Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that causes cryptococcal meningoencephalitis, particularly in immunocompromised patients. The fungal cell wall is an excellent target for antifungal therapies as it is an essential organelle that provides cell structure and integrity, it is needed for the localization or attachment of known virulence factors, including the polysaccharide capsule, melanin, and phospholipase, and it is critical for host-pathogen interactions. In C. neoformans, chitosan produced by the enzymatic removal of acetyl groups from nascent chitin polymers has been implicated as an important component of the vegetative cell wall. In this study, we identify four putative chitin/polysaccharide deacetylases in C. neoformans. We have demonstrated that three of these deacetylases, Cda1, Cda2, and Cda3, can account for all of the chitosan produced during vegetative growth in culture, but the function for one, Fpd1, remains undetermined. The data suggest a model for chitosan production in vegetatively growing C. neoformans where the three chitin deacetylases convert chitin generated by the chitin synthase Chs3 into chitosan. Utilizing a collection of chitin/polysaccharide deacetylase deletion strains, we determined that during vegetative growth, chitosan helps to maintain cell integrity and aids in bud separation. Additionally, chitosan is necessary for maintaining normal capsule width and the lack of chitosan results in a “leaky melanin” phenotype. Our analysis indicates that chitin deacetylases and the chitosan made by them may prove to be excellent antifungal targets.
Cryptococcus neoformans is an opportunistic fungal pathogen that causes cryptococcosis in immunocompromised individuals. The incidence of cryptococcosis is rising in direct proportion to the spread of human immunodeficiency virus (for a review, see the work of Casadevall and Perfect [7]). In the United States, it is estimated that up to 13% of AIDS patients will develop life-threatening cryptococcal meningitis. Additionally, in some parts of Africa, this estimation increases to 40% (7). Current antifungal therapies for the treatment of cryptococcosis are inadequate: amphotericin B is effective, but toxic, fluconazole is fungistatic, having high relapse rates; and resistance to flucytosine occurs frequently (37, 39, 45, 58).
Antifungal drug design has been problematic because fungi are eukaryotes and share many biochemical processes with animals. The only new members of the class of antifungals to emerge in recent years are the echinocandins. This class of drug, which targets β-(1,3)-glucan synthase, an enzyme essential for the synthesis of a major component in the fungal cell wall, has been shown to be safe and effective for specific fungal infections, including candidiasis and aspergillosis caused by Candida albicans and Aspergillus fumigatus, respectively (21, 55). C. neoformans possesses the target enzyme β-(1,3)-glucan synthase, and in vitro assays have shown this enzyme's activity to be inhibited by the echinocandin caspofungin (30). However, C. neoformans still exhibits resistance to this class of drugs (23).
The fungal cell wall contains components that differentiate it from that of other eukaryotic cells. The cell wall is an essential organelle that provides cellular structure and integrity. Significantly, the wall and its biosynthetic components are not present in the host; therefore, drugs designed to target cell wall biosynthesis should pose fewer toxicity issues.
The C. neoformans cell wall is important for viability; it is also associated with a variety of known virulence factors and is important for host-pathogen interactions. The major virulence factor is the polysaccharide capsule, whose attachment to the outer portion of the cell wall requires α-(1,3)-glucan (16, 42). Another cell wall-associated virulence factor in C. neoformans is melanin pigment. Melanin provides the pathogen with protection against host defenses (57) and is produced by two laccase proteins, Lac1 and Lac2 (33, 41). Lac1 is responsible for producing the majority of melanin and has been shown to be localized to the cell wall (33, 59, 60).
An essential component of the fungal cell wall that contributes to its strength and integrity is chitin (1). Chitin is a linear polymer of β-(1,4)-linked N-acetylglucosamine (GlcNAc) and is formed from cytoplasmic pools of UDP-GlcNAc. Chitosan, the deacetylated version of chitin, is produced enzymatically by chitin deacetylases (EC 3.5.1.41). Chitosan polymers can possess differing degrees of deacetylation (53). Information about the biological role of chitosan comes from research using the model yeast Saccharomyces cerevisiae. In this yeast, chitosan forms a layer of the ascospore cell wall and is suggested to be in the bridges between individual spores (11). S. cerevisiae has two chitin deacetylase genes, CDA1 and CDA2, that are transcribed only during sporulation (9) and are not necessary for viability. The deletion of both genes results in spores that are more sensitive to cell wall damage (10, 32). Although both Cda1p and Cda2p deacetylate the chitin produced by the chitin synthase Chs3p and its chitin synthase regulator, Sch1p, Cda2p is the predominant deacetylase. In contrast, C. neoformans has substantial chitosan in its cell wall during vegetative growth (2).
In C. neoformans, the chitin synthase Chs3 and chitin synthase regulator Csr2 synthesize the vegetative cell wall chitin that is deacetylated to chitosan (2). Strains of C. neoformans lacking either CHS3 or CSR2 have significantly reduced chitosan and are sensitive to cell wall inhibitors and elevated temperatures. This sensitivity suggests that chitosan may be an essential factor for the proper maintenance of cell wall integrity in C. neoformans (2). We hypothesized that the deletion of the chitin/polysaccharide deacetylases would lead to the same phenotypic abnormalities as those observed in the chs3Δ and csr2Δ strains.
Interestingly, one of the C. neoformans chitin/polysaccharide deacetylases, MP98 (encoded by CDA2), was isolated in a screen for cryptococcal antigens that stimulated an immune response in C. neoformans-reactive CD4+ mouse T-cell hybridomas (27) and a second polysaccharide deacetylase, D25 (encoded by FPD1), that induces a protective immune response in mice has also been reported (3). This indicates that at least two of the potential chitin/polysaccharide deacetylases are expressed in vivo and are immunogenic.
Utilizing a collection of putative chitin/polysaccharide deacetylase deletion strains, we have begun to characterize the four deacetylases and the biological role of chitosan during vegetative growth in C. neoformans. An analysis of these deletion strains indicates that three deacetylate chitin during vegetative growth and that chitosan is important for maintaining the cell wall integrity of C. neoformans. Furthermore, the absence of cellular chitosan is detrimental to growing cells and affects the ability of C. neoformans to cope with stress.
MATERIALS AND METHODS
Fungal strains and media.
KN99α and KN99a, mating-competent congenic strains of C. neoformans serotype A (34), were used as the wild-type strains, and all deletions were made in either KN99α or KN99a (Table 2). Strains were grown on rich YPD medium (1% yeast extract, 2% Bacto peptone, and 2% dextrose). Solid medium contained 2% Bacto agar. Selective YPD medium contained 100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany), 200 U/ml hygromycin (Calbiochem, La Jolla, CA), 200 μg/ml gentamicin (G418) (Invitrogen, Carlsbad, CA), or 250 μg/ml phleomycin (InvivoGen, San Diego, CA). S. cerevisiae strain S288C (diploid) was grown vegetatively in YPD with shaking at 30°C or sporulated at room temperature in sporulation medium (1% potassium acetate, 0.1% yeast extract, and 0.05% glucose).
TABLE 2.
C. neoformans predicted chitin/polysaccharide deacetylases
| Gene | JEC21 NCBI | Broad Institute serotype A strain | TIGR | Chromosome no. | No. of predicted introns | Predicted protein (aa)c | N-terminal signal sequence | Location of ω cleavage site (aa) | C terminus hydrophobic domain |
|---|---|---|---|---|---|---|---|---|---|
| CDA1 | XP_571516 | CNAG_05799.1 | 180.m00336 | 6 | 5 | 470 | MSTFATFSALLVSLAGV | 443 | MGGSLIALAAVAVGMVYVA |
| CDA2 | XP_570561 | CNAG_01230.1 | 163.m06424 | 4b | 4 | 458 | MIPSTAAAALLTLTAGVA | 432 | DGLSGVGLVLSGVFAGVMLL |
| CDA3 | XP_571200 | CNAG_01239.1a | 163.m06289 | 4b | 5 | 410 | MYGHLSLSTLSLLAVV | 385 | RPSLFVIACGLALAAIMV |
| FPD1 | XP_568540 | CNAG_06291.1 | 183.m01607 | 14 | 6 | 253 | MKFITSLFAVLAILSSV |
Found by using tBLASTn.
CDA2 and CDA3 are linked ∼27 kbp apart.
aa, amino acid.
Analysis of chitin deacetylase protein sequences.
The sequences of the two S. cerevisiae chitin deacetylase proteins, Cda1p and Cda2p, were used to perform BLAST (tBLASTn) searches of the C. neoformans JEC21 genome (28). Conserved domains were determined by using a National Center for Biotechnology Information (NCBI) BLAST search analysis (http://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi). The detection of glycosylphosphatidylinositol (GPI)-anchored proteins (DGPI) (http://129.194.185.165/dgpi/DGPI_demo_en.htm) was used to determine conserved GPI anchor characteristics.
Generation of deletion constructs.
Overlap PCR gene deletion technology (12) was used to generate gene-specific deletion cassettes of CDA2, CHS3, and CSR2; each included a nourseothricin cassette (31), and CDA1, CDA3, and FPD1 contained a hygromycin cassette (19), a phleomycin cassette (19), and a G418 cassette (19), respectively (Table 1). For the primers used to disrupt the genes in KN99 or KN99a, see Table S2 in the supplemental material. The amounts of coding sequence deleted for CDA1, CDA2, CDA3, and FDP1 were 1,147, 1,311, 1,233, and 673 bp, respectively. CHS3 and CSR2 deletions were carried out in KN99 as previously described for deletions in H99 (2).
TABLE 1.
Deletion strains created in this study
| Deletion(s) | Resistance marker(s) | Mating type | Strain background | Strain designation |
|---|---|---|---|---|
| cda1Δ | HygR | MATα | KN99 | LBCN369 |
| cda2Δ | NatR | MATa | KN99 | LBCN372 |
| cda3Δ | PheR | MATa | KN99 | LBCN392 |
| fpd1Δ | G418R | MATα | KN99 | LBCN365 |
| cda1Δ cda2Δ | HygR/NatR | MATα | cda1Δ | LBCN401 |
| cda1Δ cda3Δ | HygR/PheR | MATα | F1 cda1Δ X cda3Δ | LBCN496 |
| cda2Δ cda3Δ | NatR/PheR | MATa | cda3Δ | LBCN458 |
| cda1Δ fpd1Δ | HygR/G418R | MATα | F1 fpd1Δ X cda1Δ | TMCN593 |
| cda2Δ fpd1Δ | NatR/G418R | MATα | fpd1Δ | LBCN404 |
| cda3Δ fpd1Δ | G418R/PheR | MATa | cda3Δ | LBCN462 |
| cda1Δ cda2Δ cda3Δ | HygR/NatR/PheR | MATα | cda1Δ cda3Δ | LBCN632 |
| cda1Δ cda2Δ fpd1Δ | HygR/NatR/G418R | MATα | cda1Δ fpd1Δ | LBCN634 |
| cda1Δ cda3Δ fpd1Δ | HygR/G418R/PheR | MATa | cda3Δ fpd1Δ | LBCN523 |
| cda2Δ cda3Δ fpd1Δ | NatR/G418R/PheR | MATα | cda2Δ fpd1Δ | LBCN456 |
| cda1Δ cda2Δ cda3Δ fpd1Δ | HygR/NatR/G418R/PheR | MATa | cda1Δ cda3Δ fpd1Δ | LBCN559 |
| chs3Δ | NatR | MATα | KN99 | JRCN629 |
| csr2Δ | NatR | MATa | KN99 | LBCN364 |
| cap59Δ | HygR | MATα | KN99 | LBCN420 |
| lac1Δ lac2Δ | HygR/NatR | MATα | KN99 | LBCN563 |
Transformation of C. neoformans.
KN99α or KN99a was transformed using biolistic techniques (19, 51). Cells were grown in YPD to late log phase, concentrated, and plated onto YPD agar for transformation. The cells were bombarded with 0.6 μm gold beads (Bio-Rad, Richmond, CA) that were coated with DNA of the target construct according to the manufacturer's recommendations. Following the transformation, the cells were incubated at 30°C for 4 h on nonselective medium to allow for recovery and then transferred with 0.8 ml sterile phosphate-buffered saline (PBS) to the appropriate selective medium. Transformants were observed in 3 to 5 days.
Generation of multiple deletion strains.
Multiple deletion strains were created (Table 1) either by sequential biolistic transformation (see above) (19, 51) or by mating followed by random spore selection (18).
Analysis of transformants.
To isolate stable transformants, all transformants were passaged five times on nonselective YPD medium and then tested for resistance to the appropriate selective marker. Only those transformants that grew equally well on selective and nonselective media were considered to be stable transformants. A three-primer PCR screen was used to verify homologous integration at both the 5′ and 3′ ends of the deletion cassette (34). In this manner, homologous recombinants can be distinguished from the wild type. A PCR screen using primers outside the deletion construct was used to amplify the entire integration region, demonstrating that a single copy of the transforming DNA had been inserted at the desired locus. Southern blot analyses were performed to screen for single integration in the genome. Single bands were observed on all Southern blots when the blots were hybridized with a selectable marker-specific probe. Each deletion strain generated for this work had a single-deletion construct homologously integrated at the appropriate locus and no other insertions in the genome (data not shown). At least three independent isolates for each mutant were obtained.
Genomic DNA preparation.
Genomic DNA was prepared by a modification of the glass bead DNA extraction protocol described previously (17). Briefly, C. neoformans cells were suspended in a microfuge tube in 500 μl lysis buffer (50 mM Tris-HCl [pH 7.5], 20 mM EDTA, and 1% sodium dodecyl sulfate [SDS]) with 400 mg glass beads (425 to 600 μm; G-9268; Sigma, St. Louis, MO). Cells were disrupted by vortexing for 10 min, followed by a 10-min incubation at 70°C. After brief vortexing, 200 μl 5 M potassium acetate and 150 μl 5 M NaCl were added. The tubes were placed on ice for 20 min and centrifuged at 14,000 rpm for 20 min. The supernatant was mixed with 500 μl phenol-chloroform and spun for 5 min at 14,000 rpm. The aqueous phase was then mixed with 450 μl chloroform and spun for 5 min at 14,000 rpm. The DNA was then precipitated by the addition of 200 μl ethanol, washed with 70% ethanol, dried, and resuspended in 50 μl deionized water.
Southern hybridizations.
Approximately 10 μg of genomic DNA from each strain was digested with various restriction endonucleases according to the manufacturer's recommendations. Restriction fragments were separated on a 1% agarose gel and transferred to nylon membranes using a Turboblot apparatus (Schleicher & Schuell) and 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as a transfer buffer. Probes for Southern analysis were prepared by random priming (random priming kit; Roche) using 50 μCi [α-32P]dCTP (AA0005; GE Bio-Sciences, Piscataway, NJ) according to the manufacturer's instructions. The blots were incubated in 10 ml of buffer (1× phosphate buffer and 7% SDS) solution for 1 h at 65°C, and then the probe was added to this solution and the blots were hybridized at 65°C overnight. The blots were washed twice in 2× SSC-0.1% SDS at room temperature for 10 min and once for 10 min in 0.2× SSC-0.1% SDS that had been prewarmed to 65°C.
RNA extraction and poly(A) RNA purification.
Each C. neoformans strain was initially grown for 24 h at 25°C, 30°C, or 37°C in liquid YPD medium. Cells were collected by centrifugation at 1,800 × g for 5 min, washed once with distilled water, and lyophilized overnight. The lyophilized pellet was then vortexed with 3 ml glass beads (1-mm diameter; BioSpec, Inc., Bartlesville, OK) and resuspended in 4 ml TRIzol reagent (Invitrogen, Carlsbad, CA). After sitting at room temperature for 5 min, 800 μl of chloroform was added and the mixture was shaken for 30 s. This cell lysate was then centrifuged at 4,000 rpm for 10 min, and the supernatant was transferred to a new tube. Two milliliters of isopropanol was added, incubated for 10 min at room temperature, and centrifuged at 4,000 rpm for 10 min. After washing the pellet with 75% ethanol, it was resuspended in water and incubated with DNase I at 37°C for 1 h. The RNA was extracted again with TRIzol and chloroform and precipitated with isopropanol as described above. The dried pellet was resuspended in 300 μl RNase-free water (Invitrogen, Carlsbad, CA). Poly(A) RNA was purified from the total RNA sample by using an oligo-Tex RNA purification kit (QIAGEN, Valencia, CA) following the manufacturer's specifications.
Northern hybridizations.
The procedure we used was adapted from that of Sambrook and Russell (46). Approximately 0.5 μg of poly(A) RNA from each strain was mixed with 2 μl of diethyl pyrocarbonate (DEPC) (Sigma, St. Louis, MO)-treated 5× formaldehyde gel running buffer (0.1 mM MOPS [morpholinepropanesulfonic acid] [Fisher] [pH 7.0], 40 mM sodium acetate [Sigma, St. Louis, MO], 5 mM EDTA [pH 8.0]), 3.5 μl of 37% formaldehyde (Sigma, St. Louis, MO), and 10 μl of formamide (Sigma, St. Louis, MO). Samples were then incubated at 65°C for 15 min and placed directly on ice. Two microliters of DEPC-treated formaldehyde gel loading buffer (50% glycerol [Fisher], 1 mM EDTA [Fisher], and 0.25% bromophenol blue [Sigma, St. Louis, MO]) was added, and samples were then loaded onto a 1.5% agarose (Roche) gel containing formaldehyde (DEPC-treated, 1× formaldehyde gel running buffer, 6.6% formaldehyde) submersed in 1× formaldehyde gel running buffer. An RNA size marker (catalog no. 3191A; Promega, Madison, WI) sample was prepared in the same way; however, ∼1 μg ethidium bromide (Sigma, St. Louis, MO) was added before the sample was loaded onto the agarose gel. A positive binding control ladder, containing 100 pg of each gene-specific DNA of interest, was also loaded on the agarose gel. For the primers used to amplify the gene-specific DNA fragments for the positive binding control ladder, see Table S1 in the supplemental material. Gels were prerun in 1× formaldehyde gel running buffer for 5 min at approximately 5 V/cm before samples were loaded and run at approximately 2 V/cm overnight (∼14 h), with constant recirculation of running buffer, until the bromophenol blue had traveled 8 to 9 cm. Gels were rinsed in several volumes of DEPC-treated water, soaked in several volumes of DEPC-treated 50 mM NaOH (Fisher) for 20 min, and soaked in several volumes of 20× SSC for 45 min. Fragments were then transferred to charged nylon membranes by using a Turboblot apparatus with 20× SSC as transfer buffer. Membranes were then UV cross-linked. Probes for Northern analysis were prepared by random priming (random priming kit; Roche) by using 50 μCi [α-32P]dCTP according to the manufacturer's instructions. The membranes were incubated in 20 ml of ULTRAhyb buffer (Ambion, Austin, TX) for 3 h at 55°C, the probe was added to this solution, and the membranes were hybridized at 55°C overnight. The membranes were washed one time in ∼50 ml of a solution of 1× SSC-0.1% SDS at 25°C for 10 min. Membranes were then washed three times in ∼50 ml of a solution of 0.5× SSC-0.1% SDS at 68°C for 10 min for each wash. When necessary, membranes were stripped of probe by incubating twice in ∼50 ml of 10 mM Tris-Cl [pH 7.4] and 0.2% SDS, heated to boiling for 1.5 h, and subsequently reprobed following instructions above.
Cell wall stress plates.
Solid YPD medium was made with designated amounts of SDS, NaCl, caffeine, Congo red (Congo red stock made in 50% ethanol), or calcofluor white (CFW) (Sigma, St. Louis, MO). C. neoformans strains were grown to mid-log phase in YPD and diluted to 1 × 107 cells/ml, and 10-fold dilutions were made. Five microliters each of the 10-fold dilutions of cultures for each strain was spotted onto the solid medium and grown at 25°C, 30°C, 37°C, or 40°C.
Eosin Y staining.
Cells were grown in 5 ml YPD, pelleted, and washed twice with 1 ml McIlvaine's buffer (0.2 M Na2HPO4 and 0.1 M citric acid [pH 6.0]). The pellet was resuspended in 500 μl McIlvaine's buffer and stained with 30 μl eosin Y (5 mg/ml stock; Sigma, St. Louis, MO). Cells were incubated at room temperature in the dark for 10 min. Excess dye was washed twice with 1 ml McIlvaine's buffer and resuspended in 500 μl McIlvaine's buffer. Cells were examined with an Olympus Vanox AHBT3 microscope using a fluorescein isothiocyanate filter.
Calcofluor white staining.
Cells were grown in 50 ml YPD, diluted in YPD to an optical density at 600 nm (OD600) of 0.1, and allowed to grow for 8, 16, 24, and 48 h. Aliquots were taken at each time point, fixed, stained with CFW (Sigma, St. Louis, MO), and mounted following the protocol of Pringle et al. (40). Cells were examined with an Olympus Vanox AHBT3 microscope using a DAPI (4′,6′-diamidino-2-phenylindole) filter.
Cellular chitin and chitosan content assay.
To measure the chitin and chitosan content of cells, samples were divided into two aliquots. One aliquot was treated with acetic anhydride to measure chitin plus chitosan, and the second aliquot remained untreated to measure chitin. The difference between the two measurements was an estimate of the amount of chitosan. Cultures were initially grown for 24 h in liquid YPD medium and then diluted to an OD650 of 0.05 in fresh medium and incubated at 25°C with shaking at 225 rpm for 68 to 72 h. The two 0.5- to 1.0-ml aliquots of each culture were transferred to tared 2-ml microfuge tubes. Cells were collected by centrifugation at 14,000 rpm for 2 min, the medium was removed, and tubes were spun again at 14,000 rpm for 1 min so as to remove residual medium. The weight of the cell pellet of each sample was determined and defined as wet weight, typically 30 to 40 mg. Dry weights were measured following 2 to 3 days of evaporation at 37°C. One aliquot of pelleted cells was resuspended in 1.0 ml 1 M sodium bicarbonate, followed by the addition of 50 μl acetic anhydride. The acetylation reaction proceeded for 20 min at room temperature, with occasional mixing, followed by 5 min at 100°C. Cells were pelleted as described above. Both aliquots of cells were subsequently extracted with 1 ml 6% KOH at 80°C for 90 min. Samples were centrifuged at 14,000 rpm for 20 min, and the supernatants were discarded. Each pellet was suspended in 1 ml PBS and spun again, and the buffer was discarded. Finally, each pellet was suspended in 0.2 ml of McIlvaine's buffer (0.2 M Na2HPO4, 0.1 M citric acid [pH 6.0]) and frozen at −20°C. Upon thawing, 5 μl of purified Streptomyces plicatus chitinase-63 (5 mg/ml in PBS) was added to hydrolyze chitin to GlcNAc; samples were incubated for 2 to 3 days at 37°C and then stored at −20°C. For colorimetric determination of GlcNAc, the Morgan-Elson method was adapted for microplate readers essentially as previously described (6). Chitinase-treated samples were spun at 14,000 rpm for 1 min and each 10 μl of sample supernatant was combined with 10 μl 0.27 M sodium borate [pH 9.0] in 0.2 ml PCR strip tubes. Samples were heated to 99.9°C in a thermocycler (Techne, Inc., Princeton, NJ) for about 60 s, mixed gently, and incubated further at 99.9°C for 10 min. Immediately upon cooling to room temperature, 100 μl of freshly diluted DMAB solution (Ehrlich's reagent [10 g p-dimethylaminobenzaldehyde in 12.5 ml concentrated HCl and 87.5 ml glacial acetic acid] diluted 1:10 with glacial acetic acid) was added, followed by incubation at 37°C for 20 min. One hundred microliters of each sample was transferred to 96-well, low-evaporation microtiter plates, and absorbance at 585 nm was recorded. Standard curves were prepared from stocks of 0.075 to 2.0 mM GlcNAc (Sigma, St. Louis, MO). The data were analyzed using a one-way analysis of variance (ANOVA) test with a Bonferroni post hoc test.
Analysis of melanin production.
Cells of each strain were taken from solid YPD medium and resuspended in 2 ml glucose-free asparagine medium (1 g/liter l-asparagine, 0.5 g/liter MgSO4·7H2O, 3 g/liter KH2PO4, and 1 mg/liter thiamine) plus 1 mM l-3,4-dihydroxyphenylalanine (l-DOPA) at a concentration of 5 × 107 cells/ml. Cells were shaken at 30°C for 24 h, spun down at 652 × g for 10 min, and photographed. The OD400 was measured for the supernatants from three independent cultures. We used a two-tailed equal-variance Student t test to determine significant differences between supernatant OD400 values (P < 0.001).
Analysis of capsule formation.
Strains were streaked onto DME plates (13.4 g/liter Dulbecco's modified Eagle's medium [Sigma, St. Louis, MO], 25 mM MOPS [pH 7.0], and 1.8% agar) and incubated for 2 or 5 days at 30°C. Individual isolates were resuspended in a 1:4 India ink-to-H2O solution. Cells were observed through an Olympus AHBT3 microscope at ×1,000 magnification, and the capsule diameter was measured for more than 200 cells/strain. We used ANOVA with a Bonferroni post hoc test to compare the mean capsule widths (P < 0.001).
RESULTS
Identification of chitin/polysaccharide deacetylases in C. neoformans.
The genome of C. neoformans var. neoformans strain JEC21 has been sequenced and annotated (28). An analysis revealed four potential chitin/polysaccharide deacetylase genes (Fig. 1; Table 2). Each of these genes was also found in the genome of H99, a C. neoformans var. grubii strain that has been jointly sequenced by Duke University (http://cgt.genetics.duke.edu) and the Broad Institute (http://www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans). Based on the data presented in this report, we are naming three of these genes CDA for chitin deacetylase and one FPD for fungal polysaccharide deacetylase. Two of the putative deacetylases, CDA2 and FPD1, encode proteins named MP98 and D25, respectively, which have been previously described as immunoreactive (3, 27). All of the chitin/polysaccharide deacetylase proteins are homologs of both the Cda1p and Cda2p proteins of S. cerevisiae with E values of <e−13.
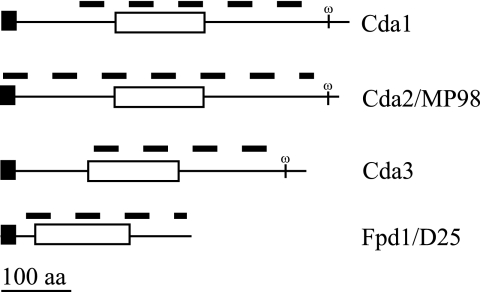
FIG. 1.
Predicted protein structure of C. neoformans deacetylases. The name of each protein is indicated to the right of the protein structure. Domains were identified by BLAST searches of the NCBI database (www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi). The N-terminal signal sequence and GPI anchor were determined. Domains are identified as follows: black rectangles, N-terminal signal sequence; white rectangles, polysaccharide deacetylase domain; omega, predicted GPI cleavage site. The dashed line above each protein indicates the extent of deletion in the corresponding genomic region for each deacetylase gene. aa, amino acids.
An NCBI BLAST search analysis of each of the C. neoformans putative chitin/polysaccharide deacetylase proteins revealed that all four contain a conserved polysaccharide deacetylase domain, Pfam01522 (http://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi). This domain was also found in at least one protein of all sequenced and annotated fungal genomes (www.broad.mit.edu/annotation/fgi/). In addition, an analysis using the DGPI program (http://129.194.185.165/dgpi/DGPI_demo_en.htm) indicated that three of the proteins, Cda1, Cda2, and Cda3, have signature characteristics of GPI-anchored proteins: an N-terminal signal sequence and an omega (ω) site located between hydrophilic and hydrophobic domains at the carboxyl terminus (Fig. 1). The attachment of a GPI moiety to the carboxyl terminus at the omega site of the polypeptide occurs after cleavage of the C-terminal hydrophobic domain (14); therefore, we predict these chitin/polysaccharide deacetylase proteins to be GPI anchored in the plasma membrane and/or cross-linked to α-glucan in the cell wall. Although Fpd1 has an N-terminal signal sequence, it does not contain the distinctive GPI anchor traits and has been identified as a secreted protein (4).
Relative CDA expression level during vegetative growth.
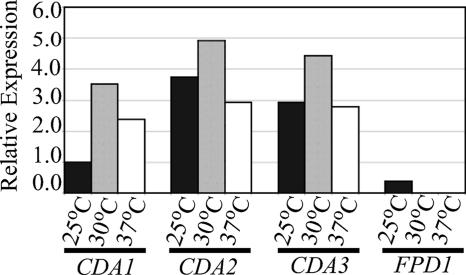
Northern blot analysis was performed to determine the expression profiles of the four putative chitin/polysaccharide deacetylases during vegetative growth in YPD medium. The native transcript expression patterns of CDA1, CDA2, and CDA3 were similar. Each was expressed at all temperatures tested, 25°C, 30°C, and 37°C and was expressed highest at 30°C. All of the chitin/polysaccharide deacetylases were expressed during vegetative growth at 25°C. However, the expression pattern of FPD1 differed from the others, as its expression was not detected during growth at either 30°C or 37°C (Fig. 2). This result is somewhat surprising since, using similar growth conditions, others have detected the D25 (Fpd1) protein in the fungal supernatants of an acapsular strain at 30°C (4).
FIG. 2.
Expression analysis of deacetylases during vegetative growth of C. neoformans. Liquid cultures of KN99α were grown in YPD for 24 h at 25°C, 30°C, and 37°C. Native deacetylase expression was normalized to actin expression, and relative expression values were determined by ImageQuant (version 5.1). Gene-specific probe and growth conditions are listed beneath the graph. The graph depicts one of three independent biological replicates (each having similar results).
Eosin Y stains specifically for chitosan.
The anionic dye eosin Y is typically used histologically in hematoxylin and eosin staining as a counterstain to hematoxylin. We have also found that eosin Y specifically binds to chitosan (a polycation at acid to neutral pH) of live fungal cells and can be visualized by fluorescence microscopy. In Saccharomyces cerevisiae, chitosan is found in only the spore cell wall (10) and not in vegetatively growing cells (2). Eosin Y clearly stained the developing spores of S. cerevisiae and not the budding yeast form (Fig. 3). In contrast, eosin Y readily bound to the cell wall of C. neoformans strains that contained chitosan and did not bind to C. neoformans chs3Δ or csr2Δ strains that have severely reduced chitosan levels (2). This evidence suggests that eosin Y binds specifically to the deacetylated form of chitin and that the chitosan appears to be uniformly distributed through the cryptococcal cell wall.
FIG. 3.
Eosin Y staining of chitosan in fungal cells. C. neoformans cells were grown for 24 h, S. cerevisiae cells were grown for 48 h, and all cells were then stained with eosin Y. Bright field and fluorescein isothiocyanate panels are shown for each strain. Strains or deletions are indicated at the top left of panels, and all cells were grown vegetatively except for those in the top right panel depicting sporulating S. cerevisiae. Arrows indicate cells that have taken up eosin Y, and these cells are dead, as confirmed by trypan blue staining (data not shown). All photos are the same magnification (×1,000).
A series of chitin/polysaccharide deacetylase deletion strains were generated as described in Materials and Methods (Table 1). Eosin Y staining indicated that the retention of a single wild-type copy of CDA1, CDA2, or CDA3 is sufficient for staining similar to the wild type. An example is the cda1Δcda2Δfpd1Δ deletion strain shown in Fig. 3. However, neither cda1Δcda2Δcda3Δ nor the cda1Δcda2Δcda3Δfpd1Δ quadruple-deletion strains stained with eosin Y, suggesting that these strains had reduced chitosan levels similar to those of chs3Δ and csr2Δ strains. Three independent isolates of the triple cda1Δcda2Δcda3Δ and quadruple-deletion strains were analyzed in this and subsequent experiments, indicating that the phenotypes were associated with the specified deletions.
Single CDA or FPD1 deletions do not adversely affect C. neoformans.
Eosin Y staining for determining chitosan content is a qualitative rather than quantitative method. Therefore, to establish whether the chitin-chitosan ratio is an important factor for maintaining the cell wall integrity of vegetatively growing C. neoformans, we measured how the deletion of the individual deacetylase genes affected cellular chitin and chitosan levels. The strains deleted for a single deacetylase were assayed for chitin and chitosan content at 70 h of growth in liquid YPD at 25°C (Table S1). We hypothesized that the loss of a chitin/polysaccharide deacetylase would result in a reduction in the chitosan level and perhaps an increase in the chitin level. The deletion of each of the potential deacetylase genes did not substantially impact either chitin or chitosan content (Table S1). Although the deletion of single deacetylase genes did not severely impact the ability of C. neoformans to produce chitosan, it is possible that the deletion of individual chitin/polysaccharide deacetylases might impact cell wall integrity. However, when the single-deletion strains were subjected to various cell wall inhibitors (see Materials and Methods) their growth levels were equivalent to that of the wild-type strain on these media (see Fig. 6 and 7; also data not shown).
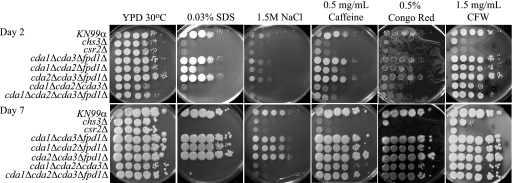
FIG. 6.
Analysis of KN99α and deletion strains for in vitro sensitivity to cell wall stressors. Strains or deletion mutants were grown in YPD medium to mid-log phase, serially diluted 10-fold (starting with 1 × 105 cells in the left column of each panel), and plated on YPD solid medium and YPD solid medium supplemented with various cell wall stressors. The times of incubation are indicated at the far left. Strains are indicated at the left of the panels. Incubation conditions are indicated at the top of the panels. All plates containing cell wall stressors were incubated at 30°C.
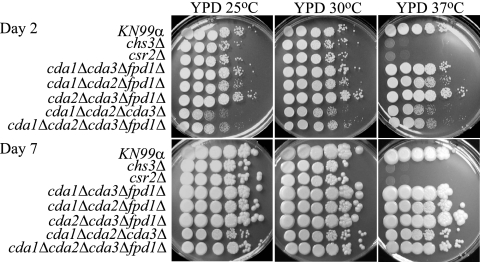
FIG. 7.
Analysis of KN99α and deletion strains for in vitro sensitivity to temperature. The experiment was carried out as described in the legend for Fig. 6.
Chitosan production is abolished by quadruple-deacetylase deletion.
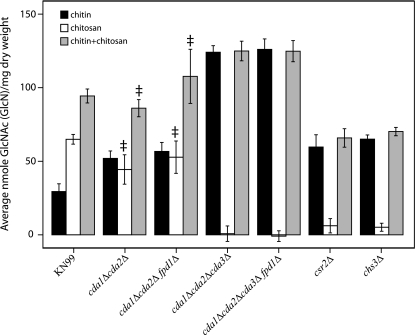
In order to assess whether we have identified all of the potential chitin/polysaccharide deacetylases, we generated three independently isolated strains that were deleted for all four potential deacetylases. Wild-type KN99α as well as the deletion strains was assayed for chitin and chitosan content at 70 h of growth in liquid YPD. We were unable to detect any chitosan production in any of the three quadruple (cda1Δcda2Δcda3Δfpd1Δ)-deletion strains (Fig. 4 and data not shown). The lack of chitosan in the quadruple-deletion strains demonstrates that we have identified all genes that encode proteins that convert chitin to chitosan during vegetative growth. The biochemical data, i.e., chitin/chitosan measurements, directly correlated with the ability of eosin Y to stain the cells (Fig. 3 and 4).
FIG. 4.
Chitin and chitosan contents of C. neoformans KN99α and selected deletion strains. Measurements of strains were taken at 70 h of growth in liquid YPD at 25°C and reported as average nanomoles per milligram of dry weight of chitin, chitosan, and the total chitin plus chitosan as indicated. Each measurement represents the average of three or more independent cultures for each strain. Double daggers indicate measurements that were not significantly different from that of the wild type (P > 0.001). Strains or deletions are indicated at the bottom of the graph. Error bars indicate standard deviations.
Fpd1 does not convert chitin to chitosan during vegetative growth.
To maintain normal or adequate levels of chitosan during vegetative growth, the individual chitin/polysaccharide deacetylases may have redundancy or act synergistically to convert chitin to chitosan. Chitosan production was abolished in the quadruple-deacetylase deletion strains; however, whether each of the putative deacetylases contributed to the conversion of chitin to chitosan during vegetative growth of C. neoformans was still unknown. To determine this contribution, we used a series of four triple-deletion strains that contained only a single deacetylase gene (Table 1). The triple-deletion strains were tested for chitin-chitosan content, and strains containing a wild-type copy of CDA1, CDA2, or CDA3 maintained the ability to deacetylate chitin into chitosan (Fig. 4 and see Table S1 in the supplemental material). This result suggests that each of these genes encodes a chitin deacetylase. In contrast, chitosan production in the cda1Δcda2Δcda3Δ triple-deletion strain was abolished, as observed in the quadruple-deletion strains (Fig. 4). This result suggests that Fpd1/D25 is not directly involved in chitin deacetylation during vegetative growth and that Cda1, Cda2/MP98, and Cda3 are responsible for the deacetylation of chitin into chitosan. Additionally, Fpd1/D25 either does not deacetylate chitin during the tested growth phase or is not a chitin deacetylase. Therefore, we conservatively named it FPD1 for fungal polysaccharide deacetylase.
Chitin content is increased in the cda1Δcda2Δcda3Δ and quadruple-deacetylase deletion strains.
In addition to the lack of cellular chitosan, the amount of the chitin component in the cda1Δcda2Δcda3Δ and the quadruple-deacetylase deletion strains was elevated, being three- to fourfold higher than that in the wild type (Fig. 4). Interestingly, the chs3Δ and csr2Δ strains also showed a reduction in their abilities to produce chitosan, but the chitin levels of these strains did not increase to the same extent as those of the cda1Δcda2Δcda3Δ or quadruple-deacetylase deletion strains (Fig. 4). Selective deacetylation of the chitin specifically made by Chs3, with its regulator Csr2, is consistent with these observations.
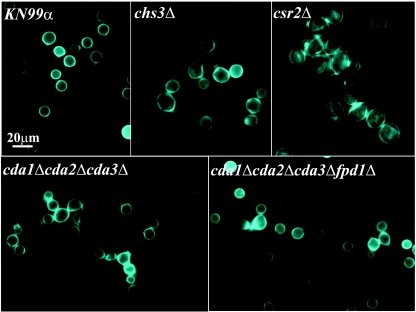
Strains lacking chitosan have abnormal bud separation.
To examine the cell morphologies of the chitin/polysaccharide deacetylase deletion strains, the strains were grown in YPD medium at 30°C, stained with CFW (a fluorescent brightener that binds to chitin), and examined using UV microscopy. The chs3Δ and csr2Δ cells are larger than the wild type, have an irregular shape and nonuniform CFW staining, and exhibit incomplete separation of the daughter cell from the mother cell (2). The cda1Δcda2Δcda3Δ and the quadruple-deacetylase deletion strains, which were unable to convert chitin to chitosan, also had an incomplete mother-daughter cell separation phenotype but were closer in size to the wild type and had a more regular shape (Fig. 5). To determine whether the observed bud separation phenotype was due to incomplete mother-daughter separation and not cell surface flocculation, the cells were sonicated prior to visualization. Sonication, which disrupts cells that are loosely associated, did not affect the abnormal bud separation phenotype, suggesting that this phenotype is not due to flocculation (data not shown). Other morphological phenotypes of the chs3Δ and csr2Δ strains, as mentioned above, were not shared with any of the CDA deletion strains, which were wild type in appearance (Fig. 5 and data not shown). The data indicated that chitosan is necessary for proper separation of daughter and mother cells.
FIG. 5.
CFW staining of C. neoformans KN99α and deletion strains. Cells were grown in YPD at 30°C and stained with CFW, a dye that binds to chitin. Strains or deletions are indicated top left of each frame. Scale bar = 20 μm. All photos are at the same magnification (×1,000).
Chitosan-deficient strains are sensitive to select cell wall inhibitors.
The majority of the cell wall chitin in vegetatively growing C. neoformans is converted to chitosan. chs3Δ and csr2Δ, both of which have dramatically reduced chitosan levels, have been shown to be sensitive to cell wall inhibitors in the H99 background (2). We show here that these deletions have similar sensitivities in the KN99 background (Fig. 6). We tested the deacetylase deletion strains on five cell wall inhibitors that assess different aspects of cell wall integrity. SDS has been used extensively to test cell wall integrity, caffeine has been used to test signal transduction and cell integrity phenotypes in S. cerevisiae (29), Congo red is a dye that inhibits microfibril assembly of the cell wall (44, 54), NaCl is used to test for osmotic stability, and CFW disrupts the assembly of chitin microfibrils (44) (Fig. 6). After 2 days on inhibitor plates, several of the deacetylase deletion strains showed growth defects. However, after 7 days of growth, only the triple-deletion cda1Δcda2Δcda3Δ strain and the quadruple-deacetylase deletion strain had phenotypes similar to those of the chs3Δ and csr2Δ strains on SDS and NaCl but not Congo red, CFW or caffeine (Fig. 6). On SDS, the growth of cda1Δcda2Δcda3Δ and the quadruple-deacetylase deletion strains was completely inhibited and growth on NaCl was substantially less than that of the wild type. The data indicate that chitosan is necessary for maintaining normal cell wall integrity during specific stress conditions.
Chitin deacetylases are not vital for growth at elevated temperatures.
The ability to grow at the host's temperature, i.e., 37°C, is an important virulence factor for any human pathogen. Therefore, the deacetylase deletion strains were assessed for temperature sensitivity on YPD medium (Fig. 7). The chs3Δ and csr2Δ strains are temperature sensitive at 37°C, suggesting that chitosan is important for growth at 37°C (2). However, the cda1Δcda2Δcda3Δ and the quadruple-deacetylase deletion strains initially appeared to have a slight growth defect at 25°C and 37°C compared to their growth at 30°C. Importantly, after 7 days, the deletion strains approached wild-type growth levels at all temperatures tested, including 40°C (Fig. 7 and data not shown). These data indicate that chitosan or the deacetylases are not necessary for growth at host temperature; therefore, the temperature sensitivity of the chs3Δ and csr2Δ strains is not due solely to the loss of chitosan (Fig. 7).
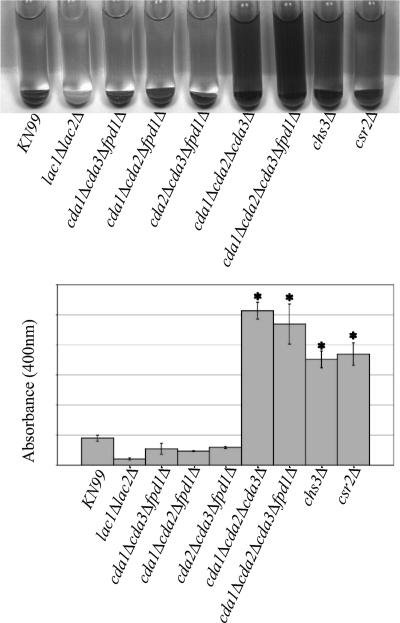
Lack of chitosan is responsible for the “leaky melanin” phenotype.
C. neoformans has the ability to produce melanin, a known virulence factor (24, 25, 38, 43). We previously reported that chs3Δ and csr2Δ strains produce a dark pigment in the medium. It is possible that these strains have lost the ability to completely retain melanin, a melanin-like pigment, or the laccase protein within the cell wall (2, 56). The term “leaky melanin” has been used to describe this phenotype and is apparent when cells are resuspended in a glucose-free asparagine medium containing the laccase substrate l-DOPA. We examined the deacetylase deletion strains to determine whether reduced chitosan content caused the “leaky melanin” phenotype. The deletion of the individual deacetylases did not affect the ability of C. neoformans to produce melanin or significantly change the OD400 of the supernatant (data not shown). However, strains lacking chitosan production, including the cda1Δcda2Δcda3Δ strain, the quadruple-deacetylase deletion strain, the chs3Δ strain, and the csr2Δ strain, had pigment in the supernatant consistent with this phenotype. Furthermore, the cell pellet remained pigmented, indicating that the deletion strains had retained at least a portion of the pigment in the cell (Fig. 8). The supernatant OD400 of the cda1Δcda2Δcda3Δ, quadruple-deacetylase deletion, chs3Δ, and csr2Δ strains was significantly increased (P < 0.001) compared to the OD400 of wild-type and other deacetylase deletion strains (Fig. 8 and data not shown). The differences of the OD400 among the “leaky melanin” strains were not significant.
FIG. 8.
Melanin production of chitin synthesis and deacetylation deletion strains. (Top panel) Cells were grown 2 days on YPD at 30°C, resuspended to identical optical densities in glucose-free asparagine medium containing l-DOPA, and allowed to shake at 30°C. Cells were centrifuged to visualize the secreted pigment and the color of the pellets. Pictures were taken at 24 h. The strain or deletion is indicated at the bottom of the panel. The photo is representative of four independent experiments. A lac1Δlac2Δ strain is included as a negative control. (Bottom panel). Quantification of the OD400 of the supernatants from the cultures as shown in the top panel. Asterisk indicates significant differences between supernatant at OD400 compared to that of the wild type by using a two-tailed equal-variance Student t test. P < 0.001. Error bars indicate standard deviations.
Strains with a native CDA1, CDA2, or CDA3 retained melanin within the pellet fraction. Only when all three of these genes were deleted in the same strain was the “leaky melanin” phenotype manifested. The deletion of FPD1 did not increase “leakiness” between the cda1Δcda2Δcda3Δ and quadruple-deletion strains (Fig. 8). Hence, Fpd1/D25 does not impact either melanin production or pigment retention (data not shown).
CDA deletions affect polysaccharide capsule size.
A major virulence factor associated with C. neoformans is its ability to produce a polysaccharide capsule, which may help it evade phagocytosis (5). Capsular sizes differ, with clinical specimens reported to range between indiscernible to 100 μm (7). Therefore, we ascertained whether changes in chitosan production had an effect on capsule size. To determine average capsular width, the capsules were measured from the outer edge of the cell wall to the outer edge of the capsule, as revealed by staining with India ink. All deacetylase deletion strains were normal in capsular width after 2 days on induction medium (data not shown). Furthermore, after 5 days on induction medium, the single-deletion strains remained similar to the wild type (data not shown). However, several of the multiple-deletion strains had increased capsule after 5 days. Compared to that of the wild-type capsule (5.4 μm), the average capsular widths were significantly increased (P < 0.001) for the chs3Δ strain; one double-deletion strain, cda1Δcda2Δ; two triple-deletion strains, cda1Δcda2Δfpd1Δ, cda1Δcda2Δcda3Δ; and the quadruple-deacetylase deletion strain, with average capsular widths of 8.2, 10.8, 9.8, 9.0, and 10.8 μm, respectively (Fig. 9 and Table 3). The capsule width of the csr2Δ strain was not uniform; therefore, we were unable to make accurate measurements. All strains with increased capsular sizes, i.e., 8.2 μm and above, including the csr2Δ strain, also displayed a mucoid appearance after 5 days of growth on DME medium, consistent with increased polysaccharide production (data not shown).
FIG. 9.
India ink staining of capsule of KN99α and deletion strains. Strains were grown for 5 days on DME medium at 30°C. Strains or deletion mutants are indicated at top left of each frame. Cells were mixed with a 1:4 dilution of India ink-to-H2O solution. Capsule widths of more than 200 cells/strain were measured from the edge of the cell wall to the distal edge of the capsule, and average capsule width is indicated on the bottom right of each frame. A cap59Δ strain is included as a negative control. Scale bar = 20 μm.
TABLE 3.
Capsule measurements, 5 days induction
| Strain | Avg capsule width (μm)a | SE (±) |
|---|---|---|
| KN99α | 5.4 | 0.14 |
| chs3Δ | 8.2 | 0.25 |
| cda1Δcda2Δ | 10.8 | 0.27 |
| cda1Δcda2Δfpd1Δ | 9.8 | 0.29 |
| cda1Δcda2Δcda3Δ | 9.0 | 0.22 |
| cda1Δcda2Δcda3Δfpd1Δ | 10.8 | 0.23 |
ANOVA with a Bonferroni post hoc test indicated significant capsule difference between deletion strains and the wild type. P was <0.001.
The individual deletion of either CDA1 or CDA2 did not alter capsule production, since strains with a wild-type copy of either of these genes had wild-type capsule width (data not shown). In contrast, all strains with both CDA1 and CDA2 deleted had increased capsular widths (Fig. 9 and Table 3). This result suggests that the chitosan produced by either Cda1 or Cda2/MP98 is sufficient for normal capsule production and retention. The capsule of the chs3Δ strain was also increased in diameter, as this deletion strain no longer produces chitin that can be converted to chitosan by the chitin deacetylases.
DISCUSSION
C. neoformans encodes eight chitin synthases and three chitin synthase regulator proteins, but only the deletion of chitin synthase CHS3 or regulator CSR2 affects the amount of chitin deacetylated to chitosan for vegetatively growing cells (2). Furthermore, the deletion of either of these two genes resulted in a plethora of phenotypes possibly associated with a striking reduction in cellular chitosan levels, i.e., a decrease in cell wall integrity and viability at 37°C (2). The current study was initiated to determine whether the cell wall defects displayed by the chs3Δ and csr2Δ strains were caused solely by their reduced cellular chitosan levels. As such, we have genetically characterized and described the effects of deleting the four putative chitin/polysaccharide deacetylase genes in C. neoformans.
Each of the predicted chitin/polysaccharide deacetylase proteins has a highly conserved polysaccharide deacetylase domain, Pfam01522, and three (Cda1, Cda2/MP98, and Cda3) have the necessary signal peptides for posttranslational modification with GPI anchors (Fig. 1). Polysaccharide deacetylase domains have been described for proteins of Rhizobium, a chitooligosaccharide deacetylase (nodulation protein B) (15), and S. cerevisiae, which has two chitin deacetylases that function during spore wall formation (9, 10, 32). GPI anchors are common posttranslational modifications of extracellular eukaryotic proteins that allow proteins to be linked to the exterior surface of the plasma membrane (14). In fungi, proteins with GPI anchors are often covalently linked to the cell wall glucans (20). Cda1, Cda2/MP98, and Cda3 are predicted to be GPI anchored, suggesting that they transverse the plasma membrane and/or attach to the cell wall to deacetylate chitin as it is being extruded through the plasma membrane. No GPI anchor is predicted for Fpd1/D25; however, it is an immunoreactive secreted protein (3, 4).
Expression analysis of the chitin/polysaccharide deacetylase genes indicates that CDA1, CDA2, and CDA3 are expressed at 25°C, 30°C, and 37°C, with expression levels being highest at 30°C (Fig. 2). Chitosan production is completely abolished when these three genes are deleted, as indicated by both the eosin Y detection and chitin/chitosan content assays (Fig. 3 and 4). The data suggest that these three chitin deacetylases function together for the conversion of chitin to chitosan during vegetative growth, but the presence of only one of these three chitin deacetylases is sufficient for substantial chitosan production. At this point, it is unclear why C. neoformans has three functionally redundant chitin deacetylases. It is possible that individual chitin deacetylases function during other specific developmental stages of growth, such as mating, filamentation, or sporulation. The predicted polysaccharide deacetylase Fpd1 did not convert chitin to chitosan in these assays, suggesting that either it does not actively deacetylate chitin during vegetative growth or it deacetylates a polysaccharide other than chitin.
The data indicate that chitosan is essential for proper bud separation (Fig. 5). Chitosan may have a role in the separation of budding cells, as C. neoformans deletion strains lacking chitosan, including chs3Δ and csr2Δ, are observed to have problems disassociating from each other. Interestingly, in S. cerevisiae, the chitin synthase Chs3p produces not only the lateral cell wall chitin but also chitin in the bud neck (26). Chs3p is the homolog to C. neoformans Chs3. If, like Chs3p, Chs3 from C. neoformans produces the bud neck chitin that is then converted to chitosan by the chitin deacetylases Cda1, Cda2/MP98, and Cda3, then the defect in separation of the daughter from the mother cell could be a direct result of the lack of chitosan in the bud neck region. Importantly, none of the double-chitin deacetylase mutants had separation defects, suggesting that all three CDAs participate in budding. However, because single cells are present in cultures, this result also indicates that cell separation can eventually occur in strains that lack chitosan. It also suggests that during bud separation, hydrolyzing enzymes digest the chitosan polymers more readily than they do chitin.
Chitosan is a more flexible and soluble polymer than chitin (50); consequently, it may increase cell wall plasticity, which could allow the cell to change in response to various environmental stimuli. The deletion strains that lack or have reduced chitosan share similar phenotypes for maintaining normal in vivo cell wall integrity, such as sensitivity to cell wall inhibitors and a “leaky melanin” phenotype (Fig. 6 and 8). Strains having at least one of the chitin deacetylases, Cda1, Cda2/MP98, or Cda3, were able to grow on YPD containing either SDS or NaCl. Ionic detergents, such as SDS, disrupt integral membrane proteins that are permanently attached to the membrane, and an osmoticum, such as NaCl, increases the external osmotic pressure. The deletion strains incapable of converting chitin to chitosan are susceptible to these reagents, indicating that cellular chitosan is important for coping with these external stresses.
Another aspect of cell wall integrity is the ability to maintain components in the cell wall. When cells lacking chitosan were grown in liquid medium and centrifuged, a dark pigment was found in both the supernatant and pellet fractions, indicating that these strains produce melanin or another pigment but cannot retain all of it in the cell wall. The two laccase enzymes that produce the pigment melanin, Lac1 and Lac2, as well as melanin itself, are localized to the cell wall (33, 59, 60). One explanation for the observed phenotype is that the chitosan component of the cell wall could provide scaffolding for the laccase proteins and/or the pigment(s). The loss of this support system may lead to partial mislocalization of cell wall elements, which results in melanin “leaking” from the cell wall fraction.
The cda1Δcda2Δ, cda1Δcda2Δfpd1Δ, and cda1Δcda2Δcda3Δ CDA deletion strains and the quadruple chitin/polysaccharide deacetylase deletion strain all have increased capsule widths (Fig. 9). Because at least two of the deletion strains, cda1Δcda2Δ and cda1Δcda2Δfpd1Δ, still contain cellular chitosan, the increase cannot be attributed solely to the lack of chitosan. The common denominator between the deletion strains with increased capsule width is that both the CDA1 and CDA2 genes were deleted. Several possibilities exist as to how Cda1 and Cda2/MP98 could affect capsule size. The chitosan produced by both proteins might mask capsule attachment sites. The polysaccharide α-(1,3)-glucan is required to anchor the capsule of C. neoformans to the cell wall (42). Strains lacking the chitosan produced by Cda1 and Cda2/MP98, including the chs3Δ strain, could allow access to α-(1,3)-glucans that may not normally be available for capsular attachment. Alternatively, the proteins Cda1 and Cda2/MP98 themselves might mask capsule attachment sites; consequently, the capsule size increases when both proteins are missing. However, the capsule width of the chs3Δ strain also is increased, and the chitin deacetylases should be present in this strain. Therefore, it is more plausible that the chitosan produced by Cda1 and Cda2/MP98 causes the observed increase in capsule in the chitin deacetylase deletion strains.
The chs3Δ, csr2Δ, and cda1Δcda2Δcda3Δ deletion strains and the quadruple-deacetylase deletion strain, all of which have drastic reductions in cellular chitosan, share several phenotypes: a defect in bud separation, sensitivity to the cell wall inhibitors SDS and NaCl, a “leaky” melanin phenotype, and an increase in capsular width. The chitin deacetylase proteins should still be present in the chs3Δ and csr2Δ strains, and likewise, Chs3 should still be present in the cda1Δcda2Δcda3Δ and the quadruple-deacetylase deletion strains. Indeed, we do see an increase of chitin in these strains, suggesting that Chs3, with its regulator Csr2, is still functional in these strains that lack chitin deacetylases. This suggests that the lack of chitosan within the cell wall is responsible for these shared phenotypes. In contrast, the chs3Δ and csr2Δ strains have additional phenotypes: an enlarged and misshapen cell morphology (Fig. 5); sensitivity to other cell wall inhibitors, i.e., caffeine, Congo red, and CFW (Fig. 6); and sensitivity to elevated temperature, i.e., growth at 37°C (Fig. 7). The decreased level of total chitin plus chitosan in the chs3Δ and csr2Δ strains may explain some of the phenotypic differences relative to the cda1Δcda2Δcda3Δ and quadruple-deacetylase deletion strains which have increased total chitin plus chitosan as well as increased chitin. If the increased cellular chitin provides additional cell wall support to the chitin deacetylase deletion strains, this may help them maintain stronger cell wall structures and normal morphologies, as observed with the CFW staining (Fig. 5). Although an increase in chitin content may explain the morphological differences between the deletion strains, in S. cerevisiae, Chs3p and Skt5p, homologs to C. neoformans Chs3 and Csr2, respectively, have multiple roles. Chs3p in S. cerevisiae produces the chitin that is converted to chitosan in the spore cell wall and interspore bridges (11, 36) and produces the chitin ring during bud emergence (13, 26) and bud neck chitin (13, 26) and in cells responding to mating pheromone, Chs3p is found throughout the mating projection (47). The chitin synthase regulator Skt5p not only recruits Chs3p to the bud neck but also activates it throughout vegetative growth and regulates it during mating (22, 35, 48, 52). Therefore, the potential for Chs3 and Csr2 in C. neoformans to have additional functions, besides making the chitin that is converted to chitosan during vegetative growth, may also explain the observed variations between the deletion strains.
Not only do the cda1Δcda2Δcda3Δ deletion strains and the quadruple-deacetylase deletions make substantially more chitin than wild-type KN99 but also the total of chitin plus chitosan is significantly different. This indicates that chitin does not negatively regulate its own synthesis in C. neoformans. However, it appears that overall chitin synthesis in these deacetylase mutants is increased. It is possible that chitosan negatively regulates chitin synthesis. The chs3Δ and csr2Δ strains, which make very little chitosan, have increased expression levels of other chitin synthases (2) and elevated chitin levels (Fig. 4) compared to that of the wild type.
The deletion of the gene encoding the putative polysaccharide deacetylase Fpd1/D25, either alone or in combination with other deacetylase deletions, did not affect chitosan levels or cell integrity. This result suggests that Fpd1/D25 does not deacetylate chitin during vegetative growth. Although Fpd1/D25 has been identified as a secreted protein that induces a protective immune response in mice (3, 4), it seems unlikely that it would function as a chitin deacetylase during an infection. Since it is secreted, it could potentially deacetylate polysaccharides other than chitin found in its environment. Because C. neoformans is an opportunistic pathogen, it is unlikely to have evolved a mechanism for specifically deacetylating mammalian polysaccharides. However, it has been proposed that C. neoformans evolved mechanisms to protect itself from predation by amoebae and other phagocytic cells (8, 49) and it is likely that it has had to compete with bacteria for resources. It is possible that Fpd1/D25 could have evolved to deacetylate polysaccharides in phagocytic microorganisms or the acetylated peptidoglycan in bacteria.
An emerging model for the conversion of chitin to chitosan in vegetatively growing C. neoformans points to the chitin synthase Chs3 and the chitin synthase regulator Csr2 producing the chitin that is deacetylated by a combination of three chitin deacetylases. The chitin deacetylases are predicted to be GPI anchored and may be covalently attached to the cell wall or anchored in the plasma membrane in close proximity to where Chs3 is producing chitin, thus allowing for the efficient conversion of chitin to chitosan. Different combinations of the chitin deacetylases may dictate the degree of deacetylation for a particular chitin polymer.
This is the first study to investigate the role of chitin deacetylases during vegetative growth of a fungus and their impact on cell wall integrity. The data indicate that more than one of the chitin deacetylases Cda1, Cda2/MP98, and Cda3 in C. neoformans are responsible for converting the chitin produced by Chs3 and Csr2 into chitosan. Finding chitosan in the vegetative cell wall and showing that it is important for cell wall integrity are novel observations among the pathogenic yeasts. This study broadens our understanding of cell wall biosynthesis in C. neoformans. Both Cda2/MP98 and Fpd1/D25 have previously been isolated as immunoreactive proteins, which would suggest that the deletion of CDA2 and FPD1 may affect C. neoformans virulence; however, it is unknown how the loss of chitosan, the loss of any of the chitin/polysaccharide deacetylase proteins (either individually or in combination), or the loss of the functions of these proteins affects virulence. However, because chitosan and chitin are not present in the mammalian host, chitin deacetylases could prove to be important new drug targets for the treatment of life-threatening fungal diseases, such as cryptococcal meningitis, through the targeting of components that produce both chitosan and chitin.
Supplementary Material
Acknowledgments
We thank Stuart M. Levitz for his contributions to this article, Tod Mattis and Jack A. Ruff for their technical assistance, and Leona Campbell and Kim Gerik for critical reading of the manuscript.
This work was supported by NIH-NIAID grants RO1-AI50184 to J.K.L. and RO1 AI025780 to S.M.L.
Footnotes
Published ahead of print on 30 March 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, D. J. 2004. Fungal cell wall chitinases and glucanases. Microbiology 150:2029-2035. [DOI] [PubMed] [Google Scholar]
- 2.Banks, I. R., C. A. Specht, M. J. Donlin, K. J. Gerik, S. M. Levitz, and J. K. Lodge. 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4:1902-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biondo, C., C. Beninati, M. Bombaci, L. Messina, G. Mancuso, A. Midiri, R. Galbo, and G. Teti. 2003. Induction of T helper type 1 responses by a polysaccharide deacetylase from Cryptococcus neoformans. Infect. Immun. 71:5412-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biondo, C., C. Beninati, D. Delfino, M. Oggioni, G. Mancuso, A. Midiri, M. Bombaci, G. Tomaselli, and G. Teti. 2002. Identification and cloning of a cryptococcal deacetylase that produces protective immune responses. Infect. Immun. 70:2383-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulik, D. A., M. Olczak, H. A. Lucero, B. C. Osmond, P. W. Robbins, and C. A. Specht. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall, A., and J. R. Perfect (ed.). 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, DC.
- 8.Casadevall, A., J. N. Steenbergen, and J. D. Nosanchuk. 2003. ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi—the Cryptococcus neoformans paradigm. Curr. Opin. Microbiol. 6:332-337. [DOI] [PubMed] [Google Scholar]
- 9.Christodoulidou, A., V. Bouriotis, and G. Thireos. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 271:31420-31425. [DOI] [PubMed] [Google Scholar]
- 10.Christodoulidou, A., P. Brizab, A. Ellingerc, and V. Bouriotisa. 1999. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS 460:275-279. [DOI] [PubMed] [Google Scholar]
- 11.Coluccio, A., and A. M. Neiman. 2004. Interspore bridges: a new feature of the Saccharomyces cerevisiae spore wall. Microbiology 150:3189-3196. [DOI] [PubMed] [Google Scholar]
- 12.Davidson, R., J. Blankenship, P. Kraus, M. Berrios, C. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 138:2607-2615. [DOI] [PubMed] [Google Scholar]
- 13.DeMarini, D. J., A. E. M. Adams, H. Fares, C. D. Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenhaber, B., P. Bork, and F. Eisenhaber. 1998. Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 11:1155-1161. [DOI] [PubMed] [Google Scholar]
- 15.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 16.Fromtling, R. A., H. J. Shadomy, and E. S. Jacobson. 1982. Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia 79:23-29. [DOI] [PubMed] [Google Scholar]
- 17.Fujimura, H., and Y. Sakuma. 1993. Simplified isolation of chromosomal and plasmid DNA from yeasts. BioTechniques 14:538-540. [PubMed] [Google Scholar]
- 18.Goins, C. L., K. J. Gerik, and J. K. Lodge. 2006. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: Absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet. Biol. 43:531-544. [DOI] [PubMed] [Google Scholar]
- 19.Hua, J., J. Meyer, and J. Lodge. 2000. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 7:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapteyn, J. C., H. Van Den Ende, and F. M. Klis. 1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426:373-383. [DOI] [PubMed] [Google Scholar]
- 21.Kauffman, C. A. 2006. Clinical efficacy of new antifungal agents. Curr. Opin. Microbiol. 9:483-488. [DOI] [PubMed] [Google Scholar]
- 22.Kozubowski, L., H. Panek, A. Rosenthal, A. Bloecher, D. J. DeMarini, and K. Tatchell. 2003. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 14:26-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnarao, T., and J. Galgiani. 1997. Comparison of the in vitro activities of the echinocandin LY303366, the pneumocandin MK-0991, and fluconazole against Candida species and Cryptococcus neoformans. Antimicrob. Agents Chemother. 41:1957-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon-Chung, J. K., I. Polacheck, and T. J. Popkin. 1982. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 150:1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon-Chung, J. K., and J. C. Rhodes. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 51:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesage, G., J. Shapiro, C. A. Specht, A. M. Sdicu, P. Ménard, S. Hussein, A. H. Y. Tong, C. Boone, and H. Bussey. 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 6:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitz, S. M., S. Nong, M. K. Mansour, C. Huang, and C. A. Specht. 2001. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T-cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 98:10422-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. M. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Saez, J. F., R. Mingo, and A. Gonzalez-Fernandez. 1982. ATP level and caffeine efficiency on cytokinesis inhibition in plants. Eur. J. Biol. 27:185-190. [PubMed] [Google Scholar]
- 30.Maligie, M. A., and C. P. Selitrennikoff. 2005. Cryptococcus neoformans resistance to echinocandins: (1,3){beta}-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 49:2851-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDade, H., and G. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 32.Mishra, C., C. E. Semino, K. J. McCreath, H. de la Vega, B. J. Jones, C. A. Specht, and P. W. Robbins. 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13:327-336. [DOI] [PubMed] [Google Scholar]
- 33.Missall, T. A., J. M. Moran, J. A. Corbett, and J. K. Lodge. 2005. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot. Cell 4:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono, N., T. Yabe, M. Sudoh, T. Nakajima, T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 2000. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology 146:385-391. [DOI] [PubMed] [Google Scholar]
- 36.Pammer, M., P. Briza, A. Ellinger, T. Schuster, R. Stucka, H. Feldmann, and M. Breitenbach. 1992. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast 8:1089-1099. [DOI] [PubMed] [Google Scholar]
- 37.Paugam, A., J. Dupouy-Camet, P. Blanche, J. P. Gangneux, and C. Tourte-Schaefer, and D. Sicard. 1994. Increased fluconazole resistance of Cryptococcus neoformans isolated from a patient with AIDS and recurrent meningitis. Clin. Infect. Dis. 19:975-976. [DOI] [PubMed] [Google Scholar]
- 38.Polacheck, I., Y. Platt, and J. Aronovitch. 1990. Catecholamines and virulence of Cryptococcus neoformans. Infect. Immun. 58:2919-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powderly, W. G., D. M. Finkelstein, J. Feinberg, P. Frame, W. He, C. van der Horst, S. L. Koletar, M. E. Eyster, J. Carey, H. Waskin, T. M. Hooton, N. Hyslop, S. A. Spector, and S. A. Bozzette for the NIAID AIDS Clinical Trials Group. 1995. A randomized trial comparing fluconazole with clotrimazole troches for the prevention of fungal infections in patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 332:700-705. [DOI] [PubMed] [Google Scholar]
- 40.Pringle, J. R., R. A. Preston, A. E. Adams, T. Stearns, D. G. Drubin, B. K. Haarer, and E. W. Jones. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357-435. [DOI] [PubMed] [Google Scholar]
- 41.Pukkila-Worley, R., Q. D. Gerrald, P. R. Kraus, M.-J. Boily, M. J. Davis, S. S. Giles, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reese, A. J., and T. L. Doering. 2003. Cell wall α-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50:1401-1409. [DOI] [PubMed] [Google Scholar]
- 43.Rhodes, J. C., I. Polacheck, and K. J. Kwon-Chung. 1982. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect. Immun. 36:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roncero, C., and A. Duran. 1985. Effect of calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saag, M. S., R. J. Graybill, R. A. Larsen, P. G. Pappas, J. R. Perfect, W. G. Powderly, J. D. Sobel, and W. E. Dismukes. 2000. Practice guidelines for the management of cryptococcal disease. Infectious Diseases Society of America. Clin. Infect. Dis. 30:710-718. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Santos, B., and M. Snyder. 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanz, M., J. A. Trilla, A. Duran, and C. Roncero. 2002. Control of chitin synthesis through Shc1p, a functional homologue of Chs4p specifically induced during sporulation. Mol. Microbiol. 43:1183-1195. [DOI] [PubMed] [Google Scholar]
- 49.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tharanathan, R. N., and F. S. Kittur. 2003. Chitin—the undisputed biomolecule of great potential. Crit. Rev. Food Sci. 43:67-87. [DOI] [PubMed] [Google Scholar]
- 51.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trilla, J. A., T. Cos, A. Duran, and C. Roncero. 1997. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast 13:795-807. [DOI] [PubMed] [Google Scholar]
- 53.Tsigos, I., A. Martinou, D. Kafetzopoulos, and V. Bouriotis. 2000. Chitin deacetylases: new, versatile tools in biotechnology. Trends Biotechnol. 18:305-312. [DOI] [PubMed] [Google Scholar]
- 54.Vannini, G., F. Poli, A. Donini, and S. Pancaldi. 1983. Effects of Congo red on wall synthesis and morphogenesis in Saccharomyces cerevisiae. Plant Sci. Lett. 31:9-17. [Google Scholar]
- 55.Walsh, T. J. 2002. Echinocandins—an advance in the primary treatment of invasive candidiasis. N. Engl. J. Med. 347:2070-2072. [DOI] [PubMed] [Google Scholar]
- 56.Walton, F. J., A. Idnurm, and J. Heitman. 2005. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol. Microbiol. 57:1381-1386. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y., P. Aisen, and A. Casadevall. 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 63:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, M. H., and D. Armstrong. 1994. Cryptococcosis. Infect. Dis. Clin. N. Am. 8:383-398. [PubMed] [Google Scholar]
- 59.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, X., J. Gibbons, J. Garcia-Rivera, A. Casadevall, and P. R. Williamson. 2001. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect. Immun. 69:5589-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.