Abstract
Chromatin remodeling enzymes contribute to the dynamic changes that occur in chromatin structure during cellular processes such as transcription, recombination, repair, and replication. Members of the chromodomain helicase DNA-binding (Chd) family of enzymes belong to the SNF2 superfamily of ATP-dependent chromatin remodelers. The Chd proteins are distinguished by the presence of two N-terminal chromodomains that function as interaction surfaces for a variety of chromatin components. Genetic, biochemical, and structural studies demonstrate that Chd proteins are important regulators of transcription and play critical roles during developmental processes. Numerous Chd proteins are also implicated in human disease.
Keywords: CHD family (Chromodomain Helicase DNA-binding); ATPase; Chromatin remodeling; SNF2 family; Transcription elongation; Development, differentiation, and disease
Introduction
The regulation of genes is a highly coordinated process that involves the ordered recruitment of transcriptional machinery components in concert with alterations in chromatin structure. Chromatin remodeling enzymes play critical roles in organizing genomic DNA within the native chromatin state. These highly conserved enzymes can be divided into two classes: those that mediate post-translational histone modifications and those that utilize energy derived from ATP hydrolysis to alter the histone-DNA contacts within the nucleosome. These modulators alter the chromatin structure by regulating the accessibility of nucleosomal DNA, shielding certain DNA regions while exposing others for interaction with the cell's regulatory machinery.
Targeted covalent modifications of specific histone residues specify particular events in multiple cellular processes, including transcriptional activation and silencing. Numerous chemical modifications of the histone proteins occur; these include acetylation, methylation, phosphorylation, ubiquitination, sumolation, and ADP-ribosylation [1]. These modifications affect local chromatin structure and also affect higher-order folding of the nucleosomal fiber. In contrast, multisubunit complexes that utilize energy derived from ATP hydrolysis alter chromatin structure by disrupting or mobilizing nucleosomes. Each of the identified ATP-dependent chromatin remodeling enzymes contains an ATPase subunit that belongs to the SNF2 superfamily of proteins [2,3]. Based on the presence of other conserved domains, these enzymes are further classified into the SWI/SNF (mating type switching/sucrose non-fermenting), ISWI (imitation switch), INO80 (inositol), and CHD (chromodomain helicase DNA-binding) families.
The SWI/SNF family contains yeast SNF2 and STH1, Drosophila melangaster brahma (BRM), and mammalian BRM and brahma-related gene 1 (BRG1). The distinguishing feature of these proteins is a bromodomain, which recognizes the acetylated lysine residues on the N-terminal tails of histones [4,5]. The ISWI family comprises yeast homologues Isw1 and Isw2 and mammalian homologues SNF2H and SNF2L. These enzymes are characterized by a SANT domain, which functions as a histone-binding module [4,6]. The INO80 member of the family is the only chromatin remodeling protein in which DNA helicase activity has been observed, and evidence indicates a role for this complex in the facilitation of DNA repair [7,8]. Lastly, the CHD family includes a number of proteins that are highly conserved from yeast to humans, though the function of many of these proteins remains unknown or poorly characterized [9,10] (Table 1). This review highlights the progress made in understanding the function of the CHD family of proteins.
Table 1.
Functions of the Chd Family Class of Chromatin Remodelers
| Subunit | Organism | Biochemical Activities | In vivo functions |
|---|---|---|---|
| Chd1 |
Saccharomyces cerevisiae |
ATPase activity and relocates nucleosomes [54-56]. Exists as a monomer or dimer [24,54]. Component of SAGA and SLIK complexes and HAT activity [24]. Interacts with H3K4me [24]. Transcription elongation [70,71]. |
transcriptional repressor [10,54,73]. Interacts with SSRP1 [18]. Transcription elongation [10,18] |
|
Drosophila melanogaster |
ATPase activity [57] | transcriptional activator [46] Interacts with SSRP1 [18] transcription elongation [72] |
|
| Mus musculus | HDAC activity [68] | transcriptional activator [32] transcriptional repressor [68] pre-mRNA splicing events[68] |
|
| Human | Interacts with H3K4me [22,25] | ||
| Chd2 | Mus musculus | Growth retardation and perinatal lethality of homozygous mutants [47]. Decreased neonatal viability, growth delay, and non- neoplastic lesions [47] |
|
| Chd3 |
Drosophila melanogaster |
nucleosome-stimulated ATPase activity and mobilizes nucleosomes [16] |
|
| Human | Component of NURD complex [60-62] |
||
| Chd4 |
Drosophila melanogaster |
DNA-dependent, nucleosome-stimulated ATPase activity [59] |
lymphocyte differentiation [76] T-cell development [76] |
| Human | Component of NURD complex [60-62]. Interacts with HDAC1 [62] |
||
| Chd9 | Human | osteogenic differentiation [39,53] |
|
Signature Motifs of the Chd proteins
The CHD family is characterized by two signature sequence motifs: tandem chromodomains located in the N-terminal region, and the SNF2-like ATPase domain located in the central region of the protein structure [9,10]. The SNF2-like ATPase domain defines the ATP-dependent chromatin remodeling proteins. This domain contains a conserved set of amino acid motifs that has been found in proteins involved in a myriad of cellular processes including chromatin assembly, transcription regulation, DNA repair, DNA replication, development and differentiation [8,11,12].
The chromodomain (chromatin organization modifier) is an evolutionarily conserved sequence motif involved in the remodeling of chromatin structure and the transcriptional regulation of genes [13-16]. This motif was originally identified as a 37 amino acid residue region of homology shared by Drosophila melangaster epigenetic repressors, heterochromatin protein 1 (HP1) and Polycomb (Pc) [17]. HP1 and Pc proteins function in heterochromatin function. The chromodomain is now recognized as a 50 amino acid region of shared homology between these polypeptides [14,17]. Additionally, chromodomains have also been found in ATP-dependent chromatin remodeling factors, histone acetyltransferases, and histone methyltransferases [13,14].
From a structural standpoint, it may be inferred that each of the sequence modules found in Chd proteins perform distinct functions. Mutations in the helicase domain and chromodomain of mouse Chd1 (mChd1) each resulted in nuclear redistribution, illustrating that both of these domains are essential for proper association with chromatin [18]. Similarly, deletion of the chromodomains in the Drosophila melangaster Chd3-Chd4 homologue impaired nucleosome binding, mobilization, and ATPase functions [19].
Functional analyses have demonstrated that the chromodomain serves as a module to mediate chromatin interactions by binding directly to DNA, RNA, and methylated histone H3 [16,19-26]. Histone H3 methylated lysine 4 (H3K4) trimethylation can be localized at transcriptional start sites of human, mouse, and Drosophila genes [27-29], and yeast Chd1 (ScChd1) was identified as a factor required for H3K4me [29]. Later it was found that ScChd1 was specifically required for an increase of H3K4 trimethylation at the 3' ends of genes when growth conditions were suboptimal [30]. Recently, the binding of ScChd1 and human CHD1 (hCHD1) to H3K4me through the chromodomain modules was suggested by several groups [22,24,25]. However, there are inconsistencies about whether the chromodomains of ScChd1 interact with the methylated lysine 4 on histone H3. Structural analyses of the related human CHD3 and CHD4 (hCHD3 and hCHD4) proteins do not predict binding to methylated lysine residues [25]. This is a likely prediction given that H3K4me is thought to be restricted to active chromatin and that Chd3 and Chd4 function is usually associated with transcriptional repression [31]. Overall, the unique combination of functional domains found in Chd proteins suggests that this family may have essential roles in chromatin based regulatory processes, including gene expression.
Chd1-Chd2 Subfamily
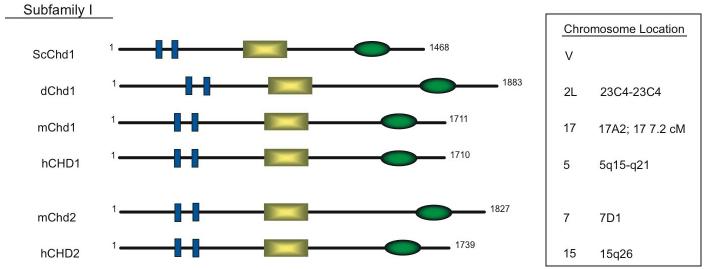
The CHD family is divided into three subfamilies according to the presence or absence of additional domains. The first subfamily contains yeast Chd1 (ScChd1 or Chd1p), which is the only Chd family member present in yeast, and the Chd1 and Chd2 proteins from higher eukaryotes (Fig. 1). The Chd1 and Chd2 proteins contain a DNA-binding domain located in the C-terminal region [9,10,32]. Currently, mChd1 is the only family member with a functionally characterized DNA-binding domain, though the high degree of homology between subfamily members makes it likely that each protein in this group has similar DNA-binding properties. The DNA-binding domain preferentially binds to AT-rich DNA motifs [9,32]. Although the Chd1-Chd2 subfamily members are highly homologous to one another, they are significantly divergent in the 3' region that contains the DNA binding domain, suggesting that Chd1 and Chd2 may possess distinct functions.
Figure 1.
Structural Domains of Subfamily I. Schematic representation of protein domains found within yeast, fly, mouse, and human Chd genes of subfamily I. Members of this subfamily are characterized by tandem chromodomains (blue rectangles), a SNF2-like ATPase domain (yellow rectangle), and a DNA binding domain (green oval). DNA activity has only been characterized for murine Chd1. All other Chd proteins in this subfamily contain putative DNA-binding domains. The chromosome location of each of the Chd genes within this subfamily is also listed. Sequence information was obtained from NCBI: ScChd1: NP_011091; dChd1: NP_477197; mChd1: NP_031716; hCHD1: NP_001261; mChd2: XP_145698; hCHD2: NP_001262. Sequence information was also obtained from Stokes et al and Woodage et al [10,32].
Chd3-Chd4 Subfamily
The second subfamily, which lacks the DNA-binding domain, includes the proteins Chd3 and Chd4 (sometimes referred to as Mi-2α and Mi-2β, respectively) (Fig. 2). Additionally, the family includes Drosophilia melangaster Mi-2 (dMi-2), a single gene encoding two transcripts [33]. These proteins harbor paired N-terminal PHD (plant homeo domain) Zn-finger-like domains that are not found in the Chd1-Chd2 subfamily [10]. The PHD Zn-finger-like domains are found in a number nuclear proteins implicated in chromatin-based transcriptional regulation [34]. Functional analyses have demonstrated that PHD domains are involved in chromatin remodeling [35,36]. A number of recent reports suggest that the recognition of methylated histone peptides is a general feature of at least a subset of PHD domains [37,38]. For example, the PHD domains of yeast and human ING family members all bound preferentially to di- and tri-methylated H3K4, whereas the PHD domains of Chd3 bound to trimethylated H3K36 [37,38].
Figure 2.
Structural Domains of Subfamily II. Schematic representation of protein domains found within fly, mouse, and human Chd genes of subfamily II. Members of this subfamily are characterized by paired PHD Zn-finger-like domains (red triangles), tandem chromodomains (blue rectangles), and a SNF2-like ATPase domain (yellow rectangle). The chromosome location of each of the Chd genes within this subfamily is also listed. Sequence information was obtained from NCBI: dChd3: NP_649154; mChd3: NP_484041; hCHD3:NP_001005271; dChd4: NP_001014591; mChd4: NP_666091; hCHD4: NP_001264.
Chd5-Chd9 Subfamily
The third subfamily contains the proteins Chd5-Chd9, which were identified on the basis of structural and sequence conservation to known Chd proteins (Fig. 3). It should be noted that Chd9 is also referred to as CReMM (chromatin-related mesenchymal modulator) [39]. This subfamily is defined by additional functional motifs in the C-terminal region, including paired BRK (Brahma and Kismet) domains, a SANT-like (switching-defective protein 3, adaptor 2, nuclear receptor co-repressor, transcription factor IIIB) domain, CR domains, and a DNA-binding domain, [39-41].
Figure 3.
Structural Domains of Subfamily III. Schematic representation of protein domains found within mouse and human Chd genes of subfamily III. Members of this subfamily are characterized by the signature sequence motifs of Chd proteins, chromodomains (blue rectangles) and a SNF2-like ATPase domain (yellow rectangle). Additional domains have also been predicted for several members of this subfamily, including paired PHD Zn-finger-like domains (red triangles), BRK domains (purple diamonds), CR1-3 domains (red rectangles), SANT domains (blue circle), and DNA-binding domains (green oval). The chromosome location of each of the Chd genes within this subfamily is also listed. Sequence information was obtained from NCBI: mChd5: XP_900117; hCHD5: NP_056372; mChd6: NP_775544; hCHD6: NP_115597; mChd7: XP_910521; hCHD7: NP_060250; mChd8: XP_619244; hCHD8: NP_065971.1; mChd9: NP_796198; hCHD9: NP_079410. Information regarding the structural domains was also obtained from Shuster et al and Shur et al [39,40].
The BRK domain is conserved in a number of SWI/SNF proteins, for example Drosophila BRM, human BRM, and human BRG1 [41-44]. Interestingly, this domain is not present in yeast chromatin remodeling factors related to BRM, including SWI2/SNF2 and STH1, suggesting that the BRK domain is involved in functions specific to higher eukaryotes or may interact with a component of chromatin unique to higher eukaryotes. The SANT domain was initially identified in nuclear receptor co-repressors and subsequently was found in the subunits of many chromatin remodeling complexes [45]. Biochemical analyses determined that SANT domains, which interact primarily with unmodified histone tails, are unique histone-interaction modules that couple histone binding to enzyme catalysis [4,6]. To date, the CR domains, CR1-CR3, are moieties of unknown function that are only found in Chd9 [39,40]. As with the Chd1-Chd2 subfamily, the DNA-binding domain of the third subfamily also shows a preference for binding to AT-rich tracts. However, DNA-binding activity has only been demonstrated experimentally for Chd9 to date [39].
Localization Properties and Tissue Expression Patterns of Chd Proteins
Studies of Drosophilia melangaster Chd proteins (dChd) on polytene chromosomes also shed light on the function of Chd proteins. dChd1 was found to localize to regions of decondensed chromatin (interbands) and regions of high transcriptional activity (puffs) in polytene chromosomes by immunostaining [18,46]. However, dChd1 did not stain all interbands and chromosome puffs. Furthermore, dChd1 signal was not detected in the chromocenter, a region that represents the heterochromatic satellite-centrosomal DNA [46]. These findings are consistent with previous observations that showed that mChd1 stained decompacted chromatin and was not present in the centromeric heterochromatin of interphase cells [32]. Collectively, these data provided evidence that Chd1 bound to genomic chromatin with locus-specific association and suggested that Chd1 might facilitate gene expression by maintaining chromatin in a transcriptionally active state.
Expression patterns have been analyzed for most of the genes within each of the three Chd subfamilies. Expression profiles for hCHD1 and hCHD2 have revealed global expression of these genes [10]. Mouse Chd2 mRNA is also ubiquitously expressed in adult tissues [47]. The absolute levels of mRNA expression in the different tissues analyzed were similar except for heart tissue, where Chd2 mRNA was highly expressed [47].
hCHD3 is ubiquitously expressed in each of the subset of tissues examined [10]. In mouse neonatal tissues, mChd4 mRNA is expressed at high levels in the thymus, the kidney, in specific areas of the brain, in hematopoietic foci in the liver, in hair follicles, and in mucosal epithelia [48]. Similar patterns of expression for mChd3 mRNA were detected in these tissues but at lower levels than mChd4, as judged by in situ hybridization [48]. In Caenorhabditis elegans, CeChd3 and CeChd4 (let-418) are expressed in most if not all cells of the embryo [49]. During larval development and in adults, expression of CeChd3 and CeChd4 was observed in the nuclei of many cells, including the ventral nerve cord cells and the vulval precursor cells (VPCs), the surrounding hypodermal cells, and cells of the head and tail regions [49]. Expression patterns of the two dMi-2 transcripts (dMi-2a and dMi-2b) were performed in five embryonic stages, in three larval stages, in pupal, and in adult stages of Drosophila. Abundant expression of the dMi-2a transcript mRNA was observed during the first 8 hours of embryogenesis and gradually decreased during later developmental stages. In the case of dMi-2b, low levels of mRNA expression were detected in the early stages of embryogenesis whereas higher mRNA levels were detected during later stages of development. Both mRNAs were grossly elevated in the ovary [33].
hCHD5 mRNA expression was restricted to neural-derived tissues whereas hCHD7 has been found to be ubiquitously expressed [50,51]. Recently, rat and human CHD9 expression at different stages of osteogenic differentiation was analyzed. Cultured primary marrow stromal cells from rats revealed higher expression of Chd9 in cells of 3-month old rats than in cells of 15-month old rats [52]. Likewise, mRNA levels of hCHD9 were higher in the primary marrow stromal cells than in trabecular bone cells (TBC) [52]. A bone marrow derived osteogenic cell line that was used to determine the kinetics of expression during cell proliferation and differentiation revealed that mammalian Chd9 was robustly expressed in proliferating cells [52]. mChd9 expression during embryonic development in the mouse skeletal system at the mRNA and protein levels was also examined; the expression of mChd9 was restricted to the marrow stromal progenitor cells, plus a small population of cells present in the bone marrow of newborn and adult mice [53].
Extensive analyses addressing RNA processing of Chd proteins have not yet been performed. Computer algorithm programs indicate that most Chd proteins will have splice variants, further increasing the diversity of possible functions. In this context, it is noteworthy that some of the alternative splicing products will not contain all of the signature motifs that distinguish each subfamily of the Chd proteins.
Chd Protein Function
Analyses of Chd proteins reveal a wide range of functions in vitro and in vivo. Table 1 provides a summary of these properties.
A Role for Chd Proteins in Chromatin Assembly and Remodeling
ATPases of the SNF2 family have been broadly implicated in many cellular processes, including nucleosome assembly, disruption, and positioning. Biochemical analyses revealed that ScChd1 has an ATPase activity that affects DNA-histone interactions within the nucleosome in a manner that is distinct from the yeast SWI/SNF complex [54]. Additionally, a partial loss of chromatin assembly activity in vitro from crude DEAE fractions derived from deletion strains of ScChd1 was observed [55]. Recently, it was shown that ScChd1 preferentially relocated nucleosomes closer to the center of DNA fragments [56]. dChd1 has the ability to assemble histone H1-deficient chromatin, but not H1-containing chromatin, suggesting that dChd1 may function in the assembly of transcriptionally active DNA into chromatin [57].
In Drosophila, recombinant dMi-2 is a nucleosome-stimulated ATPase that binds and mobilizes nucleosomes along a linear DNA fragment [58]. Highly purified, recombinant hCHD4 shows DNA-dependent, nucleosome-stimulated ATPase activity [59].
Chd Members Form Multisubunit Complexes
Most SNF2-like ATPases are components of large multisubunit complexes. Yeast two-hybrid screens and immunocytochemical analyses showed an interaction between mChd1 and a nuclear protein, SSRP1 (structure specific recognition protein 1), which is involved in transcription regulation [18]. Additionally, dChd1 was found to interact with human SSRP1 [18]. Moreover, mChd1 and SSRP1 copurified in fractionated nuclear extracts that corresponded to a complex of roughly Mr 700,000 [18]. Collectively, this provided the first evidence that a Chd protein from subfamily I may be associated with a multisubunit complex. In contrast to the mammalian system, both native and recombinant dChd1 exist predominantly as a monomer [57]. Likewise, ScChd1 was also found to fractionate at 150-340 kDa, suggesting that it may exist either as a monomer or a dimer within a cell [24,54].
However, recent work identified ScChd1 as a component of SAGA (Spt-Ada-Gcn5 acetyltransferase) and SLIK (SAGA-like), two highly homologous and conserved multi-subunit HAT (histone acetyltransferase) complexes [24]. Yeast strains bearing ScChd1 deletions compromised the HAT activity of both SAGA and SLIK complexes [24]. This activity was largely restored when an HA-tagged Chd1 protein was introduced into the ScChd1 deletion strain, demonstrating the importance of ScChd1 in proper HAT activity in these complexes [24].
Members of the CHD subfamily II, hCHD3 and hCHD4, form large protein complexes that possess both histone deacetylase and ATP-dependent chromatin remodeling activities. These complexes were identified by multiple groups and termed NURD (nucleosome remodeling and histone deacetylase). NURD contains seven proteins associated with transcriptional repression: HDAC1 and HDAC2 (type I histone deacetylases), RbAp48 and RbAp46 (Retinoblastoma-associated proteins), MTA1, MTA2, and MTA3 (metastasis-associated proteins) and MBD3 (methyl-CpG binding domain) [60-65]. It is not clear if hCHD3 and hCHD4 exist in the same or in different NURD complexes. Chd3 and Chd4 orthologs have also been identified in a complex similar to NURD in Drosophila, Xenopus laevis, and Caenorhabditis elegans [58,64,66,67]. These reports suggested that Chd3 and hChd4 couple histone deacetylation and DNA methylation with ATP chromatin remodeling.
The discovery of hCHD3 and hCHD4 as components of the NURD complex were followed by the determination that a direct association exists between Chd and HDAC proteins and that the PHD Zn-finger-like domain of hCHD4 was required, but not sufficient, for the interaction [62]. Moreover, mChd1 was observed to be associated with HDAC activity, presumably through interactions made by the first chromodomain [68]. The chromodomains of mChd1 have also been implicated in interactions with NCoR (co-repressor of nuclear hormone transcription), a protein known to associate with HDAC activity. The presence of at least one of the two chromodomains of mCHD1 was needed for an interaction between mChd1 and NCoR [68]. In summary, these observations suggest a role for Chd proteins in facilitating transcriptional repression via modification and remodeling of chromatin structure.
Chd Proteins and Transcriptional Elongation
The control of transcriptional elongation is a prominent mechanism of gene regulation. The activities of transcription elongation factors and their link to chromatin have been well established in yeast and mammalian systems [69]. Essential transcription elongation factors in yeast include Spt4-Spt5 and Spt16 and Pob3. In mammalian cells, the counterparts for these yeast factors are DSIF (DRB sensitivity inducing factor) and FACT (facilitates chromatin transcription), respectively [69].
Studies utilizing yeast deletion strains implicated ScChd1 in transcriptional elongation. As previously described, growth of ScChd1 deletion strains was significantly affected by suboptimal growth conditions, including growth medium containing 6-azauracil [10]. ScChd1, as well as other proteins implicated in transcriptional elongation, copurified with the yeast elongation factors Spt16 and Pob3 [70]. Moreover, ScChd1 was observed to interact with both Rtf1, a member of the RNA pol-II associated Paf1 complex, which is implicated in elongation, and with Spt5, a transcription elongation factor [71]. These findings are consistent with a previously described study where mChd1 was found to interact with SSRP1, the mammalian homolog of Pob3 [18]. Additionally, Drosophila studies of kismet (a thrithorax gene) mutant larvae, showed dramatically reduced levels of elongating RNA pol-II, elongation factor Spt6, and dChd1 on polytene chromosomes [72]. Collectively, these data suggest that Chd1 is likely a putative transcription elongation factor.
Chd Proteins in Development and Differentiation
Yeast strains bearing Chd1-null deletions were viable except when grown under suboptimal conditions, therby suggesting a role for ScChd1 in regulating gene expression [10,73]. Genetic analyses determined that ScChd1 interacts with SWI2 and that cells require one or the other for viability [54]. Similarly, ScChd1 also genetically interacts with ISWI1 and ISWI2 and a phenotypic survey of a triple mutant, iswi1 iswi2 chd1, indicated a moderate synthetic growth defect [73]. Together, this suggests that ScChd1 and subunits of the yeast SWI/SNF and ISWI complexes possess some parallel functions in cell growth.
Recently, a Chd2-mutant mouse model demonstrated that mutation of Chd2 dramatically affects mammalian development and long-term survival. This mutation in Chd2 resulted in a general growth delay in homozygous mutants late in embryogenesis and in perinatal lethality [47]. Animals heterozygous for the mutation showed decreased neonatal viability and increased susceptibility to non-neoplastic lesions affecting most primary organs. In particular, approximately 85% of the heterozygotes showed gross kidney abnormalities [47].
In mammalian cells, a stable interaction occurs between BCL-6, a transcriptional repressor that regulates B-lymphocyte cell fate, and components of the NURD complex [74]. In lymphoid cell lines, MTA3 and the NURD complex function as a corepressor for BCL-6 to direct transcriptional repression [74]. Moreover, the transcriptional repressor Ikaros physically interacts with mChd4 and recruits it to heterochromatic regions upon T-cell activation [48]. Subsequently, an association between Ikaros and hCHD4 was observed in adult erythroid cells [75]. Conditional mice were generated to further define the role of mChd4 in T cell development. This conditional inactivation of mChd4 showed that mChd4 is required in at least three distinct stages of T cell development: in the late double-negative stage to support the transition to the double-positive stage, in the double-positive stage for normal expression of the CD4 coreceptor, and, finally, in mature T cells for their proliferative expansion [76]. Strikingly, it was also observed that mChd4 is involved in the positive regulation of the CD4 gene during differentiation in the thymus through its interactions with the CD4 enhancer as well as the histone acetyltransferase p300 and the E box binding protein, HEB [76].
A detailed analysis of the dMi-2 gene revealed that flies carrying homozygous mutations in dMi-2 died during the larval stage of development despite a lack of any obvious structural defects [66]. This prompted researchers to generate embryos from mutant dMi-2 germ cells. However, germ cells homozygous for dMi-2 failed to develop [66]. This failure was rescued by a dMi-2 transgene, demonstrating that dMi-2 is essential for the development of germ cells [66].
Genetic analysis in C.elegans revealed that CeChd4 mRNA is maternally delivered to the early embryo. This maternal contribution is sufficient to allow some of the mutant embryos to develop into adulthood, however, 98% of these animals were sterile [49]. The remaining CeChd4 mutant embryos arrested at an early larval stage [49]. In contrast, CeChd3 mRNA is not maternally delivered and mutant embryos homozygous for this mutation develop normally with no obvious phenotype [49]. Partial rescue of CeChd4 is observed in CeChd4;CeChd3 double mutant animals, where mutants lacking functional CeChd3 and CeChd4 genes did not arrest until later stages of larval development [49]. These data suggest that CeChd4 and CeChd3 have essential and partially redundant roles in worms during development.
A Role for Chd in Human Disease
Mutations in genes encoding SNF2-like enzymes are known to cause a spectrum of disease phenotypes. The identification and characterization of these enzymes is critical for understanding the genetic events underlying the progression of disease. To date, hCHD3-hCHD5 and hCHD7 have been implicated in human disease processes (Table 2).
Table 2.
Human Diseases Associated with Chd Proteins
hCHD3 and hCHD4 have been identied as autoantigens in patients with dermatomyositis, a connective-tissue disease characterized by inflammation of both muscles and skin [77-79]. The etiology of dermatomyositis is poorly understood, but a correlation beween this disease and an elevated incidence of cancer has been established [80].
More recently, hCHD3 was associated with Hodgkin's lymphoma. A yeast two-hybrid screen identified an interaction between hCHD3 and Ki-1/57, an intracellular phospho-protein, and between hCHD3 and CGI-55, a mRNA-binding protein [81]. Although the functions of these protein is unknown, Ki-1/57 specifically detects the malignant cells in Hodgkin's lymphoma [82].
hCHD5 is associated with neuroblastoma, a malignant neoplasm of the peripheral sympathetic nervous system frequently affecting infants and children. Analysis of patient samples identified mutations in hCHD5 in the process of mapping 1p36.3, a region frequently deleted in human neuroblastomas [50,83,84].
A fourth human syndrome, CHARGE, an acronym for six of the most prevalent features of this disease: coloboma of the eye, heart defects, atresia of chonae, renal anomalies and retardation of growth and/or development, genital anomalies, and ear abnormalities or deafness, has been associated with hCHD7. [51,85]. Sequence analysis of patient samples diagnosed with CHARGE syndrome detected mutations in CHD7 in 10 of the 17 afflicted individuals [51]. In summary, these findings further support an association between dysfunction of these CHD proteins and human disease.
Conclusions and Future Directions
The hallmark of Chd proteins is their novel combination of structural domains. The signature motifs of this class of enzymes are paired chromodomains located in the N-terminal region and a SNF2-like ATPase domain located in the central region of the protein structure [9,10]. Since the discovery of murine Chd1 in 1993, other Chd genes have been identified, yielding a total of 9 highly conserved genes from diverse organisms [9,10,39,40,51]. The Chd family is divided into three subfamilies based on structural and sequence similarities [9,10,32,39,40].
The growing number of Chd genes and the links between some of these genes and human disease underscore the importance of this class of enzymes. Moreover, the complexity of gene products that may be produced from Chd loci, due to alternative splicing events remains largely unexplored but nevertheless increases the complexity of potential functions. Further studies addressing the structure/function relationship of Chd protein domains as well as continued functional analyses of each family member are required to better guage the physiological roles of these proteins. At the molecular level, additional characterization is needed to show how Chd family members are targeted to chromatin and how they alter chromatin structure to both activate and repress gene expression.
Acknowledgements
We apologize to our colleagues whose work we did not cite due to space limitations. Work in the authors' lab is supported by grants from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Koonin EV. An expanding family of helicases within the ‘DEAD/H’ superfamily. Nucleic Acids Res. 1993;21:751–752. doi: 10.1093/nar/21.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 5.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 7.Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 8.Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat Rev Mol Cell Biol. 2002;3:422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- 9.Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci U S A. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 12.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 13.Eissenberg JC. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene. 2001;275:19–29. doi: 10.1016/s0378-1119(01)00628-x. [DOI] [PubMed] [Google Scholar]
- 14.Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Koonin EV, Zhou S, Lucchesi JC. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 17.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley DE, Stokes DG, Perry RP. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma. 1999;108:10–25. doi: 10.1007/s004120050347. [DOI] [PubMed] [Google Scholar]
- 19.Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, Brehm A. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. Embo J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 21.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 23.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 25.Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Schroeder S, Fong N, Bentley DL. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress’. Embo J. 2005;24:2379–2390. doi: 10.1038/sj.emboj.7600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Stokes DG, Perry RP. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol. 1995;15:2745–2753. doi: 10.1128/mcb.15.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khattak S, Lee BR, Cho SH, Ahnn J, Spoerel NA. Genetic characterization of Drosophila Mi-2 ATPase. Gene. 2002;293:107–114. doi: 10.1016/s0378-1119(02)00698-4. [DOI] [PubMed] [Google Scholar]
- 34.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. Embo J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, AM OY, Eberharter A, Gibson TJ, Becker PB, Aasland R. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 37.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shur I, Benayahu D. Characterization and functional analysis of CReMM, a novel chromodomain helicase DNA-binding protein. J Mol Biol. 2005;352:646–655. doi: 10.1016/j.jmb.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 40.Schuster EF, Stoger R. CHD5 defines a new subfamily of chromodomain-SWI2/SNF2-like helicases. Mamm Genome. 2002;13:117–119. doi: 10.1007/s00335-001-3042-6. [DOI] [PubMed] [Google Scholar]
- 41.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 43.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, Waldrip WR, Scott MP, Kennison JA, Tamkun JW. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999;126:1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- 45.Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 46.Stokes DG, Tartof KD, Perry RP. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci U S A. 1996;93:7137–7142. doi: 10.1073/pnas.93.14.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marfella CG, Ohkawa Y, Coles AH, Garlick DS, Jones SN, Imbalzano AN. Mutation of the SNF2 family member Chd2 affects mouse development and survival. J Cell Physiol. 2006 doi: 10.1002/jcp.20718. [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 49.von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Muller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–5284. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- 50.Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 51.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 52.Marom R, Shur I, Hager GL, Benayahu D. Expression and regulation of CReMM, a chromodomain helicase-DNA-binding (CHD), in marrow stroma derived osteoprogenitors. J Cell Physiol. 2006;207:628–635. doi: 10.1002/jcp.20611. [DOI] [PubMed] [Google Scholar]
- 53.Shur I, Socher R, Benayahu D. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J Cell Physiol. 2006;207:374–378. doi: 10.1002/jcp.20586. [DOI] [PubMed] [Google Scholar]
- 54.Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. Embo J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson KM, Schultz MC. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol Cell Biol. 2003;23:7937–7946. doi: 10.1128/MCB.23.22.7937-7946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and CHD1 chromatin remodeling complexes. J Biol Chem. 2006 doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 58.Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. Embo J. 2000;19:4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang HB, Zhang Y. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res. 2001;29:2517–2521. doi: 10.1093/nar/29.12.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 61.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 63.Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 64.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 65.Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 66.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 67.Solari F, Ahringer J. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol. 2000;10:223–226. doi: 10.1016/s0960-9822(00)00343-2. [DOI] [PubMed] [Google Scholar]
- 68.Tai HH, Geisterfer M, Bell JC, Moniwa M, Davie JR, Boucher L, McBurney MW. CHD1 associates with NCoR and histone deacetylase as well as with RNA splicing proteins. Biochem Biophys Res Commun. 2003;308:170–176. doi: 10.1016/s0006-291x(03)01354-8. [DOI] [PubMed] [Google Scholar]
- 69.Belotserkovskaya R, Reinberg D. Facts about FACT and transcript elongation through chromatin. Curr Opin Genet Dev. 2004;14:139–146. doi: 10.1016/j.gde.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 70.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 73.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 75.O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, Han S, Seong RH, Park SD, Agalioti T, Munshi N, Thanos D, Erdjument-Bromage H, Tempst P, Bank A. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 77.Ge Q, Nilasena DS, O'Brien CA, Frank MB, Targoff IN. Molecular analysis of a major antigenic region of the 240-kD protein of Mi-2 autoantigen. J Clin Invest. 1995;96:1730–1737. doi: 10.1172/JCI118218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seelig HP, Moosbrugger I, Ehrfeld H, Fink T, Renz M, Genth E. The major dermatomyositis-specific Mi-2 autoantigen is a presumed helicase involved in transcriptional activation. Arthritis Rheum. 1995;38:1389–1399. doi: 10.1002/art.1780381006. [DOI] [PubMed] [Google Scholar]
- 79.Seelig HP, Renz M, Targoff IN, Ge Q, Frank MB. Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum. 1996;39:1769–1771. doi: 10.1002/art.1780391029. [DOI] [PubMed] [Google Scholar]
- 80.Airio A, Pukkala E, Isomaki H. Elevated cancer incidence in patients with dermatomyositis: a population based study. J Rheumatol. 1995;22:1300–1303. [PubMed] [Google Scholar]
- 81.Lemos TA, Passos DO, Nery FC, Kobarg J. Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 2003;533:14–20. doi: 10.1016/s0014-5793(02)03737-7. [DOI] [PubMed] [Google Scholar]
- 82.Schwab U, Stein H, Gerdes J, Lemke H, Kirchner H, Schaadt M, Diehl V. Production of a monoclonal antibody specific for Hodgkin and Sternberg-Reed cells of Hodgkin's disease and a subset of normal lymphoid cells. Nature. 1982;299:65–67. doi: 10.1038/299065a0. [DOI] [PubMed] [Google Scholar]
- 83.Law ME, Templeton KL, Kitange G, Smith J, Misra A, Feuerstein BG, Jenkins RB. Molecular cytogenetic analysis of chromosomes 1 and 19 in glioma cell lines. Cancer Genet Cytogenet. 2005;160:1–14. doi: 10.1016/j.cancergencyto.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 84.White PS, Thompson PM, Gotoh T, Okawa ER, Igarashi J, Kok M, Winter C, Gregory SG, Hogarty MD, Maris JM, Brodeur GM. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 85.Williams MS. Speculations on the pathogenesis of CHARGE syndrome. Am J Med Genet A. 2005;133:318–325. doi: 10.1002/ajmg.a.30561. [DOI] [PubMed] [Google Scholar]