Abstract
Synthetic biology involves the engineering of biological organisms by using modular and generalizable designs with the ultimate goal of developing useful solutions to real-world problems. One such problem involves bacterial biofilms, which are crucial in the pathogenesis of many clinically important infections and are difficult to eradicate because they exhibit resistance to antimicrobial treatments and removal by host immune systems. To address this issue, we engineered bacteriophage to express a biofilm-degrading enzyme during infection to simultaneously attack the bacterial cells in the biofilm and the biofilm matrix, which is composed of extracellular polymeric substances. We show that the efficacy of biofilm removal by this two-pronged enzymatic bacteriophage strategy is significantly greater than that of nonenzymatic bacteriophage treatment. Our engineered enzymatic phage substantially reduced bacterial biofilm cell counts by ≈4.5 orders of magnitude (≈99.997% removal), which was about two orders of magnitude better than that of nonenzymatic phage. This work demonstrates the feasibility and benefits of using engineered enzymatic bacteriophage to reduce bacterial biofilms and the applicability of synthetic biology to an important medical and industrial problem.
Keywords: phage therapy, synthetic biology
Over the last few years, synthetic biology has enabled the development of many engineered biological devices and cells with interesting and well modeled characteristics (1–3). At the same time, new technologies for more cost-effective DNA synthesis and sequencing have been reported (4). These advances will allow for large-scale synthetic genomes to be designed and built with much greater ease than is currently possible with traditional molecular biology methods. Synthetic biologists have begun to address important real-world problems by modifying organisms to produce artemisin precursors (5), developing bacteria that can target cancerous cells (6), and producing new antimicrobial peptides (7), to name a few examples (2). Synthetic biology is distinguished from traditional genetic engineering through the use of modularity, abstraction, and standardization to allow generalized principles and designs to be applied to different scenarios. In this work, we engineered bacteriophage with biofilm-degrading enzymatic activity to create a synthetic biology platform for eradicating bacterial biofilms.
Bacteria frequently live in biofilms, which are surface-associated communities encased in a hydrated extracellular polymeric substances (EPS) matrix that is composed of polysaccharides, proteins, nucleic acids, and lipids and helps maintain a complex heterogeneous structure (8, 9). Biofilms constitute an essential and protective lifestyle for bacteria in many different natural and man-made environments, including dental plaques, water pipes, medical devices, and industrial systems (10). Bacterial biofilms have been implicated as a source of persistent infection and contamination in medical, industrial, and food processing settings due to inherent resistance to antimicrobial agents and host defenses (8, 11–13). Thus, there exists a growing need for novel and effective treatments targeted at biofilms, particularly in light of the continually worsening problem of antibiotic resistance and the discovery that antibiotic use can even induce biofilm formation (14, 15).
Bacteriophage treatment has been proposed as one method for controlling bacterial biofilms (16). Phage have been used since the early 20th century to treat bacterial infections, especially in Eastern Europe, and have been shown to decrease biofilm formation (16, 17). For example, phage T4 can infect and replicate within Escherichia coli biofilms and disrupt biofilm morphology by killing bacterial cells (18–20). Phage have also been modified to extend their natural host range. E. coli, which produces the K1 polysaccharide capsule, is normally resistant to infection by T7 but is susceptible to T7 that has been designed to express K1-5 endosialidase (21). Enzymatic degradation of EPS components is another useful strategy for disrupting biofilms, although bacterial cells are not killed (8, 22, 23). For instance, enzymatic degradation of a cell-bound EPS polysaccharide adhesin known as polymeric β-1,6-N-acetyl-d-glucosamine by exogenously applied dispersin B (DspB) has been demonstrated to reduce biofilms of several different species of bacteria (22). DspB, an enzyme that is produced by Actinobacillus actinomycetemcomitans, hydrolyzes β-1,6-N-acetyl-d-glucosamine, a crucial adhesin needed for biofilm formation and integrity in Staphylococcus and E. coli, including E. coli K-12, as well as clinical isolates (22). Reports of natural lytic phage with phage-borne polysaccharide depolymerases have shown that phage-induced lysis and EPS degradation are used in combination in natural systems to reduce bacterial biofilms (24, 25). These depolymerases appear to be carried on the surfaces of phage and degrade bacterial capsular polysaccharides to allow access to bacterial cell surfaces (24). However, the chance that one can isolate a natural phage that is both specific for the bacteria to be targeted and expresses a relevant EPS-degrading enzyme is likely to be low (26).
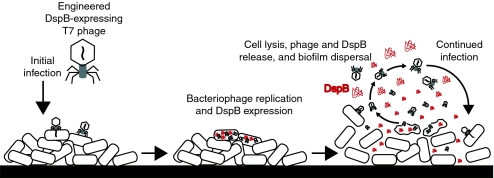
Therefore, we propose a modular design strategy in which phage that kill bacteria in a species-specific manner are engineered to express the most effective EPS-degrading enzymes specific to the target biofilm. This strategy should permit the development of a diverse library of biofilm-dispersing phage rather than trying to isolate such phage from the environment. By multiplying within the biofilm and hijacking the bacterial machinery, engineered enzymatically active phage should be able to achieve high local concentrations of both enzyme and lytic phage to target multiple biofilm components, even with small initial phage inoculations. Rapid phage replication with subsequent bacterial lysis and expression of biofilm-degrading enzymes should render this two-pronged attack strategy an efficient, autocatalytic method for removing bacterial biofilms in environmental, industrial, and clinical settings (Fig. 1). This design also removes the need to express, purify, and deliver large doses of enzyme to specific sites of infection that may be difficult to access and should improve the efficacy of phage therapy at removing biofilms. Increasingly cost-effective genome sequencing and synthetic biology technologies, which include the refactoring of phage genomes and large-scale DNA synthesis (2, 27, 28), should further enable the production of engineered enzymatic phage and significantly extend the limited repertoire of biofilm-degrading phage that have been isolated from the environment.
Fig. 1.
Two-pronged attack strategy for biofilm removal with enzymatically active DspB-expressing T7DspB phage. Initial infection of E. coli biofilm results in rapid multiplication of phage and expression of DspB. Both phage and DspB are released upon lysis, leading to subsequent infection as well as degradation of the crucial biofilm EPS component, β-1,6-N-acetyl-d-glucosamine (22).
Results
Design of Enzymatically Active Bacteriophage.
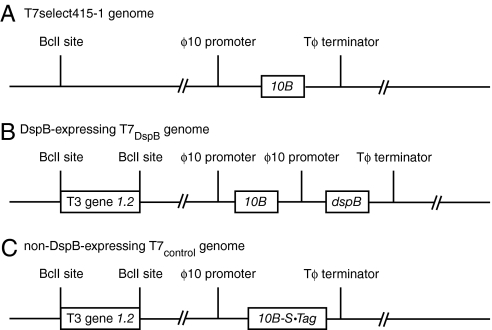
As a proof-of-principle design of artificial biofilm-degrading bacteriophage, we engineered T7, an E. coli-specific phage (29, 30), to express DspB intracellularly during infection so DspB would be released into the extracellular environment upon cell lysis (Fig. 1). We used a modified T7 strain (Novagen T7select415-1) with several deletions of nonessential genes (Fig. 2A). We cloned the gene coding for DspB (dspB) under the control of the strong T7 ϕ10 promoter so that dspB would be strongly transcribed by T7 RNA polymerase during infection (Fig. 2B). As a control, we cloned an S·Tag insert into the T7 genome so that no DspB would be produced (Fig. 2C).
Fig. 2.
Genomes of engineered phage used for biofilm treatment. (A) Genome of T7select415-1 shows a unique BclI site and capsid gene 10B. (B) DspB-expressing phage T7DspB was created by cloning T3 gene 1.2 into the unique BclI site and cloning the ϕ10-dspB construct after capsid gene 10B. (C) Non-DspB-expressing control phage T7control was created by cloning T3 gene 1.2 into the unique BclI site and cloning the control S·Tag insert (included in the T7select415-1 kit) as a fusion with the capsid gene 10B.
To test the effectiveness of our engineered phage against pregrown biofilm, we cultivated E. coli TG1(lacI::kan) biofilms in LB media on plastic pegs by using the standardized MBEC biofilm cultivation system. We used E. coli TG1 as the target biofilm strain because TG1 forms a thick, mature biofilm and contains the F plasmid (31). The F plasmid enhances biofilm maturation along with other biofilm-promoting factors in E. coli, including β-1,6-N-acetyl-d-glucosamine, flagellum, cellulose, curli, antigen 43, and other conjugative pili and cell surface adhesins (31, 32). Because T7 is unable to replicate efficiently in F-plasmid-containing E. coli, gene 1.2 from T3 phage was also cloned into the unique BclI site in our engineered T7 phage and control T7 phage to circumvent F-plasmid-mediated exclusion and extend the phage host range (Fig. 2 B and C) (33). The control phage and engineered phage were named T7control and T7DspB, respectively (Fig. 2 B and C).
Characterization of Enzymatically Active Bacteriophage.
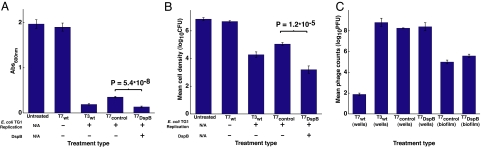
To determine whether the T7DspB phage was more effective than the T7control phage, we first used a crystal violet (CV) assay to assess the amount of biofilm on the pegs after phage treatment. Pregrown TG1(lacI::kan) biofilm was inoculated with only LB media or infected with T7control or T7DspB phage at 103 plaque forming units (PFU) per peg (Fig. 3A). To assess whether our engineered enzymatic phage was more efficacious than wild-type phage at attacking biofilm despite being made with a modified T7 phage, we also treated biofilm with wild-type T7 (T7wt) or wild-type T3 (T3wt) (Fig. 3A). After 24 h of treatment, CV staining of untreated biofilm had a 600-nm absorbance (A600) approximately equal to that for T7wt-treated biofilm (Fig. 3A). Both T3wt-treated biofilm and T7control-treated biofilm were much reduced compared with the untreated biofilm: The former had an A600 that was lower than that of untreated biofilm by a factor of 10.3, whereas the latter had an A600 that was lower than that of untreated biofilm by a factor of 5.6 (Fig. 3A). The amount of biofilm left on the T7DspB-treated pegs was the least of all of the treatment types, with an A600 that was less by a factor of 14.5 than that of untreated biofilm and less by a factor of 2.6 than that of T7control-treated biofilm (P = 5.4 × 10−8). These findings demonstrate that DspB expression in T7DspB is crucial to elevating its biofilm-removing efficacy over that of wild-type phage and nonenzymatic T7control phage (Fig. 3A).
Fig. 3.
Assays for E. coli TG1 biofilm levels and phage counts after 24 h with no treatment or with treatment with phage T7wt, phage T3wt, non-DspB-expressing phage T7control, or DspB-expressing phage T7DspB. Error bars indicate SEM. (A) Mean absorbance (600 nm) for n = 16 biofilm pegs stained with 1% CV, solubilized in 33% acetic acid, and diluted 1:3 in 1× PBS (50). (B) Mean cell densities [log10(CFU per peg)] for n = 12 biofilm pegs. Pegs treated with T7DspB resulted in a 3.65 log10(CFU per peg) reduction in viable cells recovered from E. coli biofilm compared with untreated biofilm. (C) Mean phage counts [log10(PFU per peg)] recovered from media in n = 3 microtiter plate wells (wells) or sonication of n = 3 biofilm pegs (biofilm), as indicated, after 24 h of treatment with initial inoculations of 103 PFU per well. Both T7control and T7DspB showed evidence of replication with phage counts obtained from the microtiter plate wells or with phage counts recovered from the biofilms after sonication.
To confirm that the decrease in CV staining corresponded with killing of biofilm cells, we used sonication to obtain viable cell counts (CFU per peg) for bacteria surviving in the biofilms after phage treatment. Pregrown TG1(lacI::kan) biofilm (before treatment) reached a mean cell density of 6.4 log10(CFU per peg) after 24 h of growth (Fig. 3B). After 24 h of additional growth in new LB media with no phage treatment, the untreated biofilm had a mean cell density of 6.9 log10(CFU per peg) (Fig. 3B). T3wt-treated biofilm had a mean cell density that was less than that of T7control-treated biofilm by a factor of 5.9 and greater than that of T7DspB-treated biofilm by a factor of 12 (Fig. 3B). T7control-treated biofilm had a mean cell density of 5.1 log10(CFU per peg), whereas the mean cell density for T7DspB-treated biofilm was 3.2 log10(CFU per peg), the lowest of all of the treatment types (Fig. 3B). The difference in viable cells recovered from T7control-treated biofilm and T7DspB-treated biofilm was statistically significant (P = 1.2 × 10−5). These results are consistent with the CV staining data and demonstrate that DspB-expressing T7DspB phage are substantially more effective at killing E. coli TG1 biofilm compared with T3wt, T7wt, and non-DspB-expressing T7control phage.
Our two-pronged method of biofilm eradication involves expression of DspB and rapid phage replication (Fig. 1). To confirm that our phage multiplied, we obtained PFU counts from media in the microtiter plate wells. By 24 h of treatment, T7wt had not replicated but T3wt had multiplied significantly within the biofilm (Fig. 3C). To compare the amount of phage in the microtiter plate wells with phage residing in the biofilms, we also obtained PFU counts by sonicating the biofilms. After 24 h of treatment, PFU counts for T7control and T7DspB recovered from the microtiter plate wells were several orders of magnitude greater than PFU counts recovered by sonication of the biofilms (Fig. 3C). Overall, PFU counts obtained from the wells and the biofilms were all orders of magnitude greater than the initial inoculation of 103 PFU, confirming that phage multiplication indeed took place (Fig. 3C).
Time Courses and Dose–Responses for Enzymatically Active Bacteriophage Treatment.
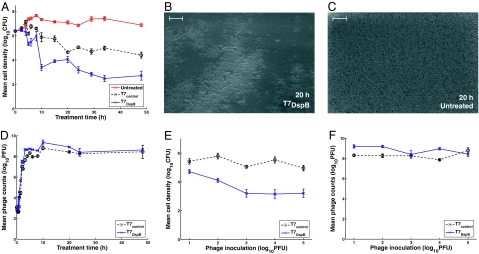
Because we determined that T7DspB had greater biofilm-removing capability than T7control after 24 h of infection, we next sought to determine the time course of biofilm destruction. As shown in Fig. 4A, by 5 h after infection, T7DspB-treated biofilm had a mean cell density that was 0.82 log10(CFU per peg) less than T7control-treated biofilm (P = 2.0 × 10−4). At 10 h after infection, T7DspB-treated biofilm began to settle at a steady-state mean cell density between 3 and 4 log10(CFU per peg), whereas T7control-treated biofilm flattened out at ≈5 log10(CFU per peg) by 20 h after infection (Fig. 4A). T7DspB-treated biofilms had mean cell densities that were approximately two orders of magnitude lower than T7control-treated biofilms, up to 48 h of total treatment (Fig. 4A), and, importantly, T7DspB treatment reduced biofilm levels by ≈99.997% [4.5 log10(CFU per peg)] compared with untreated biofilm. We found no evidence of phage resistance developing over the long time course of treatment (Fig. 4A).
Fig. 4.
Time-course curves, dosage–response curves, and SEM images for engineered phage treatment targeting E. coli TG1 biofilm. (A and E) Each data point represents the mean log10-transformed cell density of n = 12 biofilm pegs. (D and F) Each data point represents the mean log10-transformed phage counts obtained from n = 3 microtiter plate wells. Error bars indicate SEM. (A) Time course (up to 48 h) of viable cell counts for no treatment (red squares), treatment with T7control (black circles), or treatment with T7DspB (blue crosses) demonstrates that T7DspB significantly reduced biofilm levels compared with T7control. (B) SEM image of T7DspB-treated biofilm after 20 h shows significant disruption of the bacterial biofilm. (C) SEM image of untreated biofilm after 20 h shows a dense biofilm. (D) Time course of phage counts obtained after initial inoculation of E. coli TG1 biofilm with 103 PFU per well of T7control (black circles) or T7DspB (blue crosses). Both T7control and T7DspB began to replicate rapidly after initial inoculation. (E) Dose–response curves of mean cell densities (measured after 24 h of treatment) for T7control (black circles) and T7DspB (blue crosses). For all initial phage inoculations, T7DspB-treated biofilm had significantly lower mean cell densities compared with T7control-treated biofilm. (F) Dose–response curves of mean phage counts (measured after 24 h of treatment) for T7control (black circles) and T7DspB (blue crosses). For all initial phage inoculations, both T7control and T7DspB multiplied significantly. (Scale bars, 10 μm.)
We also used a SEM to image the biofilm pegs over the time course of phage treatment to directly visualize biofilm dispersal by our enzymatically active phage [Fig. 4 B and C and supporting information (SI) Fig. 5]. After 20 h of treatment, T7DspB-treated biofilm (Fig. 4B) was significantly disrupted compared with the untreated biofilm (Fig. 4C). These results confirm that T7DspB indeed causes biofilm reduction and bacterial cell killing.
To verify that phage replication was occurring over time, we obtained PFU counts in the microtiter wells. As seen in Fig. 4D, both T7control and T7DspB began to replicate within the bacterial biofilm as early as 50 min after infection. By ≈190 min, T7control and T7DspB PFU per peg approached steady-state levels of ≈8–9 log10(PFU per peg), indicating that phage replication had occurred (Fig. 4D). T7DspB PFU per peg were generally higher than T7control PFU per peg but not by orders of magnitude, as was the case for CFU counts per peg. This finding results from the T7 burst size (≈250 PFU per infective center) (34) multiplied by the number of the extra cells killed by T7DspB, compared with T7control, equaling extra PFU per peg that are insignificant compared with the PFU levels already reached by T7control. We did not note any significant differences in burst sizes and growth rates between T7DspB and T7control (data not shown).
Considering that the above experiments were carried out with initial inoculations of 103 PFU per peg, which translates to a multiplicity of infection of ≈1:103.4 (Fig. 4A), we next aimed to determine the effect of changing the initial multiplicity of infection on biofilm removal. With low phage doses, repeated rounds of phage multiplication and DspB expression should promote biofilm dispersal and allow more bacterial cells to be accessible for subsequent phage infection. With high phage doses, initial DspB production after infection should also be very disruptive to biofilm integrity. As shown in Fig. 4E, T7DspB was more effective than T7control at removing biofilm at all inoculation levels tested, ranging from 101 PFU per peg to 105 PFU per peg. A dose-dependent effect of phage inoculation on biofilm destruction was observed, with larger inoculations leading to lower mean cell densities, particularly for T7DspB (Fig. 4E). At inoculation levels greater than or equal to 102 PFU per peg, mean cell densities (CFU per peg) for T7DspB-treated biofilm were significantly lower than those for T7control-treated biofilm by factors of 49–232 (Fig. 4E). Thus, at low and high initial inoculations, DspB-expressing T7 is more efficacious at disrupting E. coli TG1 biofilm compared with non-DspB-expressing control T7. Note also that all phage dosages tested exhibited phage multiplication within the biofilm (Fig. 4F). These results together suggest that DspB-expressing phage may have improved efficacy in real-world situations for which the ability to deliver high levels of phage to biofilms may be limited or for which sustained phage replication is less likely, e.g., in the gastrointestinal tract of cholera patients (35, 36).
Discussion
In this work, we demonstrated that engineered phage that express biofilm-degrading enzymes are more efficacious at removing bacterial biofilms than nonenzymatic phage alone. Although our results were obtained for a prototype, proof-of-principle phage, we believe that our design can be adapted to work in other phage and with other biofilm-degrading enzymes to target a wide range of biofilms. Thus, engineered bacteriophage treatment should be considered as an addition to the therapies available for use against bacterial biofilms in medical, industrial, and biotechnological settings (17). Future improvements to this design may include directed evolution for optimal enzyme activity, delaying cell lysis or using multiple phage promoters to allow for increased enzyme production, targeting multiple biofilm EPS components with different proteins as well as targeting multispecies biofilm with a mixture of different species-specific engineered enzymatically active phage, and combination therapy with antibiotics and phage to improve the efficacy of both types of treatment.
Phage therapy has begun to be accepted in industrial and biotechnological settings. For example, the Food and Drug Administration recently approved the use of phage targeted at Listeria monocytogenes as a food additive (37). However, phage therapy has several challenges that must be overcome before it will be accepted in Western medicine for treating humans (17). These problems include the lack of properly designed clinical trials to date (17), development of phage resistance (26, 36, 38), phage immunogenicity in the human body and clearance by the reticuloendothelial system (26, 35), the release of toxins upon bacterial lysis (26), and phage specificity (26). Fortunately, many of these concerns are currently being studied and addressed. For example, combination therapy with antibiotics and phage may alleviate the development of phage resistance (26, 36, 38). Long-circulating phage can be isolated that can avoid reticuloendothelial system clearance to increase in vivo efficacy (35). The problem of phage clearance is an important one that needs to be solved as it may make phage therapy more useful for treating transient infections rather than chronic ones. Nonlytic and nonreplicative phage have been engineered to kill bacteria while minimizing endotoxin release (39, 40). Progress is also being made in the development of toxin-free phage preparations (41).
The specificity of phage for host bacteria is both an advantage and a disadvantage for phage therapy. Specificity allows human cells as well as innocuous bacteria to be spared, potentially avoiding serious issues, such as drug toxicity or Clostridium difficile overgrowth that can arise with antibiotic use. C. difficile infection is characterized by diarrhea and colitis and has increased in severity in recent years (42). Antibiotic therapy is believed to alter the microbial flora in the colon due to lack of target specificity, thus allowing C. difficile to proliferate and cause disease (43). Furthermore, the ability of our engineered phage to use the local bacterial synthetic machinery to produce biofilm-degrading enzymes means that exogenously applied enzymes, which could have unintended effects on off-target biofilms, are not needed. However, host specificity means that a well characterized library of phage must be maintained so that an appropriate therapy can be designed for each individual infection (26). The diversity of bacterial infections implies that it may be difficult for any particular engineered phage to be a therapeutic solution for a wide range of biofilms. Indeed, phage therapy generally requires the use of phage cocktails to cover a range of target bacteria.
Overcoming the difficulty of creating a collection of enzymatically active engineered phage is a problem that can be solved by new cost-effective, large-scale DNA sequencing and DNA synthesis technologies (2, 4, 44). Sequencing technologies will allow the characterization of collections of natural phage that have been used in phage typing and phage therapy for many years (45, 46). Once these phage have been better understood, synthesis technologies should enable the addition of biofilm-degrading enzymes to produce new, modified phage. Furthermore, rational engineering methods with new synthesis technologies can be used to broaden phage host range. For example, T7 has been modified to express K1-5 endosialidase, allowing it to effectively replicate in E. coli that produce the K1 polysaccharide capsule (21). In this study, we took advantage of gene 1.2 from phage T3 to extend our phage host range to include E. coli that contain the F plasmid, thus demonstrating that multiple modifications of a phage genome can be done without significant impairment of the phage's ability to replicate (33). Bordetella bacteriophage use an intriguing reverse-transcriptase-mediated mechanism to produce diversity in host tropism, which may provide inspiration for future designs (47, 48). In addition, using enzymes such as DspB, which target important adhesins that are common to a broad range of bacterial species, including clinical strains, should also help enzymatically active phage be applicable to a greater number of infections (22). Along these lines, the many biofilm-promoting factors required by E. coli K-12 to produce a mature biofilm are likely to be shared among different biofilm-forming bacterial strains and are thus potential targets for engineered enzymatic bacteriophage (32).
Conclusion
Because antibiotic resistance in biofilms poses a significant hurdle to eliminating biofilms with conventional antimicrobial drugs, new antibiofilm strategies, such as phage therapy, should be explored. Novel synthetic biology technologies should enable the engineering of natural phage with biofilm-degrading enzymes to produce libraries of enzymatically active phage, which could complement efforts to screen for new biofilm-degrading bacteriophages in the environment. Once bacteriophage therapy itself becomes better understood and used, engineered bacteriophage with biofilm-degrading enzymatic activity could become a viable option in meeting the challenge of biofilm control in environmental, industrial, and clinical settings.
Materials and Methods
Bacterial Strains, Bacteriophage, and Chemicals.
E. coli TG1 [F′traD36 lacIqΔ(lacZ) M15 proA+B+/supE Δ(hsdM-mcrB)5 (rk− mk− McrB−) thi Δ(lac-proAB)] was obtained from Zymo Research (Orange, CA). The strain TG1 (lacI::kan) used to grow biofilm was created by one-step inactivation of the lacI gene by a kanamycin-resistance cassette (49). E. coli BL21 was obtained from Novagen (San Diego, CA). Wild-type T7 (ATCC no. BAA-1025-B2) and T3 (ATCC no. 11303-B3) were purchased from American Type Culture Collection (Manassas, VA). Standard chemicals were obtained from sources as described in SI Materials and Methods.
Construction and Purification of Engineered Phage.
Our engineered T7 phage was created by using the T7select415-1 phage display system (Novagen) with standard molecular biology techniques. Instead of cloning DspB onto the phage surface, we designed the T7select phage to express DspB intracellularly during infection. The dspB gene was cloned from A. actinomycetemcomitans genomic DNA (ATCC no. 700685D) under the control of the strong T7 ϕ10 promoter downstream of the T7select415-1 10B capsid gene and stop codons in all three reading frames to create T7DspB-precursor (Fig. 2B). Packaging of the modified genome was done with the T7select packaging extracts. The control phage, T7control-precursor, was constructed by cloning the T7select control S·Tag insert into the T7select415-1 genome (Fig. 2C). Because T7wt cannot replicate normally in F-plasmid-containing E. coli, we cloned gene 1.2 from phage T3 into the unique BclI site in T7DspB-precursor and T7control-precursor to create T7DspB and T7control, respectively, which are able to escape exclusion by the F plasmid (Fig. 2 B and C) (33). The resulting phage were amplified on E. coli BL21 and plated on E. coli TG1(lacI::kan) to isolate T7DspB (Fig. 2B) and T7control (Fig. 2C), which were confirmed by PCR to have T3 gene 1.2. Details are available in SI Materials and Methods.
Before biofilm treatment, T7DspB and T7control were amplified on E. coli BL21 and purified. Twelve milliliters of BL21 overnight cultures were diluted with 12 ml of LB in 125-ml flasks, inoculated with 30 μl of high-titer phage stock, and allowed to lyse at 37°C and 300 rpm (model G25 incubator shaker, New Brunswick Scientific) for 3 h. Lysed cultures were clarified by centrifuging for 10 min at 10,000 × g and filtering the supernatants through 0.2-μm filters (catalog no. 190-2520; Nalge Nunc International, Rochester, NY). The purified solutions were centrifuged in a Beckman SW.41T rotor for 1 h at 150,000 × g to concentrate the phage. The supernatants were removed, and pellets were resuspended in 0.2 M NaCl/2 mM Tris·HCl (pH 8.0)/0.2 mM EDTA. Phage suspensions were reclarified in tabletop microcentrifuges at maximum speed (≈16,100 × g) for 10 min. The purified supernatants were finally diluted in 0.2 M NaCl/2 mM Tris·HCl (pH 8.0)/0.2 mM EDTA for treatment. Appropriate amounts of phage were added to LB plus 30 μg/ml kanamycin for treatment, as described below. Phage purified by this protocol were no more effective at reducing bacterial biofilm levels compared with phage purified by centrifugation with CsCl step gradients (data not shown).
All phage PFU counts were determined by combining phage with 300 μl of overnight E. coli BL21 culture and 4–5 ml of 50°C LB top agar [0.7% (wt/vol) agar]. This solution was mixed thoroughly, poured onto LB agar plates, inverted after hardening, and incubated for 4–6 h at 37°C until plaques were clearly visible.
Biofilm Growth and Treatment.
All experiments were performed in LB media plus 30 μg/ml kanamycin. E. coli biofilms were grown with an MBEC Physiology and Genetics Assay (MBEC BioProducts, Edmonton, Canada), which consists of a 96-peg lid that fits into a standard 96-well microtiter plate. Each well was inoculated with 150 μl of media containing 1:200 dilutions of overnight cultures that had been grown at 37°C and 300 rpm (model G25 incubator shaker). Control wells with only media but no bacteria were included. MBEC lids were placed in the microtiter plates, inserted into plastic bags to prevent evaporation, and placed in a Minitron shaker (Infors HT, Bottmingen, Switzerland) for 24 h at 35°C and 150 rpm to form biofilm on the pegs.
For all treatments except for the dose–response experiment, 103 PFU of phage were combined with 200 μl of LB plus 30 μg/ml kanamycin in each well in new microtiter plates (Costar 3370; Fisher Scientific, Pittsburgh, PA). For the dose–response experiment, 101, 102, 103, 104, or 105 PFU were combined with 200 μl of LB plus 30 μg/ml kanamycin in each well. Wells with only media but no phage were included as untreated biofilm controls. MBEC lids with 24-h pregrown E. coli biofilm were removed from their old 96-well microtiter plates and placed into the new microtiter plates and back into plastic bags in a shaker at 35°C and 150 rpm for treatment. After specified amounts of time for the time-course experiment or 24 h for all other experiments, MBEC lids were removed and the amounts of biofilm remaining were assayed by CV staining or viable cell counting, as described below.
CV Staining Assay.
After rinsing the MBEC pegs three times with 1× PBS, CV staining was carried out according to a standard, previously reported protocol as described in SI Materials and Methods (50).
Viable Cell Count Assay.
We obtained viable cell counts by disrupting biofilms on the pegs in a sonicating water bath. MBEC pegs were first rinsed three times with 200 μl of 1× PBS and placed into fresh microtiter plates (catalog no. 262162; Nunc) containing 145 μl of 1× PBS in each well, which completely covered the biofilms growing on the pegs. To prevent further infection of bacteria by phage, 20 ng of T7 Tail Fiber Monoclonal Antibody (Novagen) was added to each well. MBEC lids and plates were placed in a Ultrasonics 5510 sonic water bath (Branson, Danbury, CT) and sonicated for 30 min at 40 kHz to dislodge bacteria in biofilms into the wells. Serial dilutions were performed and plated on plates with LB agar plus 30 μg/ml kanamycin. CFU were counted after overnight incubation at 37°C.
SEM.
SEM was performed according to MBEC recommendations as described in SI Materials and Methods (51).
Phage Counts.
At indicated time points (Fig. 4D) or after 24 h of treatment (Fig. 3C and Fig. 4F), media from n = 3 microtiter wells for each treatment type were serially diluted to obtain PFU counts for phage in the liquid phase. To obtain PFU counts for phage residing in biofilms at 24 h after infection (Fig. 3C), MBEC pegs were rinsed three times with 200 μl of 1× PBS and placed into fresh microtiter plates (catalog no. 262162; Nunc) containing 145 μl of 1× PBS in each well, which completely covered the biofilm on the pegs. No T7 tail fiber monoclonal antibody was added. The MBEC lids and plates were placed in a Ultrasonics 5510 sonic water bath (Branson, Danbury, CT) and sonicated for 30 min at 40 kHz to dislodge bacteria and phage residing in biofilms into wells. Serial dilutions were performed to obtain PFU counts for phage in biofilms.
Statistical Analysis.
Student's unpaired two-sided t test was used to test for statistical significance as described in SI Materials and Methods. For the CV staining assays, the data set size for each treatment type was n = 16; for the CFU assays, n = 12 pegs per treatment type were used.
Supplementary Material
Acknowledgments
We thank Nicholas Guido, Dan Dwyer, and Philina Lee for comments and Anlee Krupp for assistance with obtaining SEM images. This work was supported by the Department of Energy and the National Science Foundation. T.K.L. was supported by a Howard Hughes Medical Institute predoctoral fellowship and a Harvard–MIT Health Sciences and Technology Medical Engineering/Medical Physics fellowship.
Abbreviations
- EPS
extracellular polymeric substance
- DspB
dispersin B
- CV
crystal violet.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704624104/DC1.
References
- 1.Endy D. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Andrianantoandro E, Basu S, Karig DK, Weiss R. Mol Syst Biol. 2006;2:2006.0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasty J, McMillen D, Collins JJ. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 4.Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 5.Ro D-K, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 7.Loose C, Jensen K, Rigoutsos I, Stephanopoulos G. Nature. 2006;443:867–869. doi: 10.1038/nature05233. [DOI] [PubMed] [Google Scholar]
- 8.Xavier JB, Picioreanu C, Rani SA, van Loosdrecht MCM, Stewart PS. Microbiology. 2005;151:3817–3832. doi: 10.1099/mic.0.28165-0. [DOI] [PubMed] [Google Scholar]
- 9.Davey ME, O'Toole GA. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolter R, Greenberg EP. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 11.Parsek MR, Singh PK. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 12.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 13.Costerton JW, Stewart PS, Greenberg EP. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 14.Stewart PS, Costerton JW. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 16.Curtin JJ, Donlan RM. Antimicrob Agents Chemother. 2006;50:1268–1275. doi: 10.1128/AAC.50.4.1268-1275.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merril CR, Scholl D, Adhya SL. Nat Rev Drug Discovery. 2003;2:489–497. doi: 10.1038/nrd1111. [DOI] [PubMed] [Google Scholar]
- 18.Doolittle MM, Cooney JJ, Caldwell DE. Can J Microbiol. 1995;41:12–18. doi: 10.1139/m95-002. [DOI] [PubMed] [Google Scholar]
- 19.Doolittle MM, Cooney JJ, Caldwell DE. J Ind Microbiol. 1996;16:331–341. doi: 10.1007/BF01570111. [DOI] [PubMed] [Google Scholar]
- 20.Corbin BD, McLean RJ, Aron GM. Can J Microbiol. 2001;47:680–684. [PubMed] [Google Scholar]
- 21.Scholl D, Adhya S, Merril C. Appl Environ Microbiol. 2005;71:4872–4874. doi: 10.1128/AEM.71.8.4872-4874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh Y, Wang X, Hinnebusch BJ, Preston JF, Romeo T. J Bacteriol. 2005;187:382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 24.Hughes KA, Sutherland IW, Jones MV. Microbiology. 1998;144(Pt 11):3039–3047. doi: 10.1099/00221287-144-11-3039. [DOI] [PubMed] [Google Scholar]
- 25.Hughes KA, Sutherland IW, Clark J, Jones MV. J Appl Microbiol. 1998;85:583–590. doi: 10.1046/j.1365-2672.1998.853541.x. [DOI] [PubMed] [Google Scholar]
- 26.Projan S. Nat Biotechnol. 2004;22:167–168. doi: 10.1038/nbt0204-167. [DOI] [PubMed] [Google Scholar]
- 27.Chan LY, Kosuri S, Endy D. Mol Syst Biol. 2005;1:2005.0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itaya M, Tsuge K, Koizumi M, Fujita K. Proc Natl Acad Sci USA. 2005;102:15971–15976. doi: 10.1073/pnas.0503868102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn JJ, Studier FW. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 30.Studier FW, Dunn JJ. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):999–1007. doi: 10.1101/sqb.1983.047.01.114. [DOI] [PubMed] [Google Scholar]
- 31.Ghigo JM. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 32.Re SD, Quéré BL, Ghigo J-M, Beloin C. Appl Environ Microbiol. 2007;73:3391–3403. doi: 10.1128/AEM.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia LR, Molineux IJ. J Bacteriol. 1995;177:4077–4083. doi: 10.1128/jb.177.14.4077-4083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Studier FW. Science. 1972;176:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- 35.Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S. Proc Natl Acad Sci USA. 1996;93:3188–3192. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summers WC. Annu Rev Microbiol. 2001;55:437–451. doi: 10.1146/annurev.micro.55.1.437. [DOI] [PubMed] [Google Scholar]
- 37.Shuren J US Food and Drug Administration, editors. Federal Register. Vol 71. Washington, DC: Natl Arch and Records Admin; 2006. pp. 47729–47732. [Google Scholar]
- 38.Schoolnik GK, Summers WC, Watson JD. Nat Biotechnol. 2004;22:505–506. doi: 10.1038/nbt0504-505. author reply 506–507. [DOI] [PubMed] [Google Scholar]
- 39.Hagens S, Blasi U. Lett Appl Microbiol. 2003;37:318–323. doi: 10.1046/j.1472-765x.2003.01400.x. [DOI] [PubMed] [Google Scholar]
- 40.Hagens S, Habel, AvAU, von Gabain A, Blasi U. Antimicrob Agents Chemother. 2004;48:3817–3822. doi: 10.1128/AAC.48.10.3817-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boratynski J, Syper D, Weber-Dabrowska B, Lusiak-Szelachowska M, Pozniak G, Gorski A. Cell Mol Biol Lett. 2004;9:253–259. [PubMed] [Google Scholar]
- 42.Bartlett JG. Ann Intern Med. 2006;145:758–764. doi: 10.7326/0003-4819-145-10-200611210-00008. [DOI] [PubMed] [Google Scholar]
- 43.Aslam S, Hamill RJ, Musher DM. Lancet Infect Dis. 2005;5:549–557. doi: 10.1016/S1473-3099(05)70215-2. [DOI] [PubMed] [Google Scholar]
- 44.Baker D, Church G, Collins J, Endy D, Jacobson J, Keasling J, Modrich P, Smolke C, Weiss R. Sci Am. 2006;294:44–51. doi: 10.1038/scientificamerican0606-44. [DOI] [PubMed] [Google Scholar]
- 45.Hickman-Brenner FW, Stubbs AD, Farmer JJ. J Clin Microbiol. 1991;29:2817–2823. doi: 10.1128/jcm.29.12.2817-2823.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wentworth BB. Bacteriol Rev. 1963;27:253–272. doi: 10.1128/br.27.3.253-272.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 48.Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, Miller JF. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 49.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. J Clin Microbiol. 1999;37:1771–1776. doi: 10.1128/jcm.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.