Abstract
Serum response has been used as a model for studying signaling transduction for many biological events such as cell proliferation and survival. Although expression of many genes is up- or down-regulated after serum stimulation, the Notch effector Hes1 displays oscillatory response. However, the precise mechanism and biological significance of this oscillation remain to be determined. Here, we identified serum-induced ultradian oscillators, including molecules in Stat and Smad signaling. Stat and Smad oscillations involve activation of Stat3 and Smad1 and delayed negative feedback by their inhibitors Socs3 and Smad6, respectively. Moreover, Stat oscillations induce oscillatory expression of Hes1 by regulating its half-life, and loss of Hes1 oscillations leads to G1 phase retardation of the cell cycle. These results indicate that coupled Stat and Hes1 oscillations are important for efficient cell proliferation and provide evidence that expression modes of signaling molecules affect downstream cellular events.
Keywords: Socs, microarray analysis, mathematical simulation
Serum response has been used as a model for studying signaling transduction for many biological events such as cell cycle and growth, neuronal excitation, and immune response (1–3). It has been shown that expression of >500 genes is changed substantially in fibroblasts after serum stimulation; some are up-regulated, whereas others are down-regulated and then gradually return to the original levels (1). It is generally thought that the duration and intensity of such changes in gene expression are important for downstream events.
We previously found that the Notch effector Hes1, a basic helix–loop–helix repressor gene, displays an oscillatory response with a 2-h period to serum stimulation (4, 5). This oscillation depends on negative feedback and rapid degradation of the protein (4). Although Hes1 is required for maintenance of neural progenitors (6–8), sustained Hes1 expression inhibits both proliferation and differentiation of these cells (6, 9), raising the possibility that oscillation is important for neural progenitors. The related gene Hes7 and its target gene lunatic fringe (Lfng) also display cyclic expression and regulate periodic somite segmentation. Sustained Hes7 or Lfng expression leads to fusion of somites, suggesting that oscillatory expression is required for this process (10–15). Importantly, in both cases, oscillatory vs. sustained expression seems to result in different outcomes in biological events. However, although transcriptional response to serum stimulation has been analyzed for many genes, previous studies failed to detect oscillatory responses, because the temporal profiling is usually intensive only for the first 1 or 2 h but not for longer periods.
Here, we analyzed temporal changes in gene expression more intensively for the first 4.5 h after serum stimulation and identified ultradian oscillators, including molecules in Stat signaling. Loss of Stat oscillations leads to inhibition of Hes1 oscillations, which retards cell cycle progression. These results provide insight into the significance of oscillatory expression in cell proliferation.
Results and Discussion
Identification of Ultradian Oscillators.
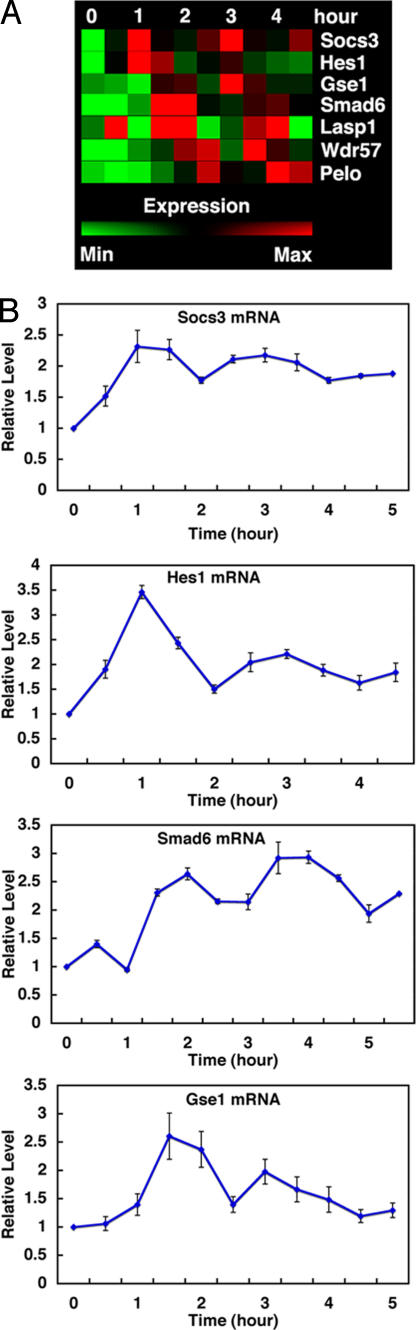
To elucidate the dynamics of regulatory networks, we searched for ultradian oscillators by performing gene expression profiling of mouse fibroblasts (C3H10T1/2) after serum stimulation. Biotinylated cRNAs were prepared every 30 min from t = 0 to 4.5 h (10 time points) and hybridized to high-density microarrays. For screening the data, we set the following three criteria. First, at least two peaks with >1.5-fold higher than the lowest signal value were required during the 4.5-h period. Second, values of two peaks were flagged with “present” and higher than 100 to exclude genes with very weak expression levels. Third, the difference between the values of two peaks and the lowest was statistically significant. These criteria resulted in seven candidate oscillators with a periodicity of ≈2 h of 45,037 probe sets [Fig. 1A and supporting information (SI) Table 1]. None of them had shorter periods, although oscillators with longer periods were not included in this study. Validation of this approach was the observation that the known ultradian oscillator Hes1 was included in this list (Fig. 1A). These candidate genes were further examined, and four of them (Socs3, Hes1, Gse1, and Smad6) were identified as oscillators by real-time PCR (Fig. 1B). Socs3 and Hes1 have the first peak at 1 h, Gse1 at 1.5 h, and Smad6 at 2 h (Fig. 1). Of particular interest are the genes for signal transduction Socs3 and Smad6, downstream molecules of Jak-Stat, and Smad signaling pathways, respectively (16–19), because these pathways are known to exhibit earliest responses to regulate many downstream genes. Jak-Stat signaling mediates the effects of various growth factors and cytokines, whereas Smad signaling mediates the effects of TGF-β/BMP, and both regulate cell proliferation and survival (16–19). Thus, we decided to further characterize these signaling pathways.
Fig. 1.
Identification of serum-induced ultradian oscillators by microarray analysis. (A) Microarray analysis of serum-induced ultradian oscillators. Seven candidate genes are found. (B) Time course of Socs3, Hes1, Gse1, and Smad6 expression in real-time PCR analysis. Means with SE of three independent experiments are shown. The patterns of microarray and real-time PCR experiments are very similar to each other.
Stat-Socs Oscillations.
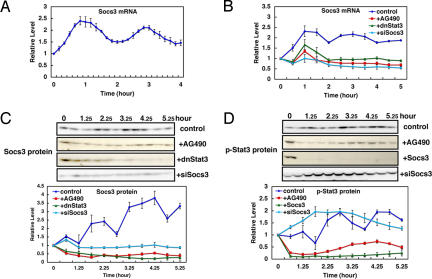
Socs3 is an inhibitor of Jak-Stat signaling (16, 17, 20): phosphorylated Stat3 (at Tyr-705, p-Stat3) induces expression of Socs3, which in turn inhibits Jak-dependent phosphorylation of Stat3. Real-time PCR analysis with shorter intervals (10 min) indicated that Socs3 mRNA expression oscillates with peaks at 50 and 170 min after serum stimulation (Fig. 2A). Western blot analysis showed that Socs3 protein expression displays oscillation with peaks ≈2 and 4 h, indicating ≈1-h delay between mRNA and protein synthesis (Fig. 2C, control).
Fig. 2.
Oscillations in Stat signaling. Cells were treated with serum at t = 0 in the absence (control) or presence of AG490, dnStat3, Socs3, or siSocs3. (A) Expression profiles of Socs3 mRNA were analyzed at 10-min intervals by real-time PCR. The peaks of Socs3 mRNA appear at 50 and 170 min after serum stimulation. (B) Expression profiles of Socs3 mRNA were analyzed by real-time PCR. Socs3 mRNA oscillation is abolished in the presence of AG490, dnStat3, or siSocs3. (C) Expression profiles of Socs3 protein in the absence (control) or presence of AG490, dnStat3, or siSocs3 were analyzed by Western blotting. Socs3 protein oscillation is abolished in the presence of AG490, dnStat3, or siSocs3. (D) Expression profiles of p-Stat3 in the absence (control) or presence of AG490, Socs3, or siSocs3 were analyzed by Western blotting. p-Stat3 oscillation is abolished in the presence of AG490, Socs3, or siSocs3. Means with SE of three independent experiments are shown in all graphs.
We next examined whether Stat3, an activator of Socs3, is also cyclic after serum stimulation. Stat3 mRNA and protein levels are almost constant after serum stimulation (SI Fig. 7 A and B). However, p-Stat3 levels were found to be oscillatory with peaks ≈1, 3, and 5 h (Fig. 2D, control), suggesting that cyclic activation of Stat3 regulates Socs3 oscillation. To show that Socs3 oscillation depends on cyclic phosphorylation of Stat3, we inhibited p-Stat3 formation. Treatment with AG490 inhibited Jak2-dependent phosphorylation of Stat3 (Fig. 2D, +AG490). Under this condition, expression of both Socs3 mRNA and protein are repressed, and their oscillations are abolished (Fig. 2 B and C, +AG490), indicating that Jak2-dependent phosphorylation is essential for Socs3 oscillations. Similarly, in the presence of dominant-negative Stat3 (dnStat3), Socs3 oscillations are abolished (Fig. 2 B and C, +dnStat3), indicating that Socs3 oscillations depend on periodic activation of Stat3.
We next examined whether periodic activation of p-Stat3 depends on Socs3 oscillations. When Socs3 is continuously expressed, periodic formation of p-Stat3 is inhibited (Fig. 2D, +Socs3), suggesting that Socs3 oscillations periodically inhibit formation of p-Stat3. To prove this suggestion, we next knocked down Socs3 expression by its specific siRNA. This siRNA successfully down-regulates both Socs3 mRNA and protein levels (Fig. 2 B and C, +siSocs3, and SI Fig. 8F). Under this condition, p-Stat3 formation is persistently up-regulated, and its oscillation is abolished (Fig. 2D, +siSocs3). These results indicate that p-Stat3 and Socs3 oscillations depend on each other and are regulated by negative feedback.
Because Jak and Stat mediate IL-6 signaling, we next examined whether Socs3 expression oscillates after treatment with IL-6. Socs3 mRNA was found to oscillate after IL-6 stimulation (SI Fig. 8A), just like serum stimulation, suggesting that Stat-Socs oscillations are induced by a single stimulator but not the results of mixed stimulators included in serum.
Smad Oscillations.
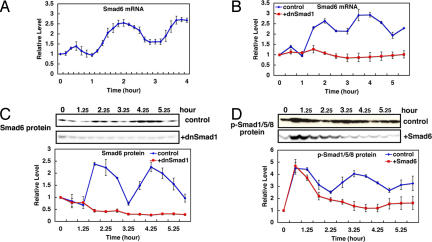
Smad6 is an inhibitor of Smad signaling (18, 19, 21): phosphorylated Smad1/5/8 (p-Smad1/5/8) induces expression of Smad6, which in turn inhibits phosphorylation of Smad1/5/8. Real-time PCR analysis with shorter intervals (10 min) indicated that Smad6 mRNA expression oscillates with peaks at 120 and 230 min after serum stimulation (Fig. 3A). Western analysis showed that Smad6 protein expression displays oscillation with peaks ≈2.5 and 4.5 h (Fig. 3C, control). mRNA and protein for Smad1, an activator of Smad6, do not oscillate after serum stimulation (SI Fig. 7 C and D). However, p-Smad1/5/8 levels were found to be oscillatory with peaks ≈1.5, 3.5, and 5.5 h (Fig. 3D, control), suggesting that periodic activation of Smad1/5/8 regulates Smad6 oscillation. To show that Smad6 oscillations depend on periodic phosphorylation of Smad1/5/8, we inhibited p-Smad1 activity. In the presence of dominant-negative Smad1 (dnSmad1), both Smad6 mRNA and protein oscillations are abolished (Fig. 3 B and C, +dnSmad1), indicating that Smad6 oscillations depend on periodic activation of Smad1. We next asked whether periodic activation of Smad1 depends on Smad6 oscillations. When Smad6 is continuously expressed, periodic formation of p-Smad1 is inhibited (Fig. 3D, +Smad6), indicating that Smad6 oscillations periodically inhibit formation of p-Smad1. These results show that p-Smad1 and Smad6 oscillations depend on each other and are regulated by negative feedback.
Fig. 3.
Oscillations in Smad signaling. Cells were treated with serum at t = 0 in the absence (control) or presence of dnSmad1 or Smad6, and mRNA and protein levels were quantified by real-time PCR and Western blots, respectively. (A) Expression profiles of Smad6 mRNA were analyzed at 10-min intervals. The peaks of Smad6 mRNA appear at 120 and 230 min after serum stimulation. (B) Expression profiles of Smad6 mRNA in the absence (control) or presence of dnSmad1 were analyzed. Smad6 mRNA oscillation is abolished by dnSmad1. (C) Expression profiles of Smad6 protein in the absence (control) or presence of dnSmad1 were analyzed. Smad6 protein oscillation is abolished by dnSmad1. (D) Expression profiles of p-Smad1/5/8 in the absence (control) or presence of Smad6 were analyzed. p-Smad1/5/8 oscillation is abolished by Smad6. Means with SE of three independent experiments are shown in all graphs.
Because Smads mediate BMP signaling, we next examined whether Smad6 expression oscillates after treatment with BMP. Smad6 mRNA was found to oscillate after BMP4 treatment (SI Fig. 9A), just like serum stimulation, suggesting that Smad oscillations are induced by a single stimulator but not the results of mixed stimulators included in serum.
Stat-Socs Oscillations Regulate Hes1 Oscillations.
The above results indicate that Stat-Socs and Smad oscillations are regulated by their own negative feedback loops, like Hes1 oscillation. We next examined whether there are any cross-talks between these oscillators. When Stat-Socs oscillations are inhibited by AG490, dnStat3, or Socs3, both Smad6 and p-Smad1 oscillations are not affected (SI Figs. 9 B–D and 10 A–C). In addition, when Smad oscillations are inhibited, both Socs3 and p-Stat3 oscillations are not affected (SI Figs. 8 B and C and 11 A and B). Thus, there seems to be no clear cross-talk between Stat-Socs and Smad oscillations. Similarly, Hes1 oscillations are not affected in the absence of Smad oscillations (SI Fig. 12), whereas Smad oscillations are not affected in the absence of Hes1 oscillations (SI Figs. 9 E and F and 10 D and E), suggesting there is no cross-talk between Smad and Hes1 oscillations.
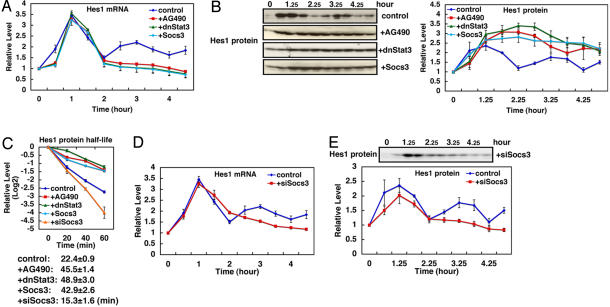
In contrast, Hes1 oscillations are significantly affected by inactivation of Stat-Socs oscillations. When p-Stat3 formation is constitutively suppressed by AG490, dnStat3, or Socs3, Hes1 mRNA is transiently induced by serum treatment, but it remains suppressed at the basal level thereafter (Fig. 4A, compare with control). We also examined Hes1 expression in individual cells by using the real-time imaging method, which used the ubiquitinated luciferase reporter gene under the control of Hes1 promoter (5). Hes1 expression oscillates after serum stimulation in individual cells (SI Fig. 13A), but this oscillation is suppressed at the single-cell level in the presence of AG490 (SI Fig. 13B). In contrast, Hes1 protein is persistently up-regulated after serum treatment in the presence of AG490, dnStat3, or Socs3 (Fig. 4B, compare with control). These results suggest that Hes1 protein is stabilized in the absence of Stat signaling, which may lead to repression of Hes1 mRNA. In accordance with this suggestion, the half-life of Hes1 protein becomes prolonged from 22.4 ± 0.9 to 40–50 min by AG490, Socs3, and dnStat3 (Fig. 4C, compare with control). Conversely, when p-Stat3 formation is constitutively up-regulated by knockdown of Socs3 with siSocs3, both Hes1 mRNA and protein oscillations are again abolished (Fig. 4 D and E, compare with control). Under this condition, Hes1 mRNA tends to be higher than the basal level, whereas the Hes1 protein is near the basal level (Fig. 4 D and E). The half-life of Hes1 protein is shortened to 15.3 ± 1.6 min by constitutive activation of Stat3 (Fig. 4C, +siSocs3). Thus, both sustained activation and inactivation of Stat signaling abolish Hes1 oscillations, suggesting that Stat-Socs oscillations regulate Hes1 oscillations by controlling the half-life of Hes1 protein. We also examined whether Stat-Socs oscillations depend on Hes1 oscillations. However, Stat-Socs oscillations are not affected by overexpression of Hes1 or dnHes1, suggesting that they are independent of Hes1 oscillations (SI Figs. 8 D and E and 11 C and D). It was previously shown that Hes1 associates with both Jak2 and Stat3 and thereby promotes Stat3 phosphorylation (22). Thus, Hes1 protein, but not Hes1 oscillation, is required for periodic Stat activation, and it is likely that coupled oscillations between Stat and Hes1 signaling pathways are occurring after serum stimulation.
Fig. 4.
Regulation of Hes1 oscillation by Stat signaling. (A) Expression of Hes1 mRNA oscillates after serum stimulation (t = 0), but this oscillation is abolished by AG490, dnStat3, or Socs3. (B) Expression of Hes1 protein oscillates after serum stimulation (t = 0) (control), but this oscillation is abolished by AG490, dnStat3, or Socs3. (C) Measurement of Hes1 protein half-life. Hes1 protein levels were measured in the presence of cycloheximide (100 μM), which blocks new protein synthesis. Hes1 protein is degraded with the half-life of 22.4 ± 0.9 min (n = 3). This half-life is elongated by AG490, dnStat3, or Socs3 and shortened by siSocs3. Thus, Stat signaling regulates Hes1 protein stability. (D) Hes1 mRNA oscillation is abolished by siSocs3. (E) Hes1 protein oscillation is abolished by siSocs3. All values are the average of three independent experiments with SE.
Hes1 Oscillations Are Important for Efficient Cell Proliferation.
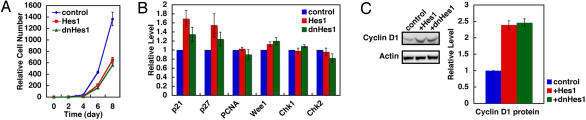
All actively dividing cultured cells that we have examined express Hes1, but without serum stimulation, Hes1 expression levels seem to be stationary on Northern and Western analyses. However, even under this condition, Hes1 expression was found to be oscillatory at the single cell level (5). Hes1 oscillation is just out of synchrony between cells without serum stimulation (5). Because Hes1 expression oscillates in proliferating cells, we next examined the effects of sustained and loss of Hes1 activity on proliferation of fibroblasts. Both cells that express Hes1 protein persistently and cells that lose Hes1 activity do not proliferate extensively, and their proliferation rates are reduced to half, compared with the control cells, where Hes1 expression oscillates (Fig. 5A). These results suggest that Hes1 oscillation is required for efficient cell proliferation.
Fig. 5.
Hes1 oscillation in cell cycles. (A) Comparison of growth curves. Cells that express Hes1 protein persistently and cells that lose Hes1 activity do not proliferate extensively, compared with the control, where Hes1 expression oscillates. (B) Regulation of cell cycle regulators by Hes1. Expression was examined by real-time PCR. Both sustained Hes1 expression (+Hes1) and knockdown of Hes1 activities (+dnHes1) increase expression of p21 and p27, G1 phase genes, compared with oscillatory Hes1 expression (control). Transfected cells with the control vector and nontransfected cells gave the same results (data not shown). (C) Expression of cyclin D1, a G1-specific marker. Both Hes1 and dnHes1 increase cyclin D1 expression, compared with control. All values are the average of three independent experiments with SE.
We next examined effects of different Hes1 expression modes on several cell cycle regulators (23). Expression of the cyclin-dependent kinase inhibitors p21 and p27, G1 phase markers, is up-regulated by persistent expression of Hes1 or dnHes1 (Fig. 5B, +Hes1 and +dnHes1), compared with the control, where Hes1 expression is oscillatory (Fig. 5B, control). Furthermore, expression of another G1-specific marker, cyclin D1, is also increased by persistent expression of Hes1 or dnHes1 (Fig. 5C, +Hes1 and +dnHes1), compared with the control (Fig. 5C, control). In contrast, expression of the S phase marker PCNA and the check point genes Wee1, Chk1, and Chk2 are not significantly affected by Hes1 or dnHes1 (Fig. 5B). Thus, both sustained Hes1 protein expression and knockdown of Hes1 protein activity increase expression of G1-specific markers, suggesting that persistent or down-regulated Hes1 expression leads to G1 phase retardation. It is likely that Stat-Socs oscillations promote cell proliferation by inducing Hes1 oscillation.
Mathematical Simulation of Hes1 Oscillations.
Hes1 oscillation has been mathematically simulated by using a negative autorepression model (4, 24–26). Because a simple negative-feedback loop is insufficient to maintain a stable oscillation, another factor or time delay has been postulated (4, 24–26). We found that p-Stat3 destabilizes Hes1 protein, suggesting that Hes1 protein half-life would be changing by p-Stat3 oscillation. With this assumption, Hes1 oscillation can now be readily simulated without postulating another factor or time delay (Fig. 6A and SI Fig. 14). To reproduce the experimental trend by numerical simulation, we adapted the framework of the negative-feedback model with the change of Hes1 protein half-life. Let m(t) and p(t) be the numbers of Hes1 mRNA and protein molecules, respectively, in a cell at time t. The rates of change of m(t) and p(t) are described as follows.
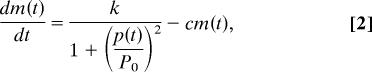 |
where the constant a [h−1] is the rate of production of new proteins, c [h−1] is the degradation rate (inverse half-life) of the mRNA molecules, and k [h−1] and P0 are the constants to represent the inhibitory process by the protein acting as a dimer. Throughout the simulations, we set a = 270.0, c = 1.6, k = 1,980.0, and P0 = 40.0 (24). To represent the change of Hes1 protein half-life by p-Stat3 oscillation, we introduced two types of time dependence to the degradation rate b(t) [h−1], sinusoidal or flip-flop, as follows.
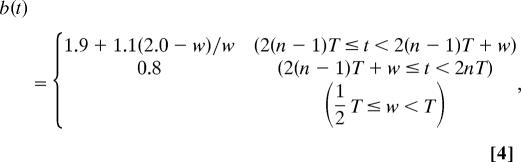 |
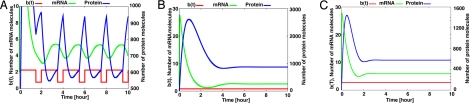
where T is the period of p-Stat3 oscillation, which is determined as 2.0 h by experiment, w in Eq. 4 is a parameter to characterize the flip-flop change of Hes1 protein half-life, and n is an integer. From the experimental value of Hes1 protein half-life induced by p-Stat3 oscillation, we can estimate the average and the trough values of b(t) as 1.9 and 0.8, respectively. With either of these b(t) (Eqs. 3 and 4), Hes1 expression shows stable oscillation (Fig. 6A and SI Fig. 14). In contrast, when the Hes1 protein degradation rate is fixed at certain values, b(t) = 0.8, which represents the absence of p-Stat3 formation, or b(t) = 2.77, which represents persistent formation of p-Stat3, Hes1 oscillation becomes damped, well mimicking the effects of sustained inhibition and activation of Stat signaling (Fig. 6 B and C).
Fig. 6.
Mathematical simulation of Hes1 oscillations. Hes1 oscillations are mathematically simulated using a negative autorepression model. (A) When Hes1 protein half-life is changing, Hes1 expression shows stable oscillation. (B and C) In contrast, when the Hes1 protein degradation rate is fixed at the value b(t) = 0.8, which represents the absence of p-Stat3 formation (B), and b(t) = 2.77, which represents persistent formation of p-Stat3 (C), Hes1 oscillation becomes damped. This simulation well mimics Stat3-dependent Hes1 oscillations.
Our study reveals an unexpected expression mode of signaling molecules in response to serum. We show that in addition to the Notch effector Hes1, Stat-Socs and Smad signaling molecules display oscillatory expression with the periodicity of ≈2 h. These oscillations are regulated by negative feedback, just like Hes1 oscillation (SI Fig. 15). Interestingly, Smad6 oscillation is delayed compared with Socs3 and Hes1 oscillations, although phosphorylation of Stat3 and Smad1 occurs simultaneously (Figs. 2D and 3D). Induction of Socs3 mRNA also occurs simultaneously with phosphorylation of Stat3, whereas that of Smad6 mRNA is delayed ≈1 h, suggesting that p-Stat3 functions faster than p-Smad1. The kinetics of these factors remains to be analyzed. Strikingly, Stat-Socs oscillations lead to Hes1 oscillations by regulating the stability of Hes1 protein. The mechanism by which Stat signaling regulates the Hes1 protein stability remains to be determined. Physical interaction with Jak and Stat (22) could lead to destabilization of Hes1 protein. Another possibility is that p-Stat3 induces expression of genes that regulate degradation of Hes1 protein. Further analysis on the E3 ligase for Hes1 protein may be required to clarify this issue.
We also show that both sustained Hes1 expression and loss of Hes1 activity reduce cell proliferation by inducing G1 phase retardation, suggesting that Hes1 oscillation is important for efficient cell proliferation. It is likely that Hes1 is required at a certain point but should be down-regulated at another point of the cell cycle, although it remains to be determined at which points Hes1 promotes and inhibits the cell cycle progression. Another important question is whether the Hes1 oscillator links the cell cycle to the somite segmentation clock. However, no obvious defect in the segmentation and the cell cycle was observed in Hes1-null mice (7), probably due to compensation by its related genes such as Hes7. Analysis of compound mutations would be required to answer this question.
In our microarray analysis, none of the downstream genes in the Notch-Hes1, Stat-Socs, and Smad pathways is found to display oscillatory expression. It is possible that oscillations in the Notch-Hes1, Jak-Stat-Socs, and Smad pathways are important just to maintain expression of downstream genes within a critical range rather than oscillatory. Alternatively, expression of downstream genes could easily become out of synchrony between cells, which might make it difficult to identify oscillatory expression. In that case, expression should be examined at the single cell level.
It has been reported that NF-κB and p53 signaling pathways also display oscillatory responses with the periods of ≈100 and 440 min, respectively (27–30). In these pathways, IκB and Mdm2 act as negative regulators for NF-κB and p53, respectively. In our present study, oscillations in the NF-κB signaling are not observed, probably because it is not activated by serum. We assume that other signaling pathways with negative feedback loops, which do not respond to serum, would also display oscillatory responses to different stimulations. Our results raise the possibility that many cellular activities are rhythmically controlled by combinations of different types of ultradian oscillators and suggest that the number of pulses as well as the duration and intensity may be important for cells to make final decisions.
Materials and Methods
Cell Culture and RNA and Protein Purification.
C3H10T1/2 mouse fibroblast cells were treated with serum at t = 0, as described (4), and RNA was prepared every 10 or 30 min. Proteins were prepared at t = 45 min (0.75 h) and every 30 min thereafter. Total RNA was purified by using TRIzol reagents (Invitrogen, Carlsbad, CA) from cells in 100-mm dishes or six-well plates. Protein was purified from cells in 100-mm dishes by Cell Lysis Buffer (50 mM Tris·Cl, pH 8.0/5 mM EDTA/150 mM NaCl/1% Nonidet P-40) with Protease Inhibitor Mixture (Nacalai Tesque, Kyoto, Japan) and phosphatase inhibitors (1 mM NaF/1 mM Na3VO4/1 mM β-glycerophosphate/1 mM sodium pyrophosphate). For treatment with AG490, the final concentration of 50 μM was added to the medium with serum stimulation. For treatment with BMP4 or IL-6, medium containing 100 ng/ml each without serum was added at t = 0.
Microarray Analysis.
Total RNA was prepared from cultured cells at indicated time points (serum stimulation at t = 0). Microarray analysis using Gene Chip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA) was done, according to the manufacturer's protocols. Data were analyzed by using GCOS (Affymetrix) and Gene Spring (Agilent Technologies, Austin, TX). In brief, per-chip normalization was done by using the value of median, and then per-gene normalization was done by using the value t = 0. To explore the candidates for new oscillators, the following three criteria were set. First, at least two peaks with >1.5-fold higher than the lowest signal value were required during the 4.5-h period. This result should be obtained in at least two independent experiments among the three. Second, the signal intensities of prospective peak points should be flagged with “Present” and higher than 100. Third, the difference between the peak and the lowest signal intensities should be statistically significant (P < 0.05, unpaired one-tail t test). The microarray data will be deposited in the Genome Network Platform (http://genomenetwork.nig.ac.jp).
Real-Time PCR.
Total RNA was reverse-transcribed by using ReverTra Ace (TOYOBO) and Random Primer (TOYOBO). Real-time PCR was done by using Applied Biosystems 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) and SYBR Premix EX Taq (TAKARA), according to the manufacturer's protocols. PCR primers are described in SI Text. GAPDH was used as a control, and data were normalized to the value of t = 0.
Western Blotting.
Antibodies used for Western blotting are as follows: anti-Hes1 (gift from Tetsuo Sudo, Toray, Japan), anti-Socs3 (Fusion Antibodies, Northern Ireland; FA1017), anti-Smad6 (IMGENEX; IMG-555), anti-phospho-Y705-Stat3 (Cell Signaling Technology, Danvers, MA; #9131), anti-Smad1 (Cell Signaling Technology; #9512), anti-phospho-Smad1/5/8 (Cell Signaling Technology; #9511), anti-Stat3 (BD Transduction Laboratories, San Jose, CA; 610189), anti-Cyclin D1 (Santa Cruz Biotechnology, Santa Cruz, CA; sc-718), anti-Actin (Sigma; A2066), anti-rabbit IgG (Amersham Biosciences, Little Chalfont, Buckinghamshire, U.K.; NA9340V) and anti-mouse IgG antibodies (Amersham Biosciences; NA9310). The signal was detected by SuperSignal West Dura Extended Duration Substrate (Pierce, Rockford, IL) and Hyperfilm ECL (Amersham Biosciences) or LAS-3000 mini (Fujifilm, Tokyo, Japan). Means with SE were calculated from three independent experiments.
Expression Vectors.
For misexpression of Hes1, Socs3, and Smad6, the coding region of each cDNA was cloned into pCI expression vector (Promega, Madison, WI). For dnHes1, mouse Hes1 cDNA with mutations of E43A, K44A, and R47A was cloned into pCI expression vector. For dnStat3, mouse Stat3 cDNA with mutation of Y705F was cloned into pCI expression vector. For dnSmad1, mouse Smad1 with mutation of A422stop was cloned into pCI expression vector. For knockdown of Socs3 by the siRNA method, the following fragments were cloned into psiRNA-h7SKneo vector, according to the manufacturer's protocol (InvivoGen, San Diego, CA): siSocs3 sense strand, acctcGCATCTTTGTCGGAAGACTGTtcaagag ACA G T CTTCCG A C A A AGATGCtt; siSocs3 antisense strand, caaaaaGCATCTTTGTCGGAAGACTGTctcttgaACAGTCT T C CG ACAAAGATGCg; siGFP sense strand, acctcGCAAGCTGACCCTGAAGTTCAccaccTGAACTTCAGGGTCAGCTTGCtt; and siGFP antisense strand, caaaaaGCAAGCTGACCCTGAAGTTCAggtggTGAACTTCAGGGTCAGCTTGCg. siGFP was used as a control.
Transfection of Expression Vectors.
Cells were transfected with expression vectors with pCI-neo by using Lipofectamine reagent and Plus reagent (Invitrogen), and transfected cells were selected by 1 mg/ml G418 (GIBCO, Carlsbad, CA).
Measurement of Hes1 Protein Half-Life.
Cells were treated with 100 μM cycloheximide and harvested at indicated time points (cycloheximide treatment at t = −20 min), and cell extracts were subjected to Western blotting using anti-Hes1 antibody.
Real-Time Imaging.
Cells containing the reporter Hes1-Ub2-Luc were cultured in DMEM/10% FBS/1 mM luciferin (Nacalai Tesque), and bioluminescence was measured as described (5).
Supplementary Material
Acknowledgments
We thank Yoshito Masamizu, Hiroshi Yamazaki, and Hiromi Hirata for technical assistance and Tetsuo Sudo for the Hes1 antibody. This work was supported by the Genome Network Project and the Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Uehara Memorial Foundation. S.Y. was supported by the 21st Century Centers of Excellence Program and the Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Genome Network Platform database, http://genomenetwork.nig.ac.jp (MicroArray Analysis for Ultradian Oscillation).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701837104/DC1.
References
- 1.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 2.Kelly K, Siebenlist U. Curr Opin Immunol. 1995;7:327–332. doi: 10.1016/0952-7915(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 3.Sheng M, Greenberg ME. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 4.Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 5.Masamizu Y, Ohtsuka T, Takashima Y, Nagahara H, Takenaka Y, Yoshikawa K, Okamura H, Kageyama R. Proc Natl Acad Sci USA. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R. Development (Cambridge, UK) 2006;133:2467–2476. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- 7.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 8.Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Development (Cambridge, UK) 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 9.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pourquié O. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- 11.Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R. Genes Dev. 2001;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, Kageyama R. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- 13.Zhang N, Gridley T. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
- 14.Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- 15.Serth K, Schuster-Gossler K, Cordes R, Gossler A. Genes Dev. 2003;17:912–925. doi: 10.1101/gad.250603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy DE, Darnell JE., Jr Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Jove R. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 18.Massagué J, Wotton D. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ten Dijke P, Miyazono K, Heldin C-H. Trends Biosci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- 20.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 21.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 22.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- 23.Sherr CJ, Roberts JM. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 24.Lewis J. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- 25.Monk NAM. Curr Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- 26.Jensen MH, Sneppen K, Tiana G. FEBS Lett. 2003;541:176–177. doi: 10.1016/s0014-5793(03)00279-5. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann A, Levchenko A, Scott ML, Baltimore D. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 28.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, et al. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 29.Bar-Or RL, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Proc Natl Acad Sci USA. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.