Abstract
The genome of the gray short-tailed opossum Monodelphis domestica is notable for its large size (∼3.6 Gb). We characterized nearly 500 families of interspersed repeats from the Monodelphis. They cover ∼52% of the genome, higher than in any other amniotic lineage studied to date, and may account for the unusually large genome size. In comparison to other mammals, Monodelphis is significantly rich in non-LTR retrotransposons from the LINE-1, CR1, and RTE families, with >29% of the genome sequence comprised of copies of these elements. Monodelphis has at least four families of RTE, and we report support for horizontal transfer of this non-LTR retrotransposon. In addition to short interspersed elements (SINEs) mobilized by L1, we found several families of SINEs that appear to use RTE elements for mobilization. In contrast to L1-mobilized SINEs, the RTE-mobilized SINEs in Monodelphis appear to shift from G+C-rich to G+C-low regions with time. Endogenous retroviruses have colonized ∼10% of the opossum genome. We found that their density is enhanced in centromeric and/or telomeric regions of most Monodelphis chromosomes. We identified 83 new families of ancient repeats that are highly conserved across amniotic lineages, including 14 LINE-derived repeats; and a novel SINE element, MER131, that may have been exapted as a highly conserved functional noncoding RNA, and whose emergence dates back to ∼300 million years ago. Many of these conserved repeats are also present in human, and are highly over-represented in predicted cis-regulatory modules. Seventy-six of the 83 families are present in chicken in addition to mammals.
The complete genome sequence of a marsupial, the short-tailed opossum Monodelphis domestica (Mikkelsen et al. 2007), provides a unique opportunity to investigate the evolutionary forces that have shaped mammalian genomes. Monodelphis is the first sequenced metatherian species, and as such, provides an important target against which to compare eutherians (placental mammals) and increase the depth of our understanding of the evolution of the Amniota. Currently, it is estimated that metatherians and eutherians diverged from a common ancestor ∼170–190 million years ago (Mya). Further back, the divergence from avian and reptile taxa occurred around ∼300 My. Thus, the positioning of Monodelphis between avians and eutherians makes it invaluable for evolutionary comparisons. Furthermore, Monodelphis is the only metatherian that is commonly maintained as a laboratory stock (VandeBerg and Robinson 1997). Insights from the unusually large (∼3.6 Gb) genome sequence should provide numerous new hypotheses for experimental investigations, and hopefully illuminate previously unresolved questions. In addition to the human genome, we now have complete sequences available for mouse, rat, dog, and chicken, among others (Waterston et al. 2002; Gibbs et al. 2004; International Chicken Genome Sequencing Consortium 2005; Lindblad-Toh et al. 2005). Initial draft sequences for the cow, wallaby, and cat are also forthcoming. One of the major features of most genomes is the presence of transposable elements (TEs). Although at times dismissed as “parasitic” residents of genomes, it is increasingly recognized that TEs have been major players in shaping genomic landscapes (Brosius and Gould 1992; Kidwell and Lisch 2001; Deininger and Batzer 2002; Brosius 2003; Deininger et al. 2003).
In addition to their effects due to insertional mutagenesis, high-copy number TEs provide a substrate for illegitimate homologous recombinations, causing rearrangements that may be deleterious or advantageous (Sen et al. 2006). Deletion of genomic segments by recombination between TEs is associated with numerous human diseases, while the complementary duplication of regions provides new material for evolutionary innovation (for example, see Deininger and Batzer 1999; Edelmann et al. 1999; Bailey et al. 2002; Babcock et al. 2003). Furthermore, TEs have been exapted by their host genomes into useful roles. In some cases, such as recruitment of a Mariner transposase into the primate gene SETMAR ∼40–58 Mya (Cordaux et al. 2006), exaptation makes direct use of the coding potential of autonomous elements (TEs that can catalyze their own transposition or retrotransposition). But an increasingly recognized phenomenon is the co-opting of nonautonomous elements as functional noncoding elements (Bejerano et al. 2006; Kamal et al. 2006). This fulfills the vision originally espoused by McClintock, Davidson, and Britten, that TEs, and repetitive DNA in general, may be critical “control elements” in modern genomes (McClintock 1961; Davidson and Britten 1979). Here we investigate the impact of TEs on the Monodelphis genome, and their possible role in mammalian evolution. We primarily focus on aspects of TEs in Monodelphis that highlight differences from other species. In addition, we discuss some commonalities such as the exaptation of ancient repeats that have been highly conserved across a remarkable phylogenetic range.
Results
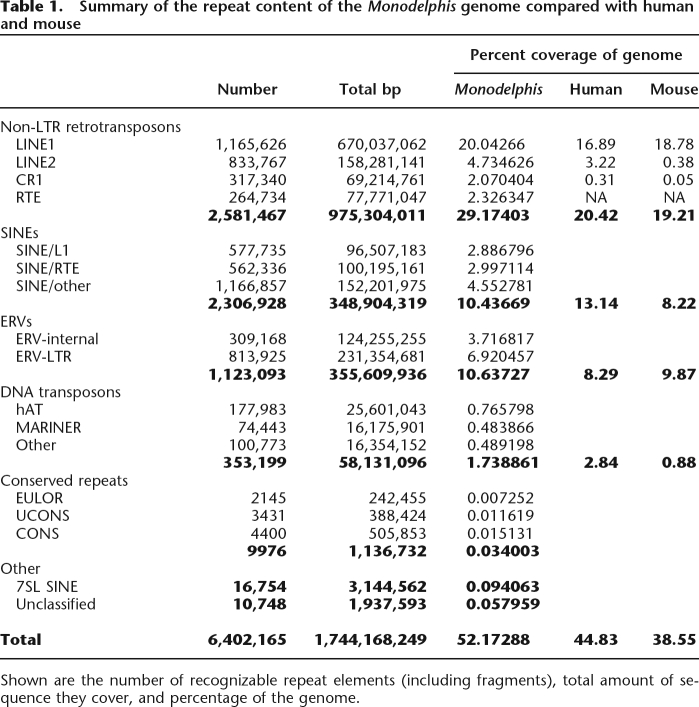
We classified nearly 500 families of interspersed repeats in the Monodelphis genome sequence data, the majority of which are newly identified. Some sequences are refinements for Monodelphis of previously identified elements included in Repbase (Jurka et al. 2005). Repeats were identified and classified using homology-based and de novo approaches, as described in the Methods. Maps of repeats were then constructed using Censor and RepeatMasker (Kohany et al. 2006; A.F.A Smit, R. Hubley, and P. Green, RepeatMasker Open-3.0 1996–2007, http://www.repeatmasker.org). Table 1 summarizes the repeat content of the current Monodelphis assembly (Mikkelsen et al. 2007), excluding contigs that lack a chromosome position. Counts and genome coverage for all families are listed in Supplemental Table 1. The human and mouse numbers are as described previously for those genomes (Lander et al. 2001; Waterston et al. 2002). The total interspersed repeat content of the Monodelphis genome identifiable by Censor is ∼52.2%, excluding simple (tandem) repeats. This is substantially higher than the corresponding proportions in human (44.83%) and mouse (38.55%). In contrast, the proportion of segmental duplications in Monodelphis (1.7% of the autosomes) is significantly lower than in human (5.2%) or mouse (5.3%). Additionally, the fraction of the genome comprising protein-coding genes is similar in Monodelphis and human (18,648 and 20,806 genes, respectively; see Mikkelsen et al. 2007, Table 3). Since human repeats are so well classified (>500 families and subfamilies in Repbase), which increases detection, the repeat content of Monodelphis relative to human may be even higher than shown.
Table 1.
Summary of the repeat content of the Monodelphis genome compared with human and mouse
Shown are the number of recognizable repeat elements (including fragments), total amount of sequence they cover, and percentage of the genome.
Non-LTR retrotransposons and associated SINEs
Monodelphis has several families of non-LTR retrotransposon that have been highly prolific, some of which have been active recently, and which may currently be retroposing in the genome. A feature of many vertebrate genomes, including human, is the high fraction generated by the activity of non-LTR retrotransposons, particularly LINE1 (L1). This domination of genomic content is also evident in Monodelphis, with an even higher proportion of the genome (20.0%) comprising L1 copies, than in human (16.9%) or mouse (18.9%). The L1–1_MD element (hereafter we drop the “_MD” from Monodelphis Repbase identifiers) shows strong evidence of recent activity: there are numerous full-length copies that are >99.5% identical to the ∼6-kb consensus sequence, and which possess intact ORF1 and ORF2 coding regions. L1–L2 is 80% similar to L1–L1, but its copies are ∼90% similar to the consensus, suggesting that it is a separate L1 that was active perhaps 60 Mya (depending on the mutation rate for Monodelphis). Furthermore, L1–L2 is the most frequently occurring of the L1 elements in the Monodelphis genome. The remaining L1s have higher divergences and are no longer active; given their higher divergence from the consensus, they are probably not Monodelphis specific, but rather were active in a common ancestor of marsupials.
The highly frequent tRNA-derived SINE element SINE-1 is most likely associated with L1, based on the fact that it has the characteristic 15–16-bp target-site duplications and poly(A) tails of L1-mediated insertions. SINE-1 has a 5′ end that is similar to leucine and serine tRNAs. Further evidence of L1 mobilization of sequences is provided by the occurrence of 16,754 7SL RNA-derived SINE loci. Trans-mobilization of other sequences, including SINE RNAs and (more rarely) mRNAs is believed to occur very soon after translation of L1 RNA on ribosomes (Wei et al. 2001). Most frequently, L1 attaches in cis to the 3′ poly(A) tail of its own mRNA transcript, which is then reverse transcribed and inserted into the genome. However, if other targets with poly(A) tails are available, L1 may capture these in trans and retrotranspose this molecule rather than its own mRNA. Localization of this process to the ribosomes naturally favors trans-mobilization of similarly located sequences, such as 7SL RNA, which is part of the signal recognition particle.
Monodelphis has at least four families of RTE-like retrotransposons, a class of non-LTR element that was originally discovered in Caenorhabditis elegans (Youngman et al. 1996) RTE is widely distributed phylogenetically, with representatives in genomes as diverse as Anopheles gambiae (mosquito), Danio rerio (zebrafish), Thalassiosira pseudonana (diatom), Strongylocentrotus purpuratus (purple sea urchin), Vipera ammodytes (Horn-nosed viper), and plants. However the phylogenetic distribution is “patchy,” with many species (including humans) entirely lacking in RTEs. Thus, the comparatively high proportion of the Monodelphis genome comprised of RTE copies (∼2.3%) is a distinguishing feature. Three of the RTE families have been clearly inactive for some time. RTE0 is an old RTE, indicated by its presence in bacterial artificial chromosome (BAC) sequences from the Tamar wallaby Macropus eugenii and high divergence of copies from the consensus sequence (∼30%). It comprises ∼1.4% of the Monodelphis genome.
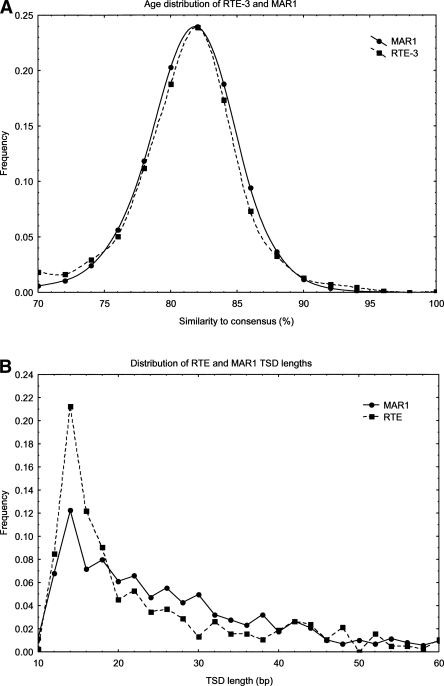
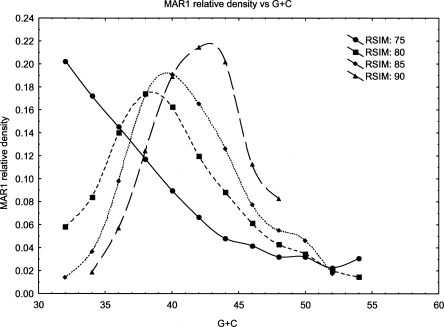
The youngest element, RTE-1 is slightly over 4 kb in length, and the consensus contains an ORF of 1108-aa length. This ORF contains domains encoding exonuclease/phosphatase activity and a reverse transcriptase; both are characteristic features of retrotransposons. There is also a small 30-aa region matching glutamyl tRNA synthetases. Full-length copies of RTE-1 average ∼95% similarity to the consensus, with a maximum identity of 96.7%. Thus, this element has probably mobilized relatively recently. The element RTE-3 has an associated SINE (MAR1) that has been highly successful in colonizing the Monodelphis genome. Mobilization of MAR1 by RTE-3 is supported by a highly similar age distribution (Fig. 1A), target-site duplication length distribution (Fig. 1B), and similarity of TSD (target site duplication) composition (∼28% G+C). TSD lengths are heterogenous for RTE elements and range in size from ∼10 bp to several hundred base pairs. Their occurrence flanking MAR1 insertions leads us to conclude that MAR1 is a true SINE element mobilized by RTE, rather than simply a deletion product of full-length RTEs, as is the case for many previously postulated RTE SINEs (Malik and Eickbush 1998). Identification of MAR1/MAR1b as deletion products of RTE-3 is further contraindicated by the fact that full-length insertions of these SINEs contain unique sequences that are not present in RTE-3. MAR1 and RTE-3 have a shared region of 100% identity between 50 bp at the 5′ end of RTE-3 and 50 bp in the central region of MAR1. The subfamily MAR1b has a region of 69 bp that is 98% identical to RTE-3, encompassing the 50-bp fragment of MAR1. We further characterized this similarity and its relationship to other known RTEs and SINE elements. The 69-bp region of near identity is shared with other BOVB-type RTE elements from Vipera ammodytes (95% identity) and cow (90% identity). Additionally, it is shared with several SINE elements from cow, notably, the Bov-tA SINEs, BTALU, and BDDF family elements. Comparison of Bov-tA2 with MAR1b_MD showed 79% identity over 60% of the SINE sequence. Outside of RTE elements and SINEs, there do not appear to be other significant matches to this 69-bp sequence. It coincides with a region of BDDF elements that has been proposed to be involved in their site-specific integration (Szemraj et al. 1995). Furthermore, we identified a smaller region of lower homology between these varied elements toward the 3′ end of the sequences. The alignments of the 5′ and 3′ regions for all elements are shown in Supplemental Figure 1.
Figure 1.
(A) Age distribution of RTE-3 and MAR1. RTE-3 and MAR1 insertions were separately split into groups according to their similarity to consensus, in bins of width 2% (horizontal axis). The vertical axis shows the proportion of RTE-3 (MAR1) elements of that age, calculated as the number of base pairs of sequence covered by elements in that similarity range divided by the total genome base pairs covered by RTE-3 (MAR1). (B) Distribution of target site duplication lengths of RTE-3 and MAR1. Length of target site duplication is shown on the horizontal axis. The vertical axis shows the frequency of TSDs of that length for RTE-3 and MAR1.
Possible horizontal transfer of RTE elements
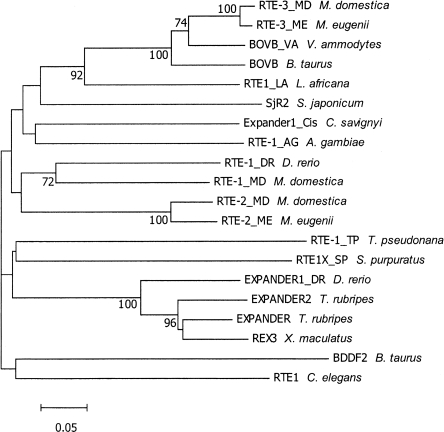
RTE-1, RTE-2, and RTE-3 have extremely different G+C contents (52.2%, 43.3%, and 39.1%, respectively), and low overall sequence similarities (maximum 60% between RTE-1 and RTE-2). This led us to investigate whether there was any evidence for horizontal transfer of this non-LTR retrotransposon, as has been hypothesized in other species (Zupunski et al. 2001). We extracted 20 reconstructed consensus sequences for RTE elements from Repbase Update. As described in the Methods, we built a multiple alignment of these sequences using DIALIGN2 (Morgenstern 1999). We generated a phylogenetic tree from this alignment using MrBayes (Ronquist and Huelsenbeck 2003) under a GTR model with gamma-distributed rate variation across sites. Convergence was achieved with a standard deviation of split frequencies <0.02, and potential scale-reduction factors of all branches deviating by <0.01 from 1.0. The resulting tree is displayed in Figure 2, which shows the Repbase name of each sequence, the originating species, and estimates of the Bayesian posterior probabilities for each branch. All labeled branch points had support of 70% or better. The topology is consistent with that previously reported for RTE elements based on protein alignments of individual intact RTE elements (Zupunski et al. 2001). RTE-3 lies within the BOVB group of RTEs, while RTE-1 and RTE-2 cluster in a distinct clade with RTEs from sea urchin and zebrafish.
Figure 2.
Phylogenetic relationship between reconstructed RTE consensus sequences. The tree was reconstructed using MrBayes, as described in Methods. Numbers at nodes indicate bootstrap support for that node (%); only support of >70% is shown.
Close relatives of RTE0, RTE-2, and RTE-3 are found in the Tamar wallaby Macropus eugenii (RTE0_ME, RTE-2_ME and RTE-3_ME). The corresponding consensus sequences in Monodelphis and Macropus (reconstructed from 22 Mb of BAC sequences available in GenBank) are ∼90% similar to each other. However, we could not find copies of RTE-1 in the available wallaby genome sequence data. We performed BLASTN searches of all available Macropus sequences in GenBank, including WGS and trace archives (totaling >4.1 Gb of sequence), but no significant hits were found. Conclusive support for the absence of RTE-1 from wallaby requires experimental assays; however, the copy number would have to be extremely low. Furthermore, if RTE-1 was active in a common ancestor of opossum and wallaby, we should be able to detect decayed copies in the wallaby genome, as for the much older RTE-0. These data are consistent with a relatively recent origin and expansion of RTE-1 in the opossum genome. Phylogenetic reconstruction of the history of L1 elements shows a pattern of clear vertical inheritance, where elements from each species are more closely related to other L1s within that species than to L1s in other species (data not shown).
Other old non-LTR elements and associated SINEs that are mammalian-wide, and represented in Monodelphis, include L2A and L2B, MIR and MIR3, and L3 (CR-1). Both L2 and L3 have been significantly more prolific in Monodelphis than in human or mouse. In particular, 2.1% of the opossum genome is identified as copies of L3 elements, which is seven times higher than the 0.3% found in human.
Endogenous retroviruses
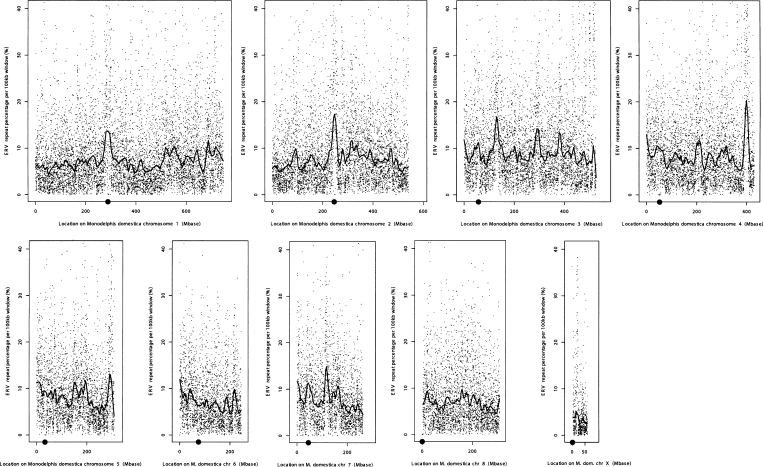
We identified at least 45 families of endogenous retroviruses (ERV; LTR retrotransposons) that have recognizable internal coding regions and complete, or largely complete, long terminal repeats. A few have been active quite recently, indicated by high similarity to their consensus sequences and intact ORFs. The total number of ERVs is not determined, but there are at least 20 other families that have significant copy numbers, together with a large number of genomic insertions showing similarity to retroviral reverse transcriptases. Thus, there may be as many as several hundred families in total. We also identified nearly 200 families and subfamilies of solo LTRs in the genome, some of which are likely to be associated with specific internal ERV elements that have not yet been identified. Neither complete ERV nor LTR insertions are correlated with local G+C content of the genome (correlation coefficient −0.06, P < 0.05). However, density of ERV insertions along chromosomes appears to be nonrandom, with several regions that are highly enriched for ERVs (Fig. 3). Notably, distinctive peaks in local ERV density on chromosomes 1 (∼288.7–291.7 Mb) and 2 (246.7–249.8 Mb) are adjacent to known positions of centromeres. The centromere locations for the genome assembly were fixed from FISH data. Whenever markers transitioned from p-arm to q-arm, the centromere was designated to lie between the last p-arm mapped scaffold and the first q-arm mapped scaffold (Mikkelsen et al. 2007). Centromeric regions themselves are generally not sequenceable, at least using WGS assembly methods, because of the high proportion of simple repeat sequences. A weaker density peak occurs on chromosome 3 adjacent to the centromeric region around 56.8–59.9 Mb. ERV densities on the remaining chromosomes do not appear to colocalize with centromeres in the current genome assembly. Interestingly, however, comparison of our ERV densities with cytological determination of centromere positions in Marsupials (Rens et al. 2003), indicated that peak ERV densities on chromosomes 3, 4, and 5 do appear to occur at the approximate locations of active centromeres. However, the resolution of the cytological data is low, and this association can only be visually estimated. The reason for the discrepancy between cytological centromere positions and those determined from the genome sequence is unclear. Densities in the distal telomeric regions of chromosomes 3 (500 Mb to end) and 4 (∼400 Mb) show extended regions that are nearly entirely comprised of fragments of ERV internal and LTR sequences. The regions of enhanced ERV density typically span 10–20 Mb (Fig. 3). Chromosomes 5, 6, and 7 also show some enrichment near the telomeres. Finally, the X chromosome shows an extended 10-Mb region of high ERV density centered around ∼21 Mb.
Figure 3.
Density of ERV insertions across Monodelphis chromosomes. The density shown is the percentage of sequence that is identified as internal ERV or LTR sequence in 100-kb segments spanning each chromosome. Centromere positions (determined from FISH data, see text) are indicated by a gray circle on the horizontal axis. Position along chromosomes is shown in megabases. The gray dots are values for each individual 100-kb segment. Black lines are a smoothed running mean. Peaks in ERV density on chromosomes 1 and 2 correspond to centromere locations. Prominent peaks are also found on chromosomes 3–5, but do not correspond to centromeric regions in the genome assembly (Mikkelsen et al. 2007); however, they are roughly consistent with locations of cytologically determined centromere activity reported in the literature (Rens et al. 2003).
Among internal ERV regions that are substantially intact, the most common and youngest is that corresponding to ERV2. This element has 244 copies with >85% of the full consensus length intact, with average similarity to the consensus of 98.2%. Five other ERVs have insertions of internal regions with copy number of 40 or above, and average similarity to their consensus of >96%. ERV1, ERV2, ERV3, ERV4, ERV9, ERV11, and ERV16 have portions of intact ORFs exceeding 1000 aa in length, and a total of 42 ORF fragments of >500 aa are identifiable from other consensus sequences of ERV. There is evidence for exchange of LTR sequences between different ERVs. The same LTR is sometimes found in separate full-length ERV insertions, with alternative internal coding regions. Conversely, coding parts of ERVs may utilize more than one LTR sequence. For example, the internal sequence of ERV18 (6250 bp) occurs in full-length insertions with two different LTRs of lengths 322 and 336 bp; similarly, ERV12 has alternative LTRs of 769 and 841 bp. There is one example, ERV6, where the element appears to be able to utilize three different LTRs, of lengths 510, 576, and 700 bp. Such chimeric structures have been observed in a few human ERVs, but the extent of its occurrence in Monodelphis seems to be novel.
It was recently reported that the koala retrovirus (KoRV) appears to be currently invading the host koala genome as an endogenous retrovirus (Tarlinton et al. 2006). We were interested to see whether this process could also be occurring in Monodelphis. Unfortunately, no complete sequences of exogenous retroviruses are available for Monodelphis; however, we found that the internal part of ERV10 spans the whole of a 932-bp fragment sequenced from the RV Opossum retrovirus (GenBank accession no. AJ236123), with 94% nucleotide identity (see alignment in Supplemental Figure 2). RVOp is a Gammaretrovirus, most closely related to Murine Leukemia Virus (translated BLAST search E-value of 2.10−64).
Ancient LINE/SINE repeats and DNA transposons
Together with the L2, L3, and MIR elements, old DNA transposons, particularly of Mariner and hAT classes, occur frequently, comprising a small, but significant percentage of the genome. Since these are generally mammalian-wide sequences, most have already been characterized in Repbase. L2 has been slightly more active in the marsupial lineage, covering 4.7% of the genome (compared with 3.2% in human and 0.38% in mouse). L3/CR1 is significantly more prominent in Monodelphis, with recognizable insertions comprising 2.1% of the genome. This is seven times higher than in human (0.3%), and 42 times higher than in mouse (0.05%). There is evidence of Mariner activity (both historical and recent) in Monodelphis, with at least 70,000 insertion loci of nonautonomous elements. In addition, there appear to be at least two autonomous Mariners; one of which has a largely intact ORF, although the TIRs (terminal inverted repeats) appear to be damaged, and it is not clear whether it is still mobile. In total, Mariner copies account for 0.5% of the genome (∼74,400 insertions). Two putative families of autonomous hAT DNA transposons are present in the genome, with mean identity to their consensus sequences of 93% (Hat1) and 94% (Hat2). Together with nonautonomous elements of varying age, there are nearly 178,000 hAT transposons insertions in Monodelphis (0.77% of the genome). Many are mammalian-wide, such as the CHARLIE and CHAPLIN elements (Smit and Riggs 1996), and are represented only by heavily mutated copies. In addition to Mariner and hAT, we identified 100,773 apparent nonautonomous DNA transposon insertions, whose superfamily could not be identified. Their classification is based on the presence of terminal inverted repeats (TIRs) and 2-bp target site duplications (TSDs). The total genomic content of DNA transposons is ∼1.73%. This is lower than the 2.84% found in humans, which is likely to be due to the fact that many more low copy-number elements (typically with less than a few hundred insertion loci) have been reconstructed in human. Finally, we found seven families of interspersed repeat (10,748 insertions), which we were unable to classify.
Distributions
L1s and their associated SINE-1/SINE-2 elements in Monodelphis show a very similar pattern of integration to human L1/Alu elements. Human L1 has a preferred target site for integration (TT-AAAA), and preferentially integrates into A+T rich regions of the genome. Alus mirror this distribution upon initial integration, but over time, accumulate in more G+C rich regions (Lander et al. 2001). Human L1s do not shift in G+C with age. L1 and its counterparts SINEs, SINE1, and SINE2, demonstrate the same behavior in Monodelphis as in human (Gu et al. 2007); namely, L1 integrates preferentially into A+T-rich regions and remains there, while SINE1 and SINE2 accumulate in G+C-rich regions of the genome. MAR1, surprisingly, behaves in an opposite manner to Alu i.e., young elements are biased toward G+C regions, and shift to more A+T-rich DNA with time (see Fig. 4). Also, whereas this shift has already occurred for Alus that are 2%–3% diverged from their consensus, the process with MAR1 appears more gradual and progressive. We believe that the SINE MAR1 is mobilized by RTE-3, as discussed above. In order to check that associations between TE densities and local G+C content were not tautological (L1 is itself A+T rich, SINEs tend to be G+C rich), we also examined TE density as a function of (1) local G+C content of genomic sequence that was not masked out as repetitive, and (2) G+C content at the third codon position of genes. In all cases, the density distribution of TEs was essentially unchanged relative to G+C (data not shown).
Figure 4.
Distributions of the RTE-mobilized SINE MAR1 across G+C ranges in Monodelphis. Distribution across G+C regions of the Monodelphis genome of MAR1 (putatively RTE-3 -mobilized). The horizontal axis shows G+C content in 5% bins, while the vertical axis shows the normalized densities of the TEs in that bin. For each TE, we categorized elements by age according to their similarity to their consensus sequence (“RSIM” in the legend) and plotted the distribution separately for each. RSIM = 70% indicates similarity to the consensus of 70%–75%, RSIM = 75% indicates 75%–80%, etc. Normalization of TE densities is described in Methods.
Conserved repeats
There has been considerable recent interest generated by the discovery that several ancient TEs have been exapted as noncoding functional elements in vertebrate genomes (Bejerano et al. 2006; Kamal et al. 2006; Nishihara et al. 2006). We identified 76 previously unknown families of repetitive sequences that are present in mammals and chicken. Within these, there are four major groups: Eulor (20 families) containing conserved secondary structures, UCONS (31 families) without any additional diagnostic features apart from multicopy number, and 12 MER elements (MER123, 125, 126, 127, 129, 130, 131, 132, 133A, 113B, 134, and 136), which appear to be derived from putative nonautonomous DNA transposons, and ancient SINE elements. The remaining 13 families are fragments of diverse LINE elements with common names X*_LINE, where the asterisk stands for family or subfamily identification. In addition, we found seven new families present in mammals, but not in chicken (MER124, 128, 135, MARE1–3, and one LINE derived family X3_LINE). All of these families have been deposited in Repbase (see also Supplemental Table 2).
These 83 elements are present in 18,290 copies in Monodelphis, compared with 11,488 copies in the human genome, with 3512 copies localizing in previously identified evolutionarily conserved regions in vertebrate genomes (Siepel et al. 2005). The conserved regions identified by Siepel et al. represent 4.75% of the human genome and encompass >30% of all repeat insertions from the newly described families. The genomic copy numbers can vary somewhat with different search parameters, but they are systematically higher in Monodelphis than in the human genome by 40%–60%, and their corresponding proportions in the evolutionarily conserved regions remain five to six times higher than expected for the human genome. All identified families are dispersed on different chromosomes, which strongly suggests that they spread by transposition. This is underscored by the finding that 14 of them either preserved ORFs of LINE families or are significantly similar to LINE-derived families. Two previously identified families are classified as SINE elements (Bejerano et al. 2006; Nishihara et al. 2006)
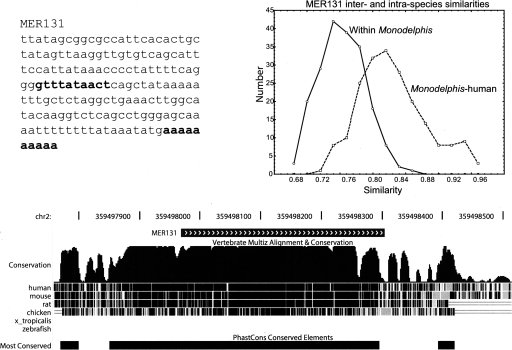
We performed a more detailed analysis of a new putative t-RNA SINE element, MER131, which contains an internal RNA polIII Box-B promoter sequence (consensus GWTYRANNCY), and a poly(A) tail (Fig. 5). These are typical characteristics of a LINE-mobilized SINE, although the age of the repeat copies precludes identification of target-site duplications. There are 885 copies of MER131 in the Monodelphis genome, with mean pairwise similarity between copies of 73%. The March 2006 assembly of the human genome (NCI Build 36.1) has 517 MER131 insertions. To examine the degree of conservation of MER131 at syntenic positions of human and Monodelphis, we extracted MER131 copies plus 100 bp flanking their 5′ and 3′ ends for the 517 human and 885 Monodelphis sequences. Pairwise alignments were constructed for each possible pair of sequences within Monodelphis, and between Monodelphis and human, using SWAT (P. Green, unpubl.). We extracted the 200 highest scoring alignments for the inter- and intraspecies alignments, and plotted the distribution of similarities between aligned sequences (Fig. 5). We found that MER131s are more highly conserved between their syntenic positions in the Monodelphis and human genomes (mean similarity 82%) than they are within Monodelphis (73%), which is consistent with non-neutral evolutionary constraints. Synteny was inferred based on the preservation between species of the 100-bp flanking sequences, which are unique to each insertion within a particular genome. The highest pairwise similarity between elements within Monodelphis was only 82%, and does not include flanking sequences. The pairwise comparisons between Monodelphis and human showed 68 instances where between-species similarities of MER131 insertions plus their flanking sequence exceeded 82%, with a maximum identity of 94.3%. A total of 20 insertions plus their flanking regions were >90% identical between human and Monodelphis. We compared the above 68 MER131 sequences from Monodelphis to chicken. In five cases, the element had split in the middle and dispersed on different chromosomes in chicken. There were 38 cases in which the Monodelphis sequence was found in chicken with at least one of the 100-bp flanking sequences intact (usually at the 5′ end). In 10 cases, the similarity between chicken and Monodelphis MER131s was higher than the similarity between Monodelphis and human.
Figure 5.
Sequence and conservation of the exapted SINE element MER131. (Top left) The MER131 consensus sequence. The putative Box-B promoter and poly(A) tail are highlighted in bold. (Top right) The distribution of pairwise similarities of the 200 most conserved MER131 sequences both within Monodelphis, and syntenic regions of Monodelphis-human. (Bottom) A MER131 insertion on chromosome 2, with 100-bp flanking sequence either side and degree of conservation across Monodelphis, human, mouse, rat, and chicken (the region shown is chr2: 359,497,570–359,498,703 from the UCSC genome browser Opossum January 2006 assembly). The MultiZ alignment score across all species is shown in black. Gray shaded areas are phastCons scores between Monodelphis and the individual species. The blocks labeled “Most Conserved” are predicted by phastCons (Siepel et al. 2005).
A specific instance of a MER131 insertion and its flanking sequence from Monodelphis chromosome 2 (spanning positions 359,497.98–359,498.30 Mb) is shown in Figure 5, with conservation to other species. It forms part of a region that is conserved, with high phastCons score (Siepel et al. 2005), across Monodelphis, human, mouse, rat, and chicken, but is absent from Xenopus tropicalis and Zebrafish. Additional searches of NCBI whole genome shotgun (WGS) sequences with discontiguous megablast revealed that this MER131 insertion is preserved in other mammalian species, with similarly high conservation (data not shown). However, MER131 is completely absent from available sequence data for zebrafish, pufferfish, and Tetraodon nigriviridis. To investigate whether MER131 tended to be associated with genes, we examined whether they were unusually likely to occur within 10 kb upstream of predicted coding regions (Mikkelsen et al. 2007). We were not able to find any such enrichment of MER131 in proximity to genes (data not shown).
In addition to MER131, the less-ancient SINE element, MARE3 reported here, is also abundant, and is present in >1400 copies in Monodelphis and >500 copies in the human genome. It is present in mammals only and its density in human conserved regions is approximately five times higher that the overall human genomic density (see Supplemental Table 2).
Conserved repeats in cis-regulatory modules (CRMs)
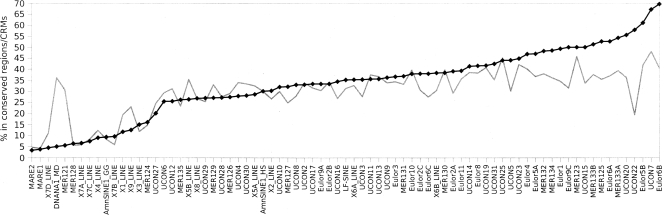
We analyzed the distribution of the repetitive families described above amongst potential cis-regulatory regions and overlapping with evolutionarily conserved regions. First, we selected 77 elements present in at least 15 copies in the human and Monodelphis genomes. They include a subset of 72 sequences from those described above (see also Supplemental Table 2), and five previously reported in the literature: LF-SINE, MER121 (DNANA1_MD), AmnSINE1_G, AmnSINE1_H (Nishihara et al. 2006). Using Censor (Kohany et al. 2006), we screened these repeats against the Monodelphis and human genome sequences, as well as against a data set of evolutionarily conserved sequences identified using the program phastCons (Siepel et al. 2005) and a recently published database of computationally predicted CRMs (Blanchette et al. 2006), representing 4.75% and 2.9% of the human genome, respectively. Around 41% of sequences predicted as CRMs lie within phastCons predicted regions, and 31% of phastCons predicted sequences lie within predicted CRMs (Blanchette et al. 2006). Overall, the 77 families are represented by 2617 copies in the CRMs, and 4312 in the evolutionarily conserved regions. Given the 13,287 copies of these repeats in the entire human genome and assuming a uniform genomic distribution, the corresponding expected numbers are 385 and 631. The distribution of repeats from individual families ranges from barely above expectation to >20 times higher than expected (Fig. 6). Some of the least abundant repeats in CRMs (e.g., MER121, X9_LINE) are relatively abundant in conserved regions and vice versa (e.g., UCON22, Eulor6B). This points to the potential diagnostic value of certain repeats in distinguishing between CRMs and other conserved regions.
Figure 6.
Interspersed repetitive elements in cis-regulatory modules (CRMs) and evolutionarily conserved regions. The Y-axis shows the percentage of 77 human interspersed repeats (listed below the X-axis) in CRMs (black diamonds/line), compared with normalized proportions of the same repeats (gray line) in evolutionarily conserved regions (Siepel et al. 2005).
Discussion
In comparison to other mammalian genome assemblies, Monodelphis has been subjected to even greater bombardment by TEs. The total identifiable genomic contribution of TEs is ∼52.2% in the opossum, compared with 44.8% for human and 38.6% for mouse (Table 1). The difference is largely due to the proliferation of LINE-type transposons in Monodelphis (29.1% of the genome, compared with 20.4% and 19.2% in human and mouse, respectively). Two percent of this contribution is due to the presence of RTE non-LTR transposons, which are not found in many other species (including human, mouse, rat, and dog). The fraction of the Monodelphis genome composed of segmental duplications is significantly lower (1.7%) than in human (5.2%), while the protein coding component is similar, or slightly less in opossum. The reason that Monodelphis is such fertile ground for TEs is unknown, but they appear to be able to account for a significant part of the excess size of the ∼3.6-Gb genome (compared with ∼3.1 Gb for human, 2.6 Gb for mouse, and 3.0 Gb for the platypus, Ornithorynchus anatinus). It has been shown previously that the recombination rate in Monodelphis is low compared with other mammalian species (Samollow et al. 2004), and it has been postulated that this may, in part, account for the extremely low CpG relative abundance (0.13 averaged across opossum chromosomes, compared with 0.23 in human). We believe that the low rate of crossing-over likely plays a significant role in the retention or preservation of TE insertions in Monodelphis. Deletions by direct homologous recombination, and the general genomic “churning” produced by the process lead to removal or obfuscation of TEs. Therefore, their longer persistence in Monodelphis would be a reasonable corollary of reduced recombination rates.
TEs have undoubtedly played a major role in shaping vertebrate genomes, and continue to do so. They are responsible for numerous human diseases and syndromes, due to their potential for mutagenic insertions (for example, retroviral induction of oncogenes such as v-src), and for providing a substrate for illegitimate homologous recombination (Deininger and Batzer 1999). In an evolutionary context, it has been shown that they can provide the material for emergence of new genes (Schmitz et al. 2004; Kapitonov and Jurka 2005; Cordaux et al. 2006) and have been utilized to more finely dissect species phylogenies (Kriegs et al. 2006). Increasingly, however, it seems that their major role may have been in influencing genetic control mechanisms such as transcription. There are now several instances reported of TEs being exapted as functional noncoding RNAs (Bejerano et al. 2006; Kamal et al. 2006; Nishihara et al. 2006), and we found additional examples of exaptation in the process of annotating the Monodelphis genome (see also Mikkelsen et al. 2007). The 83 new families of repeats from the MER, Eulor, XLINE, and UCONS families total 18,290 genomic insertions. As shown in Figure 6, a significant, but family-dependent proportion of these insertions overlap with previously identified evolutionarily conserved regions and predicted cis-regulatory modules.
For example, the ancient SINE MER131 (Fig. 5), shows strong evidence of having been exapted into a functional role. The insertion locations of MER131 are highly conserved among human, Monodelphis, and chicken genomes, but are completely absent from more distant species such as zebrafish and frog. This is consistent with the emergence of the MER131 element after the divergence of amphibians and Amniota, but preceding the reptile–mammalian divergence, i.e., ∼350–290 Mya (the Carboniferous era). Given the evolutionary distance among Amniota lineages (∼190 My since the divergence of metatherians and eutherians), it is remarkable that the homologies between so many copies of MER131 are identifiable, since they should be unrecognizable due to random point mutations. The fact that hundreds of copies are present and highly conserved across a range of species suggests that insertions of this SINE element may have been selected for a functional role in many genomic regions and had a broad distribution prior to exaptation. This potential exaptation might have occurred on a small number of elements and then been spread by genomic duplications. However, we observed that regions flanking MER131 insertions are conserved interspecies, but not intraspecies, which supports transposition of the elements prior to exaptation occurring and possible reduction of the element’s distribution by selection. We were not able to find any enrichment for MER131 in proximity to genes. However, it is known that enhancers and other regulatory elements can be as far as 1 Mb from the gene that they regulate, and roles in domain level processes are also possible. Therefore, elucidation of the functional role of MER131 and other conserved elements will require further experimental study.
In addition to MER131, insertions of ancient LINEs and DNA transposons are conserved across species, which is again suggestive of a selective constraint acting against their degradation or loss. It is interesting to speculate that many of the “evolutionarily conserved” regions that have been identified across a wide phylogenetic range (Siepel et al. 2005), as well as “ultraconserved” regions (Bejerano et al. 2004) (for review, see Bejerano et al. 2005) may eventually be identified as having been derived from ancient TEs. The conserved elements discussed here do not overlap with ultraconserved regions, but the ancient TE LF-SINE has been shown to act as a distal (∼500 Mb) enhancer—the first demonstration of a functional role for such an element (Bejerano et al. 2006). The potential for modulation of transcriptional control by SINEs, LINEs, and ERVs is clear, since they incorporate internal transcriptional promoter sequences and can be precursors of transcription-regulation signals (Thornburg et al. 2006). The recent demonstration of post-transcriptional gene regulation by Alu elements (Hasler and Strub 2006) shows that this is an ongoing process, not one relegated to the evolutionary past. The role of DNA transposons is not yet known; however, their sequence structure (with often large terminal-inverted repeats) leads to hairpin structures that are recognized by DNA transposases, and which could well be exapted for other purposes (Posey et al. 2006).
We propose that many ancient TEs localized in cis-regulatory modules became recruited as conserved elements due to advantageous modifications of the regulation process. They are preserved as recognizable modules that can be classified and used for further analysis of the composite structure of human transcription regulation. Many of the regulatory modules are tissue specific (Blanchette et al. 2006). This further implies a role for TEs in the evolution of multicellular organisms. Identification and classification of DNA repeats conserved in regulatory modules may help to decipher the detailed steps in evolution of vertebrate tissue structures. Intriguingly, recent work supports the idea that retrotransposon expression under stress conditions could initiate or drive speciation in hybrid plant species (Ungerer et al. 2006). This is likely to be associated with modification of regulatory sequences as proposed >30 yr ago by King and Wilson (1975). Identification and classification of DNA repeats conserved in regulatory modules may help to decipher the role of TEs in evolution of vertebrate tissue structures and possible impact on speciation.
The possibility of horizontal transfer of RTE sequences among species has been posited previously (Zupunski et al. 2001). While many families of TEs such as LTR-retrotransposons and Mariner DNA transposons are thought to be capable of exogenous movement, other non-LTR transposons such as L1 are not (Gueiros-Filho and Beverley 1997; Jordan et al. 1999). Unlike ERVs, which can potentially encode a retroviral-like envelope protein, there is no mechanism known for horizontal transfer of RTE elements. RTE-1 could simply be a younger RTE that was successful in proliferating in Monodelphis, but died out in related species including wallaby. It is highly diverged from the other RTE elements in Monodelphis, however, and would have to have arisen as a new subfamily from another RTE, then rapidly evolved away from it in sequence. Moreover, RTE-1 is more similar to RTEs in other species than to RTEs from other families in Monodelphis (Fig. 1); however, it is impossible to formally rule out the possibility of concerted evolution in different lineages. The small number of distinct RTE families compared with L1, and the fact that RTE-2 and RTE-3 clearly went extinct at different times, support the idea that RTE is less “robust” than L1 and is more prone to losing its capability to proliferate. Although care must be taken in invoking horizontal transfer of TEs (Capy et al. 1994), given the evidence, we believe it is the most parsimonious explanation.
In the human genome, Alus (as with their mobilizing L1 counterparts) are initially concentrated in A+T-rich genomic regions (but see Cordaux et al. 2006), at least in part because there are more TT-AAAA consensus integration sites available in such regions; but, over time, they accumulate in GC-rich regions. The most plausible mechanism proposed is that Alus in A+T-rich regions are preferentially removed by recombination, since gene densities are lower in A+T-rich areas of the human genome (Pavlicek et al. 2001). It is noteworthy that MAR1, which appears to have been mobilized by RTE, shows the opposite pattern to Alus in human, i.e., that older MAR1 copies are located in more A+T-rich genomic regions than younger MAR1s (Fig. 4). It is hard to see how the behavior of MAR1 can be explained by a similar recombination mechanism. One possibility is that the integration preferences of the mobilizing RTE elements have changed with time. Little is known about the integration process for RTE, in comparison with L1, which has been extensively characterized (Feng et al. 1996).
The nonrandom distribution of ERVs on Monodelphis chromosomes is highly pronounced, particularly on chromosomes 2, 3, 4, and X, where local densities for 100-kb windows can exceed 50%, with densities for 50-kb regions approaching 100% (Fig. 3). Peaks in ERV density on chromosomes 1, 2, and 3 correspond closely to locations of centromeres in the genome assembly. We also found strong local enrichment of ERV fragments in some telomeric regions, and for a 10-Mb region around position 20.1 Mb on chromosome X (Fig. 3). The observed distribution of ERV elements appears multifaceted, self-reinforcing (in that high densities appear to have spread over wide regions of the sequence), and partially stochastic. A plausible explanation for such accumulation is that once a TE occupies a specific locus that is safe from deletion, multiple additional or nested insertions in the same region are unlikely to be selected against. Centromeres (and some telomeres) are recognized to have significantly low recombination rates compared with the genome average. This was first noted nearly 80 yr ago (Beadle 1932), and more recent studies have found that recombination rates around centromeres are suppressed by factors of ∼10–40 (Centola and Carbon 1994; Jackson et al. 1996; Mahtani and Willard 1998). Heterochromatin structure around centromeres at meiotic crossing-over may be partially responsible for reduced recombination, but a general suppression by centromeric activity is also possible.
The formation and maintenance of heterochromatin at centromeres and telomeres, and its association with high TEs, has previously been noted (for review, see Grewal and Jia 2007). Recently, Ferreri et al. demonstrated that insertions of KERV (Kangaroo endogenous retrovirus) are present at all active centromeres of Macropus eugenii (Ferreri et al. 2004, 2005), and similar results are seen in human (Dehal et al. 2001). It is tempting to speculate that the regions of high ERV density in Monodelphis indicate the location of ancient centromeres and neocentromeres, or that they could play a role in centromere repositioning. Demethylation and reactivation of ERVs has been implicated in chromosome remodeling in mammalian hybrid species (O’Neill et al. 1998). Evolutionary break points and fusions may also play a role, and independent fission at ancient fusion points in different marsupial lineages suggests that repeat-element distributions may be important factors in marsupial chromosome evolution (Ferreri et al. 2004). The detailed repeat distribution provided by the Monodelphis domestica genome combined with the forthcoming tammar wallaby genome will provide the basis for the detailed comparative analysis of marsupial karyotypes required to rigorously test these theories.
Methods
Identification of TEs
We used a combination of similarity-based and de novo methods to reconstruct the TEs of Monodelphis. Approaches based on similarity to known elements is effective for autonomous (coding) elements, while de novo methods are useful for identifying nonautonomous elements with little similarity to known repeats.
Autonomous elements
The Monodelphis genome was screened against selected protein sequences from autonomous elements in Repbase using Censor with TBLASTN and default parameters. The use of TBLASTN against protein sequences, rather than TBLASTX against DNA sequences of known repeats, generally results in cleaner extraction of putative coding sequences. Fragments of repetitive elements detected with TBLASTN searches were grouped according to their major class (L1, RTE, endogenous retrovirus, Mariner, etc.) and then approximately clustered according to their similarity to each other. A simple clustering approach was used:
The set of all fragments was ordered according to their length.
The first (seed) sequence was taken as a reference, and all other sequences were compared with it using Censor (BLASTN).
Sequences that hit the initial seed sequence were grouped with it if they were at least 75% similarity over 50% of their length and removed from the overall sequence set.
The largest remaining sequence was taken as the seed for a new search, and steps 2–5 were repeated until no further clustering occurred
Majority consensus sequences for each repeat family were constructed based on multiple alignments of each cluster using MAFFT (Katoh et al. 2005). Using these Monodelphis-specific consensus sequences, the genome was then rescreened using Censor in default mode. Newly discovered sequences that significantly matched the consensus were then extracted along with flanking regions, and new alignments and updated consensus sequences determined as before. For young elements that were highly similar to their consensus sequence, this was sufficient, but (if necessary) consensus sequences were further refined using the more accurate LINSI module of MAFFT. This works well for TEs that are ∼80% or more similar to their consensus.
To improve the consensus sequence for older, more diverged repeats, a more computationally intensive approach was followed:
Each sequence, in turn, was taken as a seed to which all others were aligned using the SWAT implementation of the Smith-Waterman alignment algorithm (P. Green, unpubl.).
For each alignment, a majority-rule consensus was built.
After all possible consensus sequences had been constructed, each was, in turn, used as a reference to which the TE copies were aligned (again using SWAT), and the consensus sequences with the highest net SWAT scores were selected.
Steps 1–3 were repeated using these best consensus sequence, rather than the original TE copies, until the overall best consensus sequence was acquired.
In practice, this method sometimes identified related subfamilies of repeats for which a unique best consensus did not emerge, but rather several. Due to their domination of the TE landscape in mammals, L1s and endogenous retroviruses (together comprising nearly 30% of Monodelphis genomic DNA) were identified first, followed by RTE elements, then the less frequent DNA transposons. The genome was masked against these TEs using Censor before further processing. This ensures that fragments of these elements are not continuously re-identified in subsequent stages.
Nonautonomous elements
Some nonautonomous elements, notably SINEs and DNA transposons, can be found by similarity methods as above. These were identified by comparison to Repbase and masked from the genome. Although nonautonomous sequences lack coding regions for comparison, they still have homology with, for example, tRNAs and promoter regions (such as the SINE BOXB promoter sequences) that are characteristic of individual families of elements. However, many nonautonomous elements are expected to be specific to marsupials, or not represented in Repbase due to high levels of divergence. To find these, we used the masked genomic sequences as input to RepeatScout (Price et al. 2005). This algorithm does an initial search for over-represented DNA words and expands them in the 5′ and 3′ direction to identify the repeat of which they are part. RepeatScout has the advantage of being fast and memory efficient, and can handle relatively large amounts of genomic sequence. On a two-processor dual-core 3GHz Xeon system with 8 Gb of memory running Linux, 100 Mb of sequence could be processed overnight. One drawback of RepeatScout is that it can produce highly redundant output, and it does not always merge related fragments from the same repeat. We therefore used the output as a “library” of new repeats against which to screen the genome with Censor, and constructed consensus sequences using the same similarity-based methods as for autonomous elements.
Masking of genomic sequence, and determination of repeat copy number
In the first stage, the Monodelphis version 4 assembly was masked using Censor in normal sensitivity mode, with no identification of simple repeats, against the complete library of Monodelphis repeats, together with older mammalian-wide repeats from Repbase and additional Monodelphis-specific L1 sequences from the RepeatMasker library: censor4.2 Monodelphis_genome –lib Monodelphis_library –nosimple –nofound. In the second stage, the masked output from Stage 2 was run against this library using Censor in sensitive mode, with identification of simple repeats enabled: censor4.2 Stage2 –lib Monodelphis_library –nofound –mode sens. A two-stage approach is somewhat faster than a single run in sensitive mode, since easily identifiable and highly frequent repeats (such as L1 and SINEs) are found in the first stage and masked out for stage 2. This also ensures that no TE fragments are missed due to artifacts of the defragmentation algorithm. Finally, we screened the resulting output for additional tandem repeats using Tandem Repeats Finder with the options: trf400 stage3 2 7 7 80 10 50 2000 –h.
Reconstruction of phylogeny of RTE elements
The May 2006 release of Repbase contained 22 RTE elements, of which two (BTALU2 and CELE45) are small fragments, which we discarded. The remaining sequences were aligned using DIALIGN2–2 (Morgenstern) using the “-nt” parameter, which improves the nucleotide alignment by assuming that the sequences are potentially coding, and using information on conservation of putative peptides in open reading frames. The resulting alignment was visually inspected, and poorly aligned regions were removed. We then used MrBayes (Ronquist and Huelsenbeck 2003) to reconstruct the phylogenetic relationship between RTE elements. We used the General Time Reversal (GTR; Tavare 1986) model included in MrBayes, which allows for six substitution rates between nucleotides. The analysis was run for 150,000 generations, with sampling every 100 generations (1500 samples). Convergence was attained with standard deviation of split frequencies ∼0.015, and all branch potential scale reduction factors approached unity. A consensus tree with branch lengths and posterior estimates of branch probabilities was generated with the “sumt” command of MrBayes and “burnin” parameter of 375 (25% of 1500 samples).
Distribution of TEs across G+C regions
The TE densities were normalized as follows: We split the genome into 50-kb segments, and calculated G+C contents for each. These were then assigned to bins of 5% G+C range (30%–35%, 35%–40%, etc.). Repetitive elements were grouped by age, according to similarity to their respective consensus sequences. Their densities in each G+C range were then calculated as the percentage of sequence bases covered by that repeat/age group, relative to the total number of bases covered by the same repeat/age combination across all G+C ranges. For example, the relative density of SINE-1 with similarity >95% to the consensus (SINE195) in the G+C range 30%–35% (GC30) is the total base pairs of SINE195 in GC30 divided by the total base pairs of SINE195 in all G+C bins. This normalizes density across age and G+C contents.
Acknowledgments
We thank Evan Mauceli for clarification of determination of centromere locations in the genome assembly; the Tammar Wallaby Sequencing Consortium for permission to use their WGS sequences to investigate whether RTE-1 is present in Macropus eugenii; and three anonymous referees for their insightful comments. This research was supported by National Institutes of Health grants 5 P41 LM006252-09 (J.J.), R33GM065612 (D.D.P.) and RO1GM59290 (M.A.B.); National Science Foundation grants BCS-0218338 (M.A.B.) and EPS-0346411 (M.A.B. and D.D.P.); and the State of Louisiana Board of Regents Support Fund (M.A.B. and D.D.P.). M.J.W. is supported by an Australian Research Council APD fellowship DP0450066.
Footnotes
[Supplemental material is available online at www.genome.org]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6070707
References
- Babcock M., Pavlicek A., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Pavlicek A., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Shaffer L.G., Jurka J., Morrow B.E., Jurka J., Morrow B.E., Morrow B.E. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 2003;13:2519–2532. doi: 10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Beadle G.W. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. 1932;18:160–165. doi: 10.1073/pnas.18.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano B., Haussler D., Blanchette M., Haussler D., Blanchette M., Blanchette M. Into the heart of darkness: Large-scale clustering of human non-coding DNA. Bioinformatics. 2004;20(Suppl 1):I40–I48. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Siepel A.C., Kent W.J., Haussler D., Siepel A.C., Kent W.J., Haussler D., Kent W.J., Haussler D., Haussler D. Computational screening of conserved genomic DNA in search of functional noncoding elements. Nat. Methods. 2005;2:535–545. doi: 10.1038/nmeth0705-535. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Salama S.R., Rubin E.M., Kent W.J., Haussler D., Rubin E.M., Kent W.J., Haussler D., Kent W.J., Haussler D., Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Deblois G., Giguere V., Ferretti V., Bergeron D., Giguere V., Ferretti V., Bergeron D., Ferretti V., Bergeron D., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- Brosius J., Gould S.J., Gould S.J. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc. Natl. Acad. Sci. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P., Anxolabehere D., Langin T., Anxolabehere D., Langin T., Langin T. The strange phylogenies of transposable elements: Are horizontal transfers the only explantation? Trends Genet. 1994;10:7–12. doi: 10.1016/0168-9525(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Centola M., Carbon J., Carbon J. Cloning and characterization of centromeric DNA from Neurospora crassa. Mol. Cell. Biol. 1994;14:1510–1519. doi: 10.1128/mcb.14.2.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R., Udit S., Batzer M.A., Feschotte C., Udit S., Batzer M.A., Feschotte C., Batzer M.A., Feschotte C., Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl. Acad. Sci. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.H., Britten R.J., Britten R.J. Regulation of gene expression: Possible role of repetitive sequences. Science. 1979;204:1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Dehal P., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Land M., Terry A., Ecale Zhou C.L., Rash S., Terry A., Ecale Zhou C.L., Rash S., Ecale Zhou C.L., Rash S., Rash S., et al. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A., Batzer M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A., Batzer M.A. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H., Moran J.V., Batzer M.A., Kazazian H.H., Batzer M.A., Kazazian H.H., Kazazian H.H. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Edelmann L., Pandita R.K., Morrow B.E., Pandita R.K., Morrow B.E., Morrow B.E. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 1999;64:1076–1086. doi: 10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Moran J.V., Kazazian H.H., Boeke J.D., Moran J.V., Kazazian H.H., Boeke J.D., Kazazian H.H., Boeke J.D., Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Ferreri G.C., Marzelli M., Rens W., O'Neill R.J., Marzelli M., Rens W., O'Neill R.J., Rens W., O'Neill R.J., O'Neill R.J. A centromere-specific retroviral element associated with breaks of synteny in macropodine marsupials. Cytogenet. Genome Res. 2004;107:115–118. doi: 10.1159/000079580. [DOI] [PubMed] [Google Scholar]
- Ferreri G.C., Liscinsky D.M., Mack J.A., Eldridge M.D., O'Neill R.J., Liscinsky D.M., Mack J.A., Eldridge M.D., O'Neill R.J., Mack J.A., Eldridge M.D., O'Neill R.J., Eldridge M.D., O'Neill R.J., O'Neill R.J. Retention of latent centromeres in the Mammalian genome. J. Hered. 2005;96:217–224. doi: 10.1093/jhered/esi029. [DOI] [PubMed] [Google Scholar]
- Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scott G., Steffen D., Worley K.C., Burch P.E., Steffen D., Worley K.C., Burch P.E., Worley K.C., Burch P.E., Burch P.E., et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Grewal S.I.S., Jia S., Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Gu W., Ray D.A., Walker J.A., Barnes E., Gentles A.J., Samollow P.B., Jurka J., Batzer M.A., Pollock D.D., Ray D.A., Walker J.A., Barnes E., Gentles A.J., Samollow P.B., Jurka J., Batzer M.A., Pollock D.D., Walker J.A., Barnes E., Gentles A.J., Samollow P.B., Jurka J., Batzer M.A., Pollock D.D., Barnes E., Gentles A.J., Samollow P.B., Jurka J., Batzer M.A., Pollock D.D., Gentles A.J., Samollow P.B., Jurka J., Batzer M.A., Pollock D.D., Samollow P.B., Jurka J., Batzer M.A., Pollock D.D., Jurka J., Batzer M.A., Pollock D.D., Batzer M.A., Pollock D.D., Pollock D.D. SINEs, evolution and genome structure in the opossum. Gene. 2007 doi: 10.1016/j.gene.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho F.J., Beverley S.M., Beverley S.M. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- Hasler J., Strub K., Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2005;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Jackson M.S., See C.G., Mulligan L.M., Lauffart B.F., See C.G., Mulligan L.M., Lauffart B.F., Mulligan L.M., Lauffart B.F., Lauffart B.F. A 9.75-Mb map across the centromere of human chromosome 10. Genomics. 1996;33:258–270. doi: 10.1006/geno.1996.0190. [DOI] [PubMed] [Google Scholar]
- Jordan I.K., Matyunina L.V., McDonald J.F., Matyunina L.V., McDonald J.F., McDonald J.F. Evidence for the recent horizontal transfer of long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. 1999;96:12621–12625. doi: 10.1073/pnas.96.22.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Klonowski P., Kohany O., Walichiewicz J., Kohany O., Walichiewicz J., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kamal M., Xie X., Lander E.S., Xie X., Lander E.S., Lander E.S. A large family of ancient repeat elements in the human genome is under strong selection. Proc. Natl. Acad. Sci. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J., Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T., Kuma K., Toh H., Miyata T., Toh H., Miyata T., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M.G., Lisch D.R., Lisch D.R. Perspective: Transposable elements, parasitic DNA and genome evolution. Evolution Int. J. Org. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- King M.C., Wilson A.C., Wilson A.C. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kohany O., Gentles A.J., Hankus L., Jurka J., Gentles A.J., Hankus L., Jurka J., Hankus L., Jurka J., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs J.O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J., Kiefmann M., Jordan U., Brosius J., Schmitz J., Jordan U., Brosius J., Schmitz J., Brosius J., Schmitz J., Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Doyle M., FitzHugh W., Funke R., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Doyle M., FitzHugh W., Funke R., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Doyle M., FitzHugh W., Funke R., Nusbaum C., Zody M.C., Baldwin J., Devon K., Doyle M., FitzHugh W., Funke R., Zody M.C., Baldwin J., Devon K., Doyle M., FitzHugh W., Funke R., Baldwin J., Devon K., Doyle M., FitzHugh W., Funke R., Devon K., Doyle M., FitzHugh W., Funke R., Doyle M., FitzHugh W., Funke R., FitzHugh W., Funke R., Funke R., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Chang J.L., Kulbokas E.J., III, Zody M.C., Kulbokas E.J., III, Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Mahtani M.M., Willard H.F., Willard H.F. Physical and genetic mapping of the human X chromosome centromere: Repression of recombination. Genome Res. 1998;8:100–110. doi: 10.1101/gr.8.2.100. [DOI] [PubMed] [Google Scholar]
- Malik H.S., Eickbush T.H., Eickbush T.H. The RTE class of non-LTR retrotransposons is widely distributed in animals and is the origin of many SINEs. Mol. Biol. Evol. 1998;15:1123–1134. doi: 10.1093/oxfordjournals.molbev.a026020. [DOI] [PubMed] [Google Scholar]
- McClintock B. Some parallels between gene control systems in maize and in bacteria. Am. Nat. 1961;95:265–277. [Google Scholar]
- Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Garber M., Gentles A.J., Goodstadt L., Heger A., Gentles A.J., Goodstadt L., Heger A., Goodstadt L., Heger A., Heger A., et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: Improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Nishihara H., Smit A.F., Okada N., Smit A.F., Okada N., Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill R.J., O'Neill M.J., Graves J.A., O'Neill M.J., Graves J.A., Graves J.A. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- Pavlicek A., Jabbari K., Paces J., Paces V., Hejnar J.V., Bernardi G., Jabbari K., Paces J., Paces V., Hejnar J.V., Bernardi G., Paces J., Paces V., Hejnar J.V., Bernardi G., Paces V., Hejnar J.V., Bernardi G., Hejnar J.V., Bernardi G., Bernardi G. Similar integration but different stability of Alus and LINEs in the human genome. Gene. 2001;276:39–45. doi: 10.1016/s0378-1119(01)00645-x. [DOI] [PubMed] [Google Scholar]
- Posey J.E., Pytlos M.J., Sinden R.R., Roth D.B., Pytlos M.J., Sinden R.R., Roth D.B., Sinden R.R., Roth D.B., Roth D.B. Target DNA structure plays a critical role in RAG transposition. PLoS Biol. 2006;4:e350. doi: 10.1371/journal.pbio.0040350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Jones N.C., Pevzner P.A., Jones N.C., Pevzner P.A., Pevzner P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–i358. doi: 10.1093/bioinformatics/bti1018. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- Rens W., O'Brien P.C., Fairclough H., Harman L., Graves J.A., Ferguson-Smith M.A., O'Brien P.C., Fairclough H., Harman L., Graves J.A., Ferguson-Smith M.A., Fairclough H., Harman L., Graves J.A., Ferguson-Smith M.A., Harman L., Graves J.A., Ferguson-Smith M.A., Graves J.A., Ferguson-Smith M.A., Ferguson-Smith M.A. Reversal and convergence in marsupial chromosome evolution. Cytogenet. Genome Res. 2003;102:282–290. doi: 10.1159/000075764. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P., Huelsenbeck J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Samollow P.B., Kammerer C.M., Mahaney S.M., Schneider J.L., Westenberger S.J., VandeBerg J.L., Robinson E.S., Kammerer C.M., Mahaney S.M., Schneider J.L., Westenberger S.J., VandeBerg J.L., Robinson E.S., Mahaney S.M., Schneider J.L., Westenberger S.J., VandeBerg J.L., Robinson E.S., Schneider J.L., Westenberger S.J., VandeBerg J.L., Robinson E.S., Westenberger S.J., VandeBerg J.L., Robinson E.S., VandeBerg J.L., Robinson E.S., Robinson E.S. First-generation linkage map of the gray, short-tailed opossum, Monodelphis domestica, reveals genome-wide reduction in female recombination rates. Genetics. 2004;166:307–329. doi: 10.1534/genetics.166.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Churakov G., Zischler H., Brosius J., Churakov G., Zischler H., Brosius J., Zischler H., Brosius J., Brosius J. A novel class of mammalian-specific tailless retropseudogenes. Genome Res. 2004;14:1911–1915. doi: 10.1101/gr.2720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S.K., Han K., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Han K., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Dyer M., Cordaux R., Liang P., Batzer M.A., Cordaux R., Liang P., Batzer M.A., Liang P., Batzer M.A., Batzer M.A. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F., Riggs A.D., Riggs A.D. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemraj J., Plucienniczak G., Jaworski J., Plucienniczak A., Plucienniczak G., Jaworski J., Plucienniczak A., Jaworski J., Plucienniczak A., Plucienniczak A. Bovine Alu-like sequences mediate transposition of a new site-specific retroelement. Gene. 1995;152:261–264. doi: 10.1016/0378-1119(94)00709-2. [DOI] [PubMed] [Google Scholar]
- Tarlinton R.E., Meers J., Young P.R., Meers J., Young P.R., Young P.R. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- Tavare S. In: Some probabilistic and statistical problems on the analysis of DNA sequences. Lectures on mathematics in the life sciences. Miura R.M., editor. American Mathematical Society; Providence, RI: 1986. pp. 57–86. Vol. 17. [Google Scholar]
- Thornburg B.G., Gotea V., Makalowski W., Gotea V., Makalowski W., Makalowski W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006;365:104–110. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Ungerer M.C., Strakosh S.C., Zhen Y., Strakosh S.C., Zhen Y., Zhen Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 2006;16:R872–R873. doi: 10.1016/j.cub.2006.09.020. [DOI] [PubMed] [Google Scholar]
- VandeBerg J.L., Robinson E.S., Robinson E.S. The laboratory opossum (Monodelphis domestica) in laboratory research. ILAR J. 1997;38:4–12. doi: 10.1093/ilar.38.1.4. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Kazazian H.H., Boeke J.D., Moran J.V., Boeke J.D., Moran J.V., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman S., van Luenen H.G., Plasterk R.H., van Luenen H.G., Plasterk R.H., Plasterk R.H. Rte-1, a retrotransposon-like element in Caenorhabditis elegans. FEBS Lett. 1996;380:1–7. doi: 10.1016/0014-5793(95)01525-6. [DOI] [PubMed] [Google Scholar]
- Zupunski V., Gubensek F., Kordis D., Gubensek F., Kordis D., Kordis D. Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol. Biol. Evol. 2001;18:1849–1863. doi: 10.1093/oxfordjournals.molbev.a003727. [DOI] [PubMed] [Google Scholar]