglio P.M., Soriano P., Soriano P. A gene trap vector system for identifying transcriptionally responsive genes. Nat. Biotechnol. 2001;19:579–582. doi: 10.1038/89343. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N., Nagaoka S., Itoh M., Izawa M., Konno H., Carninci P., Yoshiki A., Kusakabe M., Moriuchi T., Muramatsu M., Nagaoka S., Itoh M., Izawa M., Konno H., Carninci P., Yoshiki A., Kusakabe M., Moriuchi T., Muramatsu M., Itoh M., Izawa M., Konno H., Carninci P., Yoshiki A., Kusakabe M., Moriuchi T., Muramatsu M., Izawa M., Konno H., Carninci P., Yoshiki A., Kusakabe M., Moriuchi T., Muramatsu M., Konno H., Carninci P., Yoshiki A., Kusakabe M., Moriuchi T., Muramatsu M., Carninci P., Yoshiki A., Kusakabe M., Moriuchi T., Muramatsu M., Yoshiki A., Kusakabe M., Moriuchi T., Muramatsu M., Kusakabe M., Moriuchi T., Muramatsu M., Moriuchi T., Muramatsu M., Muramatsu M., et al. Characterization of gene expression in mouse blastocyst using single-pass sequencing of 3995 clones. Genomics. 1998;49:167–179. doi: 10.1006/geno.1998.5209. [DOI] [PubMed] [Google Scholar]
- Schuler G.D. Pieces of the puzzle: Expressed sequence tags and the catalog of human genes. J. Mol. Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- Sharov A.A., Piao Y., Matoba R., Dudekula D.B., Qian Y., VanBuren V., Falco G., Martin P.R., Stagg C.A., Bassey U.C., Piao Y., Matoba R., Dudekula D.B., Qian Y., VanBuren V., Falco G., Martin P.R., Stagg C.A., Bassey U.C., Matoba R., Dudekula D.B., Qian Y., VanBuren V., Falco G., Martin P.R., Stagg C.A., Bassey U.C., Dudekula D.B., Qian Y., VanBuren V., Falco G., Martin P.R., Stagg C.A., Bassey U.C., Qian Y., VanBuren V., Falco G., Martin P.R., Stagg C.A., Bassey U.C., VanBuren V., Falco G., Martin P.R., Stagg C.A., Bassey U.C., Falco G., Martin P.R., Stagg C.A., Bassey U.C., Martin P.R., Stagg C.A., Bassey U.C., Stagg C.A., Bassey U.C., Bassey U.C., et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:E74. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes W.C. The identification of new genes: Gene trapping in transgenic mice. Curr. Opin. Biotechnol. 1993;4:684–689. doi: 10.1016/0958-1669(93)90050-7. [DOI] [PubMed] [Google Scholar]
- Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., Fuellen G., Gilbert J.G., Korf I., Lapp H., Gilbert J.G., Korf I., Lapp H., Korf I., Lapp H., Lapp H., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G. Stem cells. ‘Stemness’ genes still elusive. Science. 2003;302:371. doi: 10.1126/science.302.5644.371a. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., Mouse Genome Sequencing Consortium. et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wiles M.V., Vauti F., Otte J., Fuchtbauer E.M., Ruiz P., Fuchtbauer A., Arnold H.H., Lehrach H., Metz T., von Melchner H., Vauti F., Otte J., Fuchtbauer E.M., Ruiz P., Fuchtbauer A., Arnold H.H., Lehrach H., Metz T., von Melchner H., Otte J., Fuchtbauer E.M., Ruiz P., Fuchtbauer A., Arnold H.H., Lehrach H., Metz T., von Melchner H., Fuchtbauer E.M., Ruiz P., Fuchtbauer A., Arnold H.H., Lehrach H., Metz T., von Melchner H., Ruiz P., Fuchtbauer A., Arnold H.H., Lehrach H., Metz T., von Melchner H., Fuchtbauer A., Arnold H.H., Lehrach H., Metz T., von Melchner H., Arnold H.H., Lehrach H., Metz T., von Melchner H., Lehrach H., Metz T., von Melchner H., Metz T., von Melchner H., von Melchner H., et al. Establishment of a gene-trap sequence tag library to generate mutant mice from embryonic stem cells. Nat. Genet. 2000;24:13–14. doi: 10.1038/71622. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Ferro S., Gasteiger E., Huang H., Lopez R., Gasteiger E., Huang H., Lopez R., Huang H., Lopez R., Lopez R., et al. The Universal Protein Resource (UniProt): An expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz B.P., Friedrich G.A., Buxton E.C., Lilleberg S.L., Person C., Sands A.T., Friedrich G.A., Buxton E.C., Lilleberg S.L., Person C., Sands A.T., Buxton E.C., Lilleberg S.L., Person C., Sands A.T., Lilleberg S.L., Person C., Sands A.T., Person C., Sands A.T., Sands A.T. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- Zambrowicz B.P., Abuin A., Ramirez-Solis R., Richter L.J., Piggott J., Beltrandel-Rio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Abuin A., Ramirez-Solis R., Richter L.J., Piggott J., Beltrandel-Rio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Ramirez-Solis R., Richter L.J., Piggott J., Beltrandel-Rio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Richter L.J., Piggott J., Beltrandel-Rio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Piggott J., Beltrandel-Rio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Beltrandel-Rio H., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Buxton E.C., Edwards J., Finch R.A., Friddle C.J., Edwards J., Finch R.A., Friddle C.J., Finch R.A., Friddle C.J., Friddle C.J., et al. Wnk1 kinase deficiency lowers blood pressure in mice: A gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl. Acad. Sci. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]