Abstract
Objective:
To examine the association of single nucleotide polymorphisms (SNPs) in inflammation-related genes in the development of infections following esophagectomy.
Summary Background Data:
Genetic polymorphisms for immunoregulatory cytokines may explain individual variation in response to trauma. Esophagectomy is associated with a high risk of postoperative infection and sepsis, and this study explored a number of SNPs in cytokine genes and their relationship to postoperative infection.
Methods:
In a prospective analysis of 197 patients with esophageal cancer undergoing resection, 55 developed postoperative infections. DNA was extracted and genotyping was performed for polymorphisms in genes encoding TNF-α, IL-1β, IL-1 receptor antagonist, IL-10, and Toll-like receptor 4 (TLR-4) using Taqman chemistry and PCR/RFLP. In a blinded analysis, the cohort with infections was compared with the no complication cohort (n = 114) and a cohort that had noninfective complications (n = 28).
Results:
No differences in polymorphisms for IL-1β, IL-1 RN, IL-10, and TLR-4 genes were observed across groups. The frequency of TNF-α −308 GG homozygotes was significantly (P = 0.021) higher in the postoperative infection group. The G allele was significantly higher in the postoperative infection group compared with the no complication group (P = 0.017) and other complication group (P = 0.013). By multivariate analysis, this polymorphism as well as age and body mass index were predictors of infection.
Conclusion:
The TNF-α −308A allele has been shown to be associated with higher circulating levels of TNF-α and the −308 G allele is a comparative low secretor allele. We propose that the polymorphism in the promotor region of TNF-α gene may lead to altered expression and a possible suboptimal activity of TNF-α in persons with GG genotypes, and these data suggest a link with infection following major surgery.
Patients undergoing esophagectomy have a substantial risk of developing postoperative infections. DNA sequence variations may account for differences in individual immune responses to infectious insults. This study examines gene polymorphisms in 197 postesophagectomy cases and highlights a significantly higher prevalence of TNF-α −308 GG genotype in patients developing postoperative infections.
There has been a marked increase in the incidence of esophageal adenocarcinoma in the last 3 decades.1 Surgery is the mainstay of treatment of resectable esophageal tumors, and in recent years neoadjuvant therapy with chemotherapy alone or combined with radiation therapy is increasingly utilized. The outcomes of surgery have improved through increasing subspecialization, improved perioperative care, and the trend toward greater centralization of complex cancer surgery in centers achieving the optimum outcomes. Notwithstanding these advances, there remains no other elective cancer operation that carries the same risk of morbidity and mortality, particularly relating to infection and sepsis.1
The development of infection postoperatively is influenced by many factors, including the size of the bacterial or fungal challenge, specific bacterial virulence factors, and patient phenotypic characteristics, such as age and nutritional status.2 A key host determinant is the response of an individual's immune system to invasion by pathogens. The activation and clinical effect of an immune response to invading organisms are orchestrated by a number of pro-inflammatory and anti-inflammatory cytokines and receptors. Following severe trauma, including major surgery, the immune cell and cytokine balance is predominantly pro-inflammatory in the early stage, often clinically manifest as a systemic inflammatory response syndrome, and later by an anti-inflammatory response, the so-called compensatory anti-inflammatory response syndrome. Many factors impact on the magnitude and balance of cytokine responses in these clinical scenarios, but there is emerging consensus that gene polymorphisms mediate interindividual variation that may be clinically relevant.3–5
Tumor necrosis factor-alpha (TNF-α) and IL-1β are major mediators and amplifiers of the immune response to infectious challenge. The gene encoding TNF-α, TNFA, is located on the short arm of chromosome 6 in the HLA class III locus of the major histocompatibility complex. Raised levels of TNF-α have been found in patients with sepsis, but TNF-α inhibition has proved unsuccessful in the treatment of sepsis.6 TNFA gene polymorphisms at −308 locus have been associated with increased mortality in sepsis and poor outcome in some surgical patients, suggesting that this may be useful in screening for high-risk patients.7 IL-1β acts synergistically with TNF-α and channels a variety of cellular activities toward initiating and sustaining the immune response, including cell proliferation, differentiation, and apoptosis. The IL-1 gene cluster, including the IL-1 RN gene, has been associated with a number of medical disorders, and allele A2 (consisting of 2 copies of 86 bp tandem repeat in the polymorphic intron 2) of IL-1RN gene has been associated with sepsis.8 IL-10 is a pleiotropic cytokine that inhibits the production of IL-1β and TNF-α, it is centrally involved in the regulation of inflammation and influences the infectious process by its overall anti-inflammatory effect.9–12 Toll-like receptor (TLR) 4 is a pattern recognition receptor that has been evolutionarily conserved; it initiates an intracellular signaling cascade following the binding of gram-negative lipopolysaccharide,13 resulting in the activation of NFkB and the production of a spectrum of cytokines including TNF-α, IL-1 and IL-6.14 A mutated variant of the TLR 4 gene with a G nucleotide at position 896 (rs4986790) has been associated with a higher risk of gram-negative septic shock15 and is hyporesponsive to inhaled LPS.16
The purpose of this study was to assess whether polymorphisms in genes for the key regulatory cytokines were associated with infection following esophagectomy, and we highlight herein the significant impact of polymorphisms in the TNFA gene on these outcomes.
MATERIALS AND METHODS
The study was approved by St. James's Hospital, Federated Dublin Voluntary Hospitals and Trinity College Joint Research Ethics Committee. All study subjects were white of Irish ancestry.
In a population with high-risk allele frequency between 50% and 70%, a sample size of approximately 200 is adequate to detect an effect size of odds ratio 3+ with a power of 80% and a 2-sided significance level of 5%.17 Patients (n = 197) who underwent esophagectomy for localized cancer were studied. A total of 105 who underwent esophagectomy were recruited prospectively between 2002 and 2004, and 92 cases were selected randomly from the pathology department archives of 270 paraffin-embedded tissue blocks dating between 1990 and 2001. In all cases, the diagnoses of esophageal cancer were confirmed preoperatively on the basis of histopathologic reports, and all patients had localized disease treated with curative intent.
A prospective esophageal cancer database (Dendrite, UK) records all demographic, staging, and clinical parameters, including all complications from surgery to discharge. The records of archived paraffin-embedded tissues were a part of this database. Respiratory failure was defined as the requirement for mechanical ventilation beyond 24 hours after surgery. Adult respiratory distress syndrome and multiple organ failure were defined as per Bone et al,18 sepsis required evidence of systemic inflammatory response syndrome with microbiologic evidence of infection in blood cultures, and the diagnosis of pneumonia required either positive culture or clear clinical or radiologic evidence of consolidation.
Patients were divided into 3 groups: no postoperative complications group (also used as control), noninfectious postoperative complications group, and infectious postoperative complications group. A postoperative infection was defined as a systemic response to the presence of an infectious agent or toxin that was not present preoperatively and supported by clinical and laboratory evidence of the same. Laboratory evidence means positive culture results.19,20 Pathologic and clinical data were obtained from the pathology department database and patients’ documents and electronic database maintained in the unit by a full-time data manager. To ensure accuracy of the electronic database, 20% of the records were cross-checked with the original patients’ records.
A control cohort of 226 individuals was also studied: 140 healthy individuals and 86 hospital controls undergoing endoscopy for dyspepsia where no pathology was evident.
DNA Preparation and Analysis
DNA was extracted either from fresh blood or paraffin-embedded tissue using Qiagen (Qiagen Ltd., UK)21 and Puregene (Gentra systems, Minneapolis, MN)22 kits, respectively, according to the manufacturer's protocols. Genotyping was done using PCR RFLP,23–26 Taqman allelic discrimination assays,27,28 and amplifluor SNP genotyping technology. Details of primers and PCR conditions have been described elsewhere in the literature and are also available with the authors.
Statistical Analysis
The distribution of alleles/genotypes in the 3 postoperative outcome groups and the healthy control group was analyzed using Pearson χ2 and likelihood-ratio χ2 tests of independence; 2 × 2 tables were used to compare allele/haplotype distribution between any 2 groups and Fisher exact test was used where expected frequency was less than 5 in any cell. Epi info, version 3.3.2 (http://www.cdc.gov/epiinfo/), was used to calculate odds ratios at successive exposure levels using Extended Mantel-Haenszel χ2 and P value. Hardy-Weinberg equilibrium was determined using Genepop software (http://wbiomed.curtin.edu.au/genepop/). Hitagene (www.hitagene.com) and Phase software (version 2.1.1),29 which allows for missing genotype data, was used for haplotype inference. Best reconstructions were assumed as correct and used for further analysis. SPSS, version 12.0 (SPSS Inc., Apache Software Foundation), was used to develop a binary logistic regression model. Forward likelihood ratio and hierarchical blockwise entry methods were used to identify the predictive factors for postoperative outcome. Postoperative outcome was dichotomized using postoperative infections as one group and merging the other 2 groups. Since there was no TNF-α −308 AA genotype in the postoperative infection group, a combined AA and GA genotype was used as a reference category against GG genotype. Hosmer and Lemeshow's test was done to check the goodness of fit of the model.
RESULTS
All SNP loci were in Hardy-Weinberg equilibrium (P > 0.3 in all cases). There was near-complete linkage disequilibrium between IL-1B −511T (rs3087258) and −31C (rs1143627) alleles and vice versa, and IL-10 −819 (rs1800871) and −592 alleles (rs1800872) (−819C with −592C and −819T with −592 A). Along with the third SNP at position −1082 (rs1800896), 3 haplotypes (GCC, ACC, and ATA) in IL-10 gene represented 100% of the chromosomes.
In all 197 patients were studied, of whom 114 (58%) did not develop any postoperative complication, 55 (28%) patients developed infections (pneumonia 33, sepsis 17, and wound infection 5) and 28 (14%) had other complications [cardiac complications (12; arrhythmias, cardiac failures), anastomotic leaks (4), atelectasis (6), organ failure (3), others (3; seizures, bowel obstruction)]. The 3 groups under study did not differ from the healthy population of 226 study subjects in the distribution of genotypes and haplotypes.
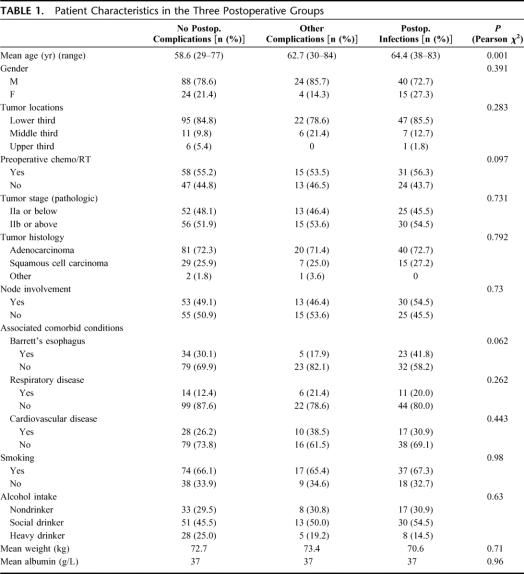
The patient characteristics in the 3 groups are shown in Table 1. Patients in the group with postoperative infective complications were significantly (P < 0.05) older than the other groups. Tumor site, stage, neoadjuvant therapy, smoking or alcohol intake history, or existing respiratory or cardiac function did not impact on the risk of infection; 96% of patients had a preoperative serum albumin in the normal (35–40 g/L) range.
TABLE 1. Patient Characteristics in the Three Postoperative Groups
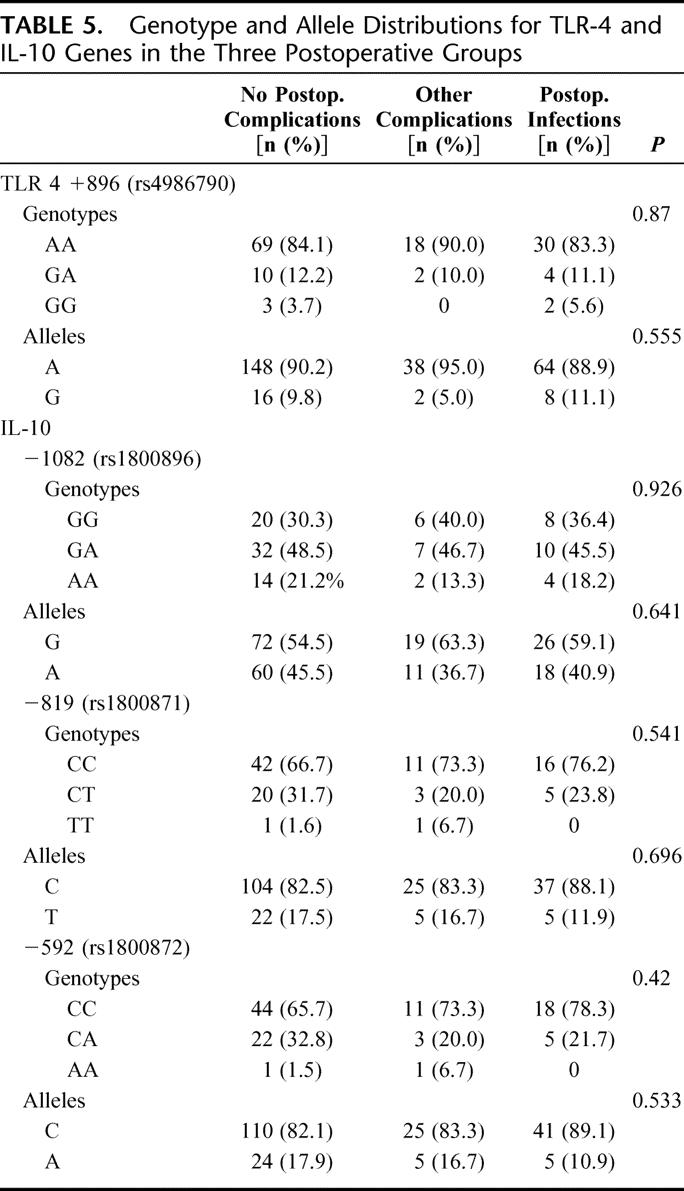
The distribution of alleles and genotypes for TNFA is shown in Table 2. Pearson χ2 analysis of the genotypes revealed a significantly (P = 0.021) higher distribution of GG genotypes in the group developing postoperative infections. Analysis of the allele distribution in the no complication group versus the group developing infections showed a significantly higher frequency of the G allele among those developing infections (P = 0.017), and this was also significantly (P = 0.013) more common than in the group developing other complications. Table 3 shows the increasing linear trend of developing postoperative infections in GG genotypes as compared with AA and AG genotypes. The genotypic pattern and allele distribution for IL-1β, IL-10, and TLR4 is shown in Tables 4 and 5, and no significant patterns of difference emerged.
TABLE 2. TNF-α Genotype and Allele Distributions in the Three Postoperative Groups
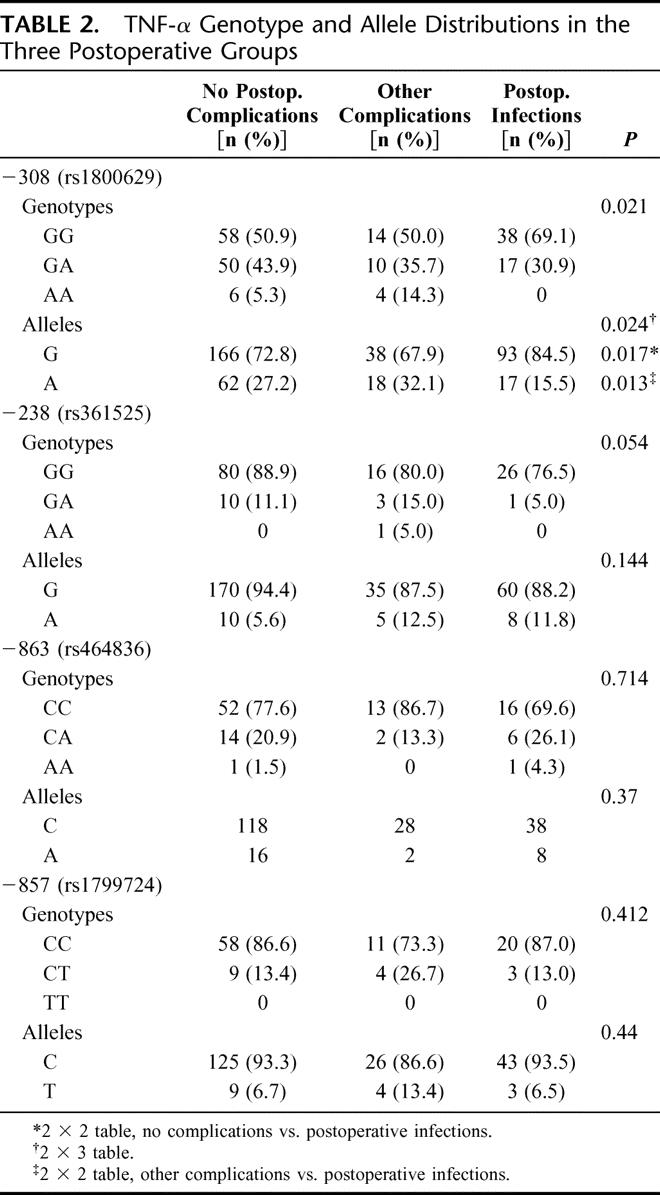
TABLE 3. Linear Trend Test for TNF-α −308 Genotypes Comparing Postoperative Infections With Other Groups
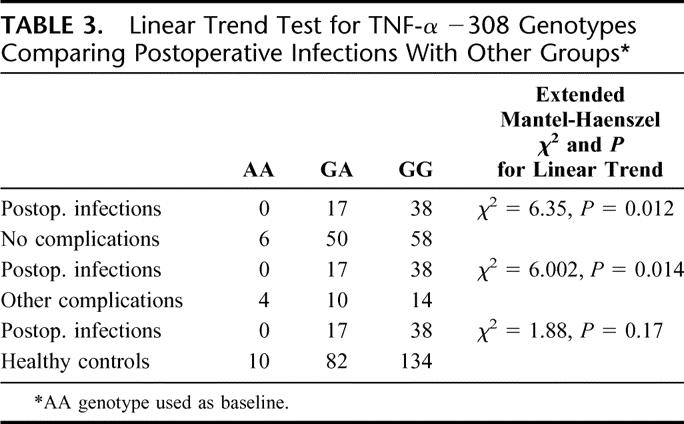
TABLE 4. Distribution of IL-1B Genotypes/Alleles and IL1RN Alleles in Postoperative Groups
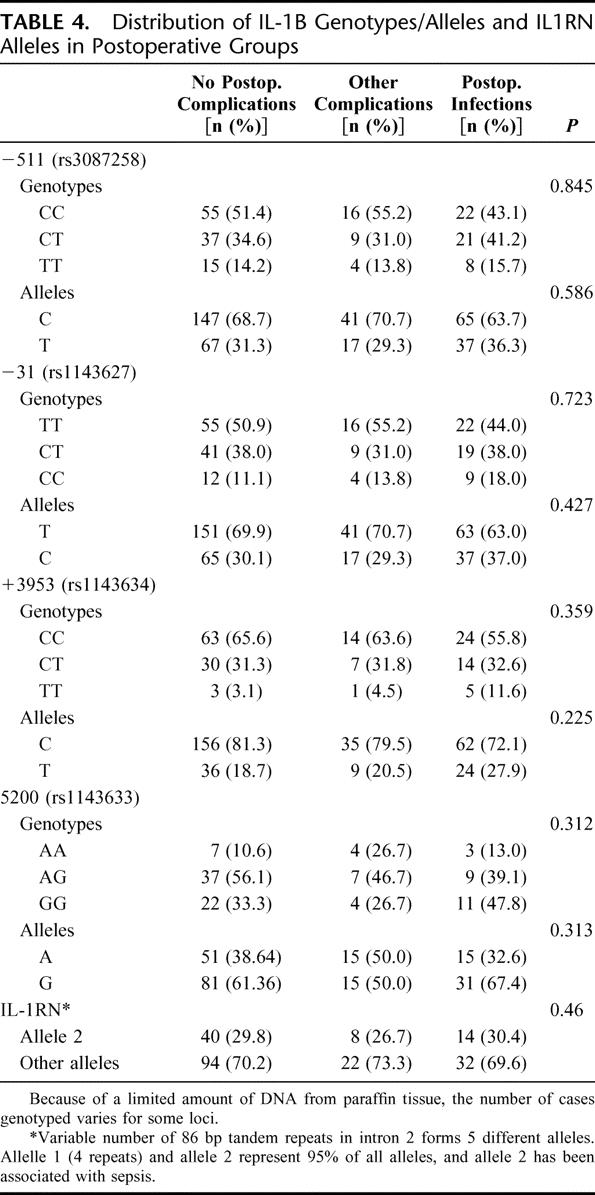
TABLE 5. Genotype and Allele Distributions for TLR-4 and IL-10 Genes in the Three Postoperative Groups
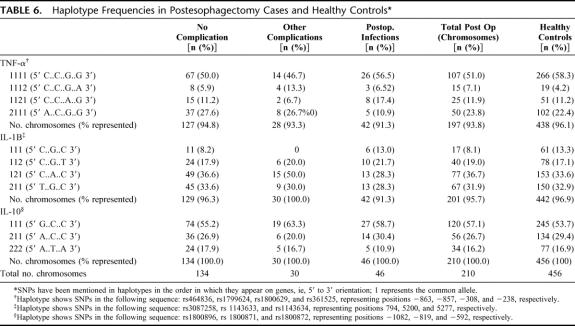
The haplotype frequencies for TNFα, IL-1β, and IL-10 are shown in Table 6. The permutation based haplotype association test implemented in Hitagene did not show any significant difference in TNFA haplotypes (P = 0.103) in the groups under study. No patterns emerged for IL-1B or IL-10 genes.
TABLE 6. Haplotype Frequencies in Postesophagectomy Cases and Healthy Controls
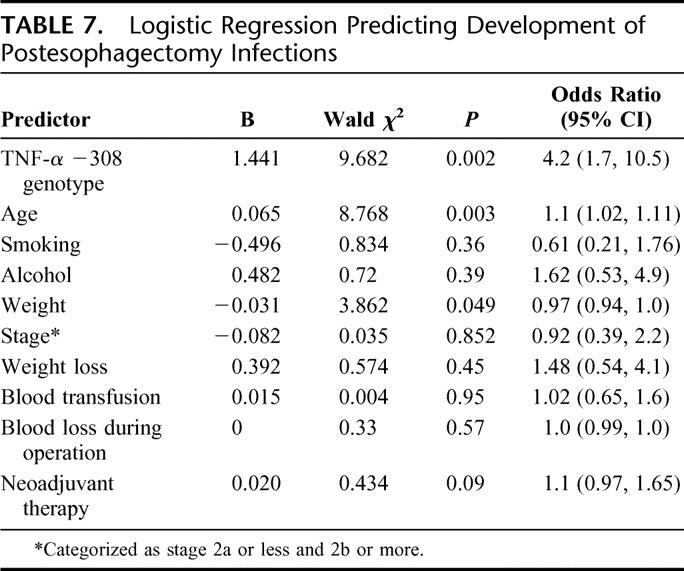
Multivariate logistic regression coefficients, Wald test, and the odds ratio for a number of factors are shown in Table 7. Only age, body mass index, and TNF-α −308 genotype were significant. The Hosmer and Lemeshow's test statistic was 0.367, implying that the model's estimates fitted the data at an acceptable level. TNF-α GG genotype significantly increased the odds of developing postoperative infections with an odds ratio (confidence interval) of 4.2 (1.7, 10.8).
TABLE 7. Logistic Regression Predicting Development of Postesophagectomy Infections
DISCUSSION
This study revealed that there is a significantly higher distribution of TNF-α −308 GG genotype in a cohort of patients that developed infections following esophagectomy compared with other groups. No association with any other single nucleotide polymorphism for IL-1β, IL-1RN, IL-10, or TLR-4 was evident. Adjusting for age, TNF-α −308 GG genotype individuals are 4.2 times more likely to develop infections following esophagectomy compared with other genotypes. The study thus supports the hypothesis that interindividual variation in the TNF-α gene may impact on the risk of infection following complex major surgery. With respect to other factors, low body weight was marginally significant as a predictor of postoperative infection, but recent weight loss was not associated with postoperative infections. Preoperative serum albumin and postoperative nutrition support have been shown to impact on infection risk postesophagectomy in other studies.2,30,31 In this study, however, the majority of patients had normal serum albumin levels preoperatively, and over 95% of patients tolerated full enteral nutritional support postoperatively; therefore, these did not represent true variables in this analysis. Other studies have linked intraoperative blood transfusions and nosocomial infections,32 but this was not significant in this study, where the median (range) units of blood administered was 0 (0–4).
TNF-α has been highly conserved across mammalian species during evolution, underscoring its critical role in innate immunity, with approximately 80% amino acid identity between human, Mus musculus, Bos taurus, and Canis familiaris TNF-α proteins. TNF-α is a pro-inflammatory cytokine that is important in the defense against infection and is produced in large amounts in response to an infectious insult. Gram-negative bacterial lipopolysaccharide is a major inducer of TNF-α33 and TNF-α knockout mice show an increased susceptibility to Candida albicans challenge.34 An increasing clinical experience with antibodies to TNF-α in conditions, including Crohn's disease, rheumatoid arthritis, and sepsis, underscores the risks associated with lower bioavailability of TNF-α. An increased risk, for instance, of pulmonary tuberculosis has been observed in patients receiving infliximab (Remicade).35 Many studies have suggested a link between TNF-α promoter polymorphism and infections.36 Moreover, the functional significance of TNF-α −308 SNP in transcriptional regulation is recognized, and the −308A allele in the TNF-α gene has been shown to be associated with higher levels of TNF-α protein by a number of studies.4,37–39
In healthy individuals, the difference in levels of expression of TNF-α due to genotypes in the promoter region would be expected to be within physiologic limits in response to stress situations. This, however, may not pertain to conditions of significant immune cell perturbations that may follow major injury, such as esophageal cancer surgery, which is often undertaken in patients already compromised immunologically by advanced age and comorbidities, cancer itself, nutritional factors, and weakened physiologic responses. In this scenario, an impaired TNF-α response to an infectious challenge might differentiate an adequate as opposed to a clinically relevant suboptimal response to infection. This has not been addressed in this study, as TNF-α production was not measured, nor was T cell or phagocyte function, and this is the subject of current research.
It is also acknowledged that the presence of a single SNP might itself be unrelated to the specific endpoint and represent instead linkage with other genes and gene variants in close physical proximity within the MHC. The TNF-α gene shows strong linkage disequilibrium with HLA class I and class II genes and other immunoregulatory genes in the MHC region. The A allele for instance is strongly linked to A1-B8-DR3 HLA haplotype, which is associated with increased TNF production.40,41 What can be stated from this study is that a pattern emerged specifically in polymorphisms of the TNF-α gene that supports the paradigm that interindividual variation of response may determine important clinical outcomes, specifically in this case, the risk of infection. It is not possible to identify all the genetic confounding variables that might predispose to infections; nevertheless, in light of studies showing association of G allele with a lower production of TNF-α and TNF blocking studies showing higher rate of infections associated with decreased bioavailability of TNF-α, we believe these data support the hypothesis that the GG genotype could be a predictive factor for postoperative infections after esophageal resection or equivalent complex major surgery. Moreover, the study, with no pattern emerging for other cytokines, suggests that efforts to understand and apply polymorphisms in TNF-α within clinical trials and novel treatment approaches may have the greatest rationale in infection prophylaxis in the high-risk surgical patient.
Footnotes
Reprints: John V. Reynolds, MD, Department of Clinical Surgery and Dublin Molecular Medicine Centre, Trinity Centre for Health Sciences, St. James's Hospital, Dublin, Ireland. E-mail: reynoljv@tcd.ie.
REFERENCES
- 1.McCulloch P, Ward J, Tekkis PP. Mortality and morbidity in gastro-oesophageal cancer surgery: initial results of ASCOT multicentre prospective cohort study. BMJ. 2003;327:1192–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daly JM, Weintraub FN, Shou J, et al. Enteral nutrition during multimodality therapy in upper gastrointestinal cancer patients. Ann Surg. 1995;221:327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall SK, Perregaux DG, Gabel CA, et al. Correlation of polymorphic variation in the promoter region of the interleukin-1 beta gene with secretion of interleukin-1 beta protein. Arthritis Rheum. 2004;50:1976–1983. [DOI] [PubMed] [Google Scholar]
- 4.Abraham LJ, Kroeger KM. Impact of the −308 TNF promoter polymorphism on the transcriptional regulation of the TNF gene: relevance to disease. J Leukoc Biol. 1999;66:562–566. [DOI] [PubMed] [Google Scholar]
- 5.Gibson AW, Edberg JC, Wu J, et al. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915–3922. [DOI] [PubMed] [Google Scholar]
- 6.Reinhart K, Karzai W. Anti-tumor necrosis factor therapy in sepsis: update on clinical trials and lessons learned. Crit Care Med. 2001;29(suppl 7):121–125. [DOI] [PubMed] [Google Scholar]
- 7.Khalil A, Hall J, Aziz F, et al. Tumour necrosis factor: implications for surgical patients. Aust NZ J Surg. 2006;76:1010–1016. [DOI] [PubMed] [Google Scholar]
- 8.Fang XM, Schroder S, Hoeft A, et al. Comparison of two polymorphisms of the interleukin-1 gene family: interleukin-1 receptor antagonist polymorphism contributes to susceptibility to severe sepsis. Crit Care Med. 1999;27:1330–1334. [DOI] [PubMed] [Google Scholar]
- 9.Fiorentino DF, Zlotnik A, Mosmann TR, et al. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 10.Cassatella MA, Meda L, Gasperini S, et al. Interleukin 10 (IL-10) upregulates IL-1 receptor antagonist production from lipopolysaccharide-stimulated human polymorphonuclear leukocytes by delaying mRNA degradation. J Exp Med. 1994;179:1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickensheets HL, Donnelly RP. IFN-gamma and IL-10 inhibit induction of IL-1 receptor type I and type II gene expression by IL-4 and IL-13 in human monocytes. J Immunol. 1997;159:6226–6233. [PubMed] [Google Scholar]
- 12.Dickensheets HL, Freeman SL, Smith MF, et al. Interleukin-10 upregulates tumor necrosis factor receptor type-II (p75) gene expression in endotoxin-stimulated human monocytes. Blood. 1997;90:4162–4171. [PubMed] [Google Scholar]
- 13.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. [DOI] [PubMed] [Google Scholar]
- 14.Kopp E, Medzhitov R. Recognition of microbial infection by Toll-like receptors. Curr Opin Immunol. 2003;15:396–401. [DOI] [PubMed] [Google Scholar]
- 15.Lorenz E, Mira JP, Frees KL, et al. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz DA. The genetics of innate immunity. Chest. 2002;121(suppl 3):62–68. [DOI] [PubMed] [Google Scholar]
- 17.Bishop D. Inherited susceptibility to cancer. In: Knowles M, Selby P, eds. Introduction to the Cellular and Molecular Biology of Cancer. New York: Oxford University Press, 2005:45–60. [Google Scholar]
- 18.Bone RC, Sprung CL, Sibbald WJ. Definitions for sepsis and organ failure. Crit Care Med. 1992;20:724–726. [DOI] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. CDC Definitions of Nosocomial Infections. Atlanta, GA: Centers for Disease Control and Prevention. Department of Health and Human Services. 1988.
- 20.Garner JS, Jarvis WR, Emori TG, et al. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16:128–140. [DOI] [PubMed] [Google Scholar]
- 21.Blood and Body Fluid Spin Protocol. Qiagen, West Sussex, United Kingdom, 2003. [Google Scholar]
- 22.Gentra Systems. DNA purification from 5–10 mg paraffin-embedded tissue. In: PUREGENE Protocol. Gentra Systems, Oslo, Norway, 2005. [Google Scholar]
- 23.Nishimura M, Mizuta I, Mizuta E, et al. Influence of interleukin-1beta gene polymorphisms on age-at-onset of sporadic Parkinson's disease. Neurosci Lett. 2000;284:73–76. [DOI] [PubMed] [Google Scholar]
- 24.Ozen S, Alikasifoglu M, Bakkaloglu A, et al. Tumour necrosis factor alpha G–>A −238 and G–>A −308 polymorphisms in juvenile idiopathic arthritis. Rheumatology (Oxf). 2002;41:223–227. [DOI] [PubMed] [Google Scholar]
- 25.Jang WH, Yang YI, Yea SS, et al. The −238 tumor necrosis factor-alpha promoter polymorphism is associated with decreased susceptibility to cancers. Cancer Lett. 2001;166:41–46. [DOI] [PubMed] [Google Scholar]
- 26.Grimm C, Berger I, Tomovski C, et al. A polymorphism of the interleukin-1 receptor antagonist plays a prominent role within the interleukin-1 gene cluster in vulvar carcinogenesis. Gynecol Oncol. 2004;92:936–940. [DOI] [PubMed] [Google Scholar]
- 27.Read RC, Pullin J, Gregory S, et al. A functional polymorphism of toll-like receptor 4 is not associated with likelihood or severity of meningococcal disease. J Infect Dis. 2001;184:640–642. [DOI] [PubMed] [Google Scholar]
- 28.El-Omar EM, Carrington M, Chow WH, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. [DOI] [PubMed] [Google Scholar]
- 29.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daly JM, Lieberman MD, Goldfine J, et al. Enteral nutrition with supplemental arginine, RNA, and omega-3 fatty acids in patients after operation: immunologic, metabolic, and clinical outcome. Surgery. 1992;112:56–67. [PubMed] [Google Scholar]
- 31.Kudsk KA, Tolley EA, DeWitt RC, et al. Preoperative albumin and surgical site identify surgical risk for major postoperative complications. JPEN J Parenter Enteral Nutr. 2003;27:1–9. [DOI] [PubMed] [Google Scholar]
- 32.Taylor RW, O'Brien J, Trottier SJ, et al. Red blood cell transfusions and nosocomial infections in critically ill patients. Crit Care Med. 2006;34:2302–2308; quiz 2309. [DOI] [PubMed]
- 33.Westendorp RG, Langermans JA, Huizinga TW, et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet. 1997;349:170–173. [DOI] [PubMed] [Google Scholar]
- 34.Marino MW, Dunn A, Grail D, et al. Characterization of tumor necrosis factor-deficient mice. Proc Natl Acad Sci USA. 1997;94:8093–8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keane J, Gershon S, Wise RP, et al. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med. 2001;345:1098–1104. [DOI] [PubMed] [Google Scholar]
- 36.Dahmer MK, Randolph A, Vitali S, et al. Genetic polymorphisms in sepsis. Pediatr Crit Care Med. 2005;6(suppl 3):61–73. [DOI] [PubMed] [Google Scholar]
- 37.Louis E, Franchimont D, Piron A, et al. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol. 1998;113:401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkpatrick A, Bidwell J, van den Brule AJ, et al. TNFalpha polymorphism frequencies in HPV-associated cervical dysplasia. Gynecol Oncol. 2004;92:675–679. [DOI] [PubMed] [Google Scholar]
- 39.Wilson AG, Symons JA, McDowell TL, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson AG, de Vries N, Pociot F, et al. An allelic polymorphism within the human tumor necrosis factor alpha promoter region is strongly associated with HLA A1, B8, and DR3 alleles. J Exp Med. 1993;177:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knight JC, Kwiatkowski D. Inherited variability of tumor necrosis factor production and susceptibility to infectious disease. Proc Assoc Am Physicians. 1999;111:290–298. [DOI] [PubMed] [Google Scholar]