Abstract
Degenerate PCR and chromosome-walking approaches were used to identify mating-type (MAT) genes and flanking regions from the homothallic (sexually self-fertile) euascomycete fungus Neosartorya fischeri, a close relative of the opportunistic human pathogen Aspergillus fumigatus. Both putative alpha- and high-mobility-group-domain MAT genes were found within the same genome, providing a functional explanation for self-fertility. However, unlike those in many homothallic euascomycetes (Pezizomycotina), the genes were not found adjacent to each other and were termed MAT1 and MAT2 to recognize the presence of distinct loci. Complete copies of putative APN1 (DNA lyase) and SLA2 (cytoskeleton assembly control) genes were found bordering the MAT1 locus. Partial copies of APN1 and SLA2 were also found bordering the MAT2 locus, but these copies bore the genetic hallmarks of pseudogenes. Genome comparisons revealed synteny over at least 23,300 bp between the N. fischeri MAT1 region and the A. fumigatus MAT locus region, but no such long-range conservation in the N. fischeri MAT2 region was evident. The sequence upstream of MAT2 contained numerous candidate transposase genes. These results demonstrate a novel means involving the segmental translocation of a chromosomal region by which the ability to undergo self-fertilization may be acquired. The results are also discussed in relation to their significance in indicating that heterothallism may be ancestral within the Aspergillus section Fumigati.
Aspergillus fumigatus and Neosartorya fischeri are two closely related euascomycete molds (Eurotiomycetes, Pezizomycotina) in the genus Aspergillus, subgenus Fumigati, section Fumigati (12, 33, 36, 42). A. fumigatus is the most common cause of invasive aspergillosis infection in humans (8, 22) and is only known to reproduce by asexual means involving the production of masses of mitotic conidiospores. N. fischeri has also been documented as an invasive opportunistic pathogen (4, 24). However, unlike A. fumigatus, N. fischeri has a known sexual cycle and thus may reproduce by both asexual and sexual means (14, 26). Close relationships between predominantly asexual and readily sexually recombining species such as these are common among fungi (1, 2, 5, 19, 23, 37). A key challenge in present research is to understand the genetic changes that underlie such shifts in reproductive modes. It is hoped that studies may provide insights into the genetic basis of asexuality and possibly allow mating to be realized by supposedly asexual species. The ability to perform sexual crosses with A. fumigatus would provide researchers with a powerful tool to analyze the genetics of pathogenicity in this prevalent mold and would be informative for an understanding of the population biology of the species and disease management (9).
One key factor governing the sexual reproductive mode in fungi is the presence or absence of so called mating-type (MAT) genes. These are located within the genome at mating-type (MAT) loci and determine mating compatibility and incompatibility (3, 7). In euascomycetes, there is most commonly a single MAT locus (termed MAT1) (39). In obligately outbreeding heterothallic species, which require a partner to mate, two nonallelic versions of the locus, termed idiomorphs, are found in isolates with complementary mating types (15, 27). By convention, the two versions are termed MAT1-1 and MAT1-2 (abbreviated to MAT-1 and MAT-2 for convenience; for all heterothallic species, the nomenclature is like that of A. fumigatus) and are distinguished by the presence of an open reading frame (ORF) encoding a MAT protein with an alpha domain and a single ORF encoding a MAT protein with a high-mobility group (HMG) domain (16), respectively (40). These MAT proteins act as transcription factors to enable the sexual cycle (3, 43). Other ORFs may also be present at the idiomorphic MAT loci. In contrast, self-fertile homothallic species usually contain a single MAT locus with the alpha and HMG genes either fused together (as in Cochliobolus homomorphus [45]) or located in close proximity (e.g., ∼1 kb apart in Mycosphaerella zeae-maydis [44]) (7). These arrangements presumably originated from a recombinational event mediating a mating system shift from heterothallism to homothallism (44, 45). To date, only two exceptions to this arrangement are known among homothallic euascomycetes. In Cochliobolus cymbopogonis and Emericella (Aspergillus) nidulans, both alpha and HMG MAT genes are present within the same genome but are unlinked (45, 10, 11). More in-depth genome analyses of E. nidulans have revealed that the alpha and HMG MAT genes are on separate chromosomes in this species (these genes are termed MAT1 and MAT2, respectively, to recognize the different loci [10]; for all homothallic species [e.g., E. nidulans], the nomenclature is like that of N. fischeri), and it has been suggested that E. nidulans was derived from a homothallic ancestor (which contained both alpha and HMG genes at the same original locus) as a result of a translocating break (13). Intriguingly, MAT genes in certain asexual euascomycetes (i.e., species with no known sexual state) have also been reported, and the MAT loci have been characterized in detail in some instances (7, 32, 44).
Although there is no known sexual cycle in A. fumigatus, genes involved in mating have been identified in the genome (13, 31, 32, 34, 41) and the expression of both MAT-1 and MAT-2 genes, together with pheromone signaling genes, has been observed (32). The MAT locus arrangement in A. fumigatus is characteristic of a heterothallic species, with isolates having either a MAT-1 or a MAT-2 idiomorph present (32), which would hypothetically make it necessary for an A. fumigatus isolate to outcross for a successful mating to proceed. It has been suggested that A. fumigatus has also evolved from a homothallic ancestor which contained both alpha and HMG genes at the same original MAT locus and that this evolution involved the loss of MAT genes through segregation (13, 14), but this hypothesis remains unproven.
The aim of the present study was to determine the organization of MAT loci within N. fischeri in order to gain insights both into the genetic basis of the reproductive mode of N. fischeri compared to that of A. fumigatus and into the evolution of MAT loci and sexual reproductive modes among the aspergilli as a whole. Evidence from phylogenetic and genomic studies, together with the predominance of homothallism among the aspergilli, has lead to the suggestion that homothallism is the ancestral state of the aspergilli, with heterothallism being a derived characteristic (13, 14). However, it is also possible that heterothallism is the ancestral state, as evidenced by the presence of an idiomorph arrangement in A. fumigatus and Aspergillus oryzae and other asexual aspergilli (13, 32; M. Paoletti and P. S. Dyer, unpublished results). Indeed, reports elsewhere have provided convincing evidence of the evolution of homothallism from heterothallism (21, 45). Thus, it was of interest to determine whether N. fischeri had a genetic arrangement with two distinct MAT1 and MAT2 loci, similar to that of the homothallic E. nidulans, or a single MAT1 locus containing both alpha and HMG domain-encoding MAT genes, consistent with an ancestral homothallic state. Alternatively, some other novel organization might support the more recent evolution of homothallism and would not contradict the hypothesis that the heterothallic arrangement is ancient.
This study therefore involved the identification and sequencing of mating-type and flanking genes from N. fischeri and a comparison of the DNA sequences to those of homologous regions from A. fumigatus and E. nidulans. Sequence data were also obtained from an ongoing N. fischeri genome-sequencing project (conducted by The Institute for Genomic Research [TIGR] in the United States and the Wellcome Trust in the United Kingdom). Arising data allowed for an assessment of the polarity of mating system evolution in the section Fumigati and provided insights into events relating to the divergence and speciation of the A. fumigatus and N. fischeri lineages.
MATERIALS AND METHODS
Strain used and DNA isolation.
N. fischeri strain NRRL4075 was used for experimental work; it forms a monophyletic group with the type strain NRRL181 (34). The type strain NRRL181 is presently the focus of a genome-sequencing project by TIGR (Rockville, MD) and the Wellcome Trust Sanger Institute (Hinxton, United Kingdom), and so another strain was intentionally chosen for ready comparison to the project data. NRRL4075 was originally isolated in the United Kingdom from garden soil.
Genomic DNA was prepared from mycelium grown in liquid potato dextrose medium (12 g/liter) by using a protocol modified from that of Zolan and Pukkila (46). Lyophilized fragments were transferred with forceps to 1.5-ml tubes containing 500 μl of 1% sodium dodecyl sulfate extraction buffer. Mycelium was ground with a polypropylene micropestle and incubated at room temperature for 1 h. DNA was purified with phenol-chloroform (1:1, vol/vol) as described by Raeder and Broda (35). Next, 300 μl of chloroform-isoamyl alcohol (24:1, vol/vol) was added, and the solution was emulsified and centrifuged for 15 min. The DNA was precipitated from the aqueous phase by the addition of an equal volume of cold isopropanol. The pellet was washed with 70% ethanol, and the DNA was resuspended in 50 μl of water.
Degenerate PCR.
Degenerate primers were used to amplify the alpha domain-encoding sequence from MAT1 family genes (MAT5-6, 5′GIMGICCIYTIAAYWSITTYATHGC3′, and MAT3-4, 5′ARRAAICKIARIATICCISWYTT3′ [32]) and the HMG domain-encoding sequence from MAT2 family genes (MAT5-4, 5′AARRTICCIMGICCICCIAAYGC3′, and MAT3-2, 5′TTNCKIGGIGTRTAITGRTARTCNGG3′, as published here). The primers were designed from regions of conserved sequences identified in E. nidulans and other closely related taxa (10, 11; Paoletti and Dyer, unpublished). The 50-μl PCR amplification mixtures contained 100 ng of genomic DNA, 100 ng of MAT5-6 and MAT4-3 or MAT5-4 and MAT3-2, 200 μM (each) deoxynucleoside triphosphates, and 1.25 U of DNA polymerase. Cycle parameters were 5 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 55°C for MAT2 and 50°C for MAT1, and 1 min at 72°C; and a final 5 min at 72°C. Following PCR, samples were resolved by electrophoresis on 1% Tris-acetate-EDTA agarose gels, which were subsequently stained with SYBR green. PCR products were purified with the QiaQuick PCR purification kit according to the protocol of the manufacturer (QIAGEN). Amplicons of predicted sizes for the MAT1 alpha-domain region and the MAT2 HMG domain were cloned using a pCRII-TOPO TA cloning kit (Invitrogen).
Sequencing reactions were performed in a 10-μl final reaction volume containing 1 μl of BigDye (BigDye Terminator cycle sequencing kit; ABI PRISM, Perkin-Elmer, and Applied Biosystems), 3 μl of BigDye buffer, 1 μl of 10 mM primer, 2 μl of distilled water, and 3 μl of purified PCR product. Sequencing primers were the same as those used for PCR, and sequencing reactions were analyzed on an ABI 3730 DNA analyzer. Sequence fragments were assembled by using Sequencher 3.0 (Gene Code Corporations). Contigs obtained were then compared to gene and genome sequences present in the National Center for Biotechnology Information (NCBI) GenBank database by using the basic local alignment search tool (BLAST), specifically tblastx and blastn.
PCR and sequencing of MAT-flanking regions.
Genomic flanking sequences of isolate NRRL4075 were obtained using the BD GenomeWalker universal kit (BD Biosciences Clontech). DNA libraries were prepared as recommended by the manufacturer by using the blunt-end-cutting restriction enzymes EcoRV, PvuII, StuIV, and XmnI. The PCR amplification procedure used BD Advantage 2 polymerase mix (BD Biosciences Clontech) with gene-specific primers and adaptor-specific primers according to PCR protocols supplied with the BD GenomeWalker universal kit (BD Biosciences Clontech). PCR, the sequencing protocol, and contig assembly were performed as outlined above. The short (∼200-bp) DNA sequences obtained from degenerate PCR were used for primer design for the first walk. For subsequent walks, primers were designed from the sequence acquired by the previous walk. Overlap between walks enabled the assembly of the sequences obtained from all walks possible by using Sequencher 3.0 (Gene Code Corporations).
DNA and amino acid sequence comparisons.
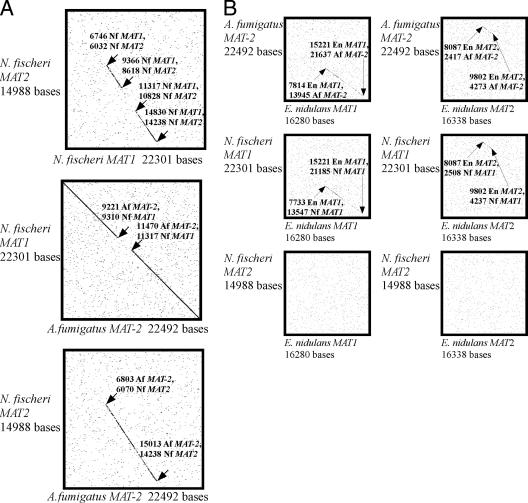
The sequences obtained for the MAT1 alpha domain- and MAT2 HMG domain-encoding genes of N. fischeri and each of their flanking genes were compared to sequences from the NCBI database and posted fungal genome project results for the species A. fumigatus (NCBI), E. nidulans (Broad Institute; http://www.broad.mit.edu, accessed 21 March 2006), and N. fischeri (NCBI), and the sequences were aligned by using MacClade 4.01 (25). Total DNA sequence similarity was estimated using the NCBI blast2seq tool. Sequence data for both MAT1 and MAT2 regions were extended by a further ∼5,000 bp on either side, or as far as possible given limited data, by finding the regions of the genome project contigs corresponding to the regions sequenced in this study and extending the continuous sequence. For A. fumigatus, a 22,492-bp sequence from the strain Af293 (MAT2) from the genome project was compared individually to the MAT1 and MAT2 sequences of N. fischeri. For E. nidulans, a 16,280-bp sequence of MAT1 and a 16,338-bp sequence of MAT2 from the strain FGSCA4 from the genome project were compared to the MAT1 and MAT2 sequences of N. fischeri and A. fumigatus, respectively. Dot plots for pairwise comparisons of the sequences were generated using the Colorado State University Web-based interactive nucleic acid dot plots program (http://www.vivo.colostate.edu/molkit/dnadot/).
Nucleotide sequence accession numbers.
Sequence data obtained for N. fischeri MAT1 and MAT2 and their respective flanking regions have been deposited in GenBank under accession numbers EF363034 and EF370391.
RESULTS AND DISCUSSION
DNA sequences of N. fischeri MAT genes and identification of flanking regions.
N. fischeri is readily self-fertile in culture, which is common for homothallic aspergilli (36). A key aim of the present study was to determine the genetic basis of homothallism in N. fischeri. Experimental work provided a DNA-based confirmation that mating-type genes encoding both putative alpha- and HMG-domain proteins are present within a single isolate of N. fischeri, providing an explanation for the homothallic breeding system since both types of protein are normally required for the sexual development of euascomycete fungi to occur (3). Homothallic euascomycete species most frequently have both alpha- and HMG-domain MAT genes tightly linked at the same locus (7). In N. fischeri, however, these MAT genes were instead found at two distinct loci, here termed MAT1 and MAT2 for the alpha- and HMG-domain-encoding genes, respectively (Fig. 1) (40). Successive genome-walking steps were used to identify a 12,300-bp sequence comprising the MAT1 gene and flanking regions and a 9,001-bp sequence comprising the MAT2 gene and flanking regions (Fig. 1). MAT1 encodes a putative 360-amino-acid protein in two exons and MAT2 encodes a putative 363-amino-acid protein in four exons (Fig. 1). The DNA sequence of MAT1 of N. fischeri is 92% similar to that of the homologue from A. fumigatus characterized by Paoletti et al. (32), and MAT2 of N. fischeri shows 84 to 87% similarity to the A. fumigatus homologue identified by genome studies (13, 31) (Table 1).
FIG. 1.
DNA sequence arrangement of the MAT regions of N. fischeri and A. fumigatus, including MAT genes and their nearest flanking genes. The sequences obtained in this study for the MAT1 and MAT2 loci of N. fischeri were described and compared with sequences from A. fumigatus. Homologous sequences are indicated by different levels of shading, and coding regions are indicated below or above the corresponding boxes. Narrow sections of the maps represent noncoding sequences. The directions of the arrows indicate the predicted directions of gene translation.
TABLE 1.
DNA sequence similarity based on alignment and blastn data
| Comparisona | % Similarity at:
|
% Overall similarity | |||
|---|---|---|---|---|---|
| APN1 | MAT-1 | MAT-2 | SLA2 | ||
| Nf1 vs A. fumigatus | 94-98 | 92 | 95-100 | 94-100 | |
| Nf2 vs A. fumigatus | 82-87 | 84-87 | 86-92 | 82-92 | |
| Nf1 vs Nf2 | 83-85 | 75-88 | 75-88 | ||
A. fumigatus data are from the genome sequence of A. fumigatus, except for MAT-1 data, which are from reference 32. Nf1 and Nf2 indicate the N. fischeri sequence at or flanking MAT1 and MAT2, respectively.
Sequence data obtained for NRRL4075 were compared with genome project sequence data for strain NRRL181 of N. fischeri. The alignment of contigs allowed for the identification of further sequences flanking the MAT regions. A total of 22,301 bp of the sequence of the MAT1 region and 14,988 bp of the sequence of the MAT2 region was obtained in this manner.
Analysis of N. fischeri MAT1- and MAT2-flanking regions.
BLAST analysis of the flanking regions of N. fischeri MAT1 revealed the presence of putative DNA lyase (APN1) (38) and cytoskeleton assembly control (SLA2) (18) genes upstream and downstream of MAT1, respectively (Fig. 1). Both genes appeared to encode functional proteins. BLAST analysis of the flanking regions of N. fischeri MAT2 also revealed the presence of fragments of DNA lyase and cytoskeleton assembly control genes adjacent to MAT2. However, these fragments of APN1 and SLA2 contained multiple stop codons and frameshift insertions and deletions (from 1 to 46 nucleotides in size), indicative of nonfunctional pseudogenes. Therefore, the loci were termed dAPN1 and dSLA2 to recognize the disabled ORFs (17) (Fig. 1).
One of or both APN1 and SLA2 have been reported to neighbor MAT loci in lineages including the hemiascomycete yeasts (2), related Aspergillus species (13), and distant euascomycetes (39). A gene arrangement identical to the N. fischeri MAT1 architecture is known to exist in Neurospora crassa, Yarrowia lipolytica, Fusarium and Xanthoria species, A. fumigatus, and A. oryzae (39, 13). This consistent gene arrangement suggests that APN1 and SLA2 were closely linked with the ancestral ascomycete MAT locus progenitor.
The APN1 and SLA2 genes flanking MAT1 in N. fischeri have greater sequence similarity to the homologous genes from A. fumigatus than to the dAPN1 and dSLA2 pseudogenes flanking N. fischeri MAT2. The MAT1-flanking copy of APN1 has 94 to 98% sequence similarity to APN1 from A. fumigatus, whereas the two N. fischeri APN1 sequences show only 83 to 85% similarity (Table 1). The MAT1-flanking copy of SLA2 has 95 to 100% similarity to SLA2 from A. fumigatus, whereas the two N. fischeri SLA2 sequences have only 75 to 88% similarity (Table 1).
It was also observed that the sequence upstream of the N. fischeri MAT2 locus contains numerous regions with sequence similarity to both named and candidate transposase genes from A. fumigatus and other fungi, including the two candidate transposase genes TAF1 and TAF3, a Nectria species pep5 DNA transposase gene, and a putative A. fumigatus reverse transcriptase gene (31).
Comparison of N. fischeri MAT regions to those of other aspergilli and the evolution of homothallism.
Dot plot comparison of the MAT1 region of N. fischeri to the MAT locus region of A. fumigatus revealed sequence similarity and synteny over a region of at least 22,300 bp (Fig. 2). The MAT1 region of N. fischeri also showed sequence similarity to and synteny with regions upstream and downstream of the E. nidulans MAT1 and MAT2 loci (10, 11, 13). In contrast, the MAT2 region of N. fischeri exhibited limited sequence similarity to the MAT locus of A. fumigatus over a region of about 8,000 bp directly adjoining the MAT locus, with no similarity beyond this (Fig. 2A and B). The MAT2 product from N. fischeri also showed amino acid homology to the MAT2 product from E. nidulans, but fewer than 150 bp of the DNA sequences could be unambiguously and continuously aligned (11), and this small fragment is not perceptible in the dot plot alignments presented here for N. fischeri MAT2 and E. nidulans MAT1 or MAT2 (Fig. 2B). A ca. 420-bp fragment of N. fischeri MAT2 was found upstream of MAT1 in N. fischeri (Fig. 1); a similar fragment is also found in the A. fumigatus MAT-1 region (32). This MAT2 fragment in N. fischeri has one midcodon insertion found in the 3′ end and presumably is not transcribed. This fragment may have arisen through meiotic recombination at some point prior to the divergence of the A. fumigatus and N. fischeri lineages. Recombination at either end of a MAT locus has been invoked to explain variations in gene orientations in other fungal groups. Yun et al. (45) observed a fragment of MAT1 in the 5′ end of MAT2 in the homothallic species Cochliobolus kusanoi.
FIG. 2.
Dot plot comparisons of pairwise alignments of DNA sequence data for N. fischeri (Nf), A. fumigatus (Af), and E. nidulans (En). Sequence lengths are given along the axes near the sequence identifications. (A) Three dot plot alignments compare MAT loci and flanking regions, including coding regions and ∼5,000 bp upstream and downstream, obtained from the preliminary assembly of sequence contigs from the TIGR N. fischeri genome project for strain NRRL181. In the top dot plot, the MAT1 and MAT2 regions from N. fischeri are compared. In the middle dot plot, the MAT locus region of A. fumigatus (MAT-2 in the genome project strain Af293) is compared to the MAT1 region of N. fischeri. In the bottom dot plot, the MAT locus region of A. fumigatus (MAT-2 in the genome project strain Af293) is compared to the MAT2 region of N. fischeri. (B) The three dot plots on the left show comparisons of the MAT1 region of E. nidulans with A. fumigatus Af293 MAT-2 (top), N. fischeri MAT1 (middle), and N. fischeri MAT2 (bottom). The three dot plots on the right show a comparison of the MAT2 region of E. nidulans with A. fumigatus Af293 MAT-2 (top), N. fischeri MAT1 (middle), and N. fischeri MAT2 (bottom).
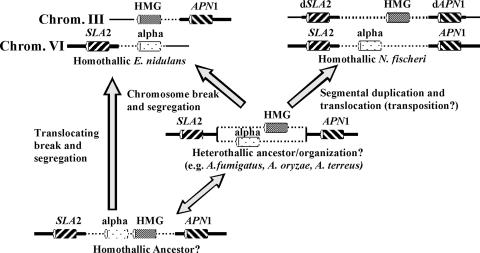
In the context of the evolution of reproductive modes, these observations lead us to speculate that N. fischeri arose from an ancestor with a MAT locus arrangement similar to that seen in extant A. fumigatus (32). The MAT2 region of N. fischeri was then incorporated into the same genome containing the MAT1 region, resulting in a homothallic breeding system. This incorporation is hypothesized to have arisen from a segmental break and translocation of a chromosomal region (Fig. 3), of significance in highlighting a novel means by which homothallism may evolve in euascomycete fungi. Furthermore, it is possible that incorporation was mediated by transposon activity. DNA transposons of euascomycete fungi are well documented (6). The possible transposon-mediated mobility of a MAT locus has not been reported before. The reasons why homothallism in the Eurotiales N. fischeri and E. nidulans has arisen in a form (the presence of two unlinked MAT loci apparently on separate chromosomes) different from that most commonly seen in other euascomycetes to date (the presence of alpha and HMG genes linked at the same single MAT locus) are unclear; it has been suggested that the separation of the MAT loci may provide a more genetically stable form of homothallism (D. M. Geiser, personal communication).
FIG. 3.
Overall gene arrangements of MAT loci, APN1, and SLA2 in the species A. fumigatus, N. fischeri, and E. nidulans. In A. fumigatus, each isolate has either a MAT-1 (encoding a MAT protein with an alpha domain) or a MAT-2 (encoding a MAT protein with an HMG domain) idiomorph located between a single genome copy each of APN1 and SLA2. In N. fischeri, MAT1 is located between APN1 and SLA2, whereas MAT2 is located between the dAPN1 and dSLA2 pseudogenes. In E. nidulans, MAT1 and MAT2 are located on different chromosomes, bordered by only one of SLA2 and APN1, respectively. Chrom., chromosome.
Evolution of reproductive modes in the section Fumigati and the genus Aspergillus.
This study also aimed to assess whether homo- or heterothallism is the ancestral mating state of (i) the most recent common ancestor of A. fumigatus and N. fischeri and (ii) the section Fumigati as a whole. Previous evidence from phylogenetic studies of the section Fumigati, combined with results from later genome analyses, suggest that homothallism is the ancestral state of the section Fumigati and the aspergilli as a whole (13, 14). However, data obtained in the present study instead suggest that the ancestral state was heterothallism, at least in the section Fumigati, based on the following grounds. Firstly, N. fischeri and A. fumigatus appear to share a conserved ancestral MAT locus, flanked by APN1 and SLA2 genes, which contains either an alpha or an HMG domain-encoding MAT gene, i.e., a heterothallic idiomorph organization (Fig. 3). A similar heterothallic organization of either the MAT-1 or MAT-2 locus has also been detected in A. oryzae, a member of the genus Aspergillus, subgenus Circumdati, section Flavi (13, 32; P. S. Dyer, M. Paoletti, K. Kitamoto, and D. B. Archer, unpublished results). The conservation of a heterothallic idiomorph organization supports the hypothesis that the shared gene arrangement (genotypic heterothallism) precedes the origins of both subgenera Fumigati and Circumdati. Given that A. fumigatus and A. oryzae are taxonomically divergent within the genus Aspergillus, their shared MAT sequence arrangement potentially represents the ancestral state.
Secondly, the organization of MAT loci seen in N. fischeri differs from that seen in E. nidulans (Fig. 3), in which homothallism is likely to have arisen from a translocating break, and that proposed in relation to an ancestral homothallic state of the aspergilli, in which the MAT genes are linked at a single MAT locus (13). Thus, there is little evidence for a consistent organization of MAT loci in homothallic aspergilli, which might be expected if homothallism was the ancestral state.
Thirdly, evidence for an ancestral heterothallic state comes from empirical studies of many genera of eusascomycetes, which have shown the evolution of a variety of MAT locus arrangements accompanying the divergence of species (21, 45). Theoretically, there is more support for the transition from heterothallism to homothallism among a fungal population (29, 30) and for a general unidirectional breeding system shift from obligate outcrossing to partial or predominant self-fertilization (20, 28). In the subsection Fumigati, there are nearly 20 additional species with a described sexual cycle, 3 of which are functionally heterothallic while the rest are homothallic. This reduced proportion of known heterothallic species in the subgenus makes an ancestral state of homothallism most parsimonious (14). However, a reconstruction of the ancestral mode of reproduction has never been evaluated using a maximum-likelihood framework to account for rate changes within the group's evolutionary history, the recent description of other taxa known to be either functionally or genetically heterothallic (32, 41), or the use of genetic confirmation of each species' mating system, including those of taxa for which no mating has yet been observed. The frequency of the heterothallic mating-type locus arrangement has been underestimated for other groups of euascomycetes in which numerous putatively asexual (anamorphic) taxa are present (21, 45). Anamorphic taxa have often been discovered to have an idiomorph arrangement characteristic of a heterothallic mating system (21, 32, 45).
Acknowledgments
This work was supported by funds from the A. W. Mellon Foundation Fund to Duke University for Plant Systematics, the Catherine Keever Fund from the Duke Biology Department, and the J. S. Karling Graduate Student Research Award from the Botanical Society of America. P.S.D. thanks the Biotechnology and Biological Research Council (United Kingdom) for financial support.
We thank Mathieu Paoletti for useful discussions. We value the thoughtful conversation and helpful comments of Thomas Mitchell, Joseph Heitman, Fred Dietrich, Rytas Vilgalys, and John Willis on an earlier draft. We thank Timothy James, James Fraser, and Heath O'Brien for useful discussion related to these data. We appreciate receiving isolate NRRL4075 from Stephen Peterson of the U.S. Department of Agriculture in Peoria, IL. We are grateful to Bill Nierman of TIGR for his ready conversation regarding the N. fischeri genome project, to TIGR and the Wellcome Trust for the availability of preliminary data from the N. fischeri genome project, and to the Broad Institute for the availability of data from the E. nidulans genome project.
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Berbee, M. L., and J. W. Taylor. 1992. Two Ascomycete classes based on fruiting-body characters and ribosomal DNA sequence. Mol. Biol. Evol. 9:278-284. [DOI] [PubMed] [Google Scholar]
- 2.Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin, and K. H. Wolfe. 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 101:1632-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coppin, E., R. Debuchy, S. Arnaise, and M. Picard. 1997. Mating-types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61:411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coriglione, G., G. Stella, L. Gafa, S. Oliver, A. A. Padhye, and L. Ajello. 1990. Neosartorya fischeri var. fischeri (Wehmer) Malloch and Cain 1972 (Anamorph: Aspergillus fischerianus Samson and Gams 1985) as a cause of mycotic keratitis. Eur. J. Epidemiol. 6:382-385. [DOI] [PubMed] [Google Scholar]
- 5.Culberson, W. L., and C. F. Culberson. 1973. Parallel evolution in the lichen-forming fungi. Science 180:196-198. [DOI] [PubMed] [Google Scholar]
- 6.Daboussi, M.-J., and P. Capy. 2003. Transposable elements in filamentous fungi. Annu. Rev. Microbiol. 57:275-299. [DOI] [PubMed] [Google Scholar]
- 7.Debuchy, R., and B. G. Turgeon. 2006. Mating-type structure, evolution, and function in euascomycetes, p. 293-323. In U. Kües and R. Fischer (ed.), The mycota. I. Growth, differentiation, and sexuality. Springer-Verlag, Berlin, Germany.
- 8.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-805. [DOI] [PubMed] [Google Scholar]
- 9.Dyer, P. S., and M. Paoletti. 2005. Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Med. Mycol. 43(Suppl. 1):S7-S14. [DOI] [PubMed] [Google Scholar]
- 10.Dyer, P. S., M. Paoletti, and D. B. Archer. 2003. Genomics reveals sexual secrets of Aspergillus. Microbiology 149:2301-2303. [DOI] [PubMed] [Google Scholar]
- 11.Dyer, P. S., M. Paoletti, and D. B. Archer. 2003. Identification of a mating-type gene in the homothallic fungus Aspergillus nidulans. Fungal Genet. Newsl. 50(Suppl.):145. [Google Scholar]
- 12.Frisvad, J. C., and R. A. Samson. 1990. Chemotaxonomy and morphology of Aspergillus fumigatus and related taxa, p. 201-208. In R. A. Samson and J. I. Pitt (ed.), Modern concepts in Penicillium and Aspergillus classification. Plenum, New York, NY.
- 13.Galagan, J., S. E. Calvo, C. Cuomo, L.-J. Ma, J. Wortman, S. Batzoglou, S.-I. Lee, M. Brudno, B. Meray, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing and comparative analysis of Aspergillus nidulans. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 14.Geiser, D. M., J. C. Frisvad, and J. W. Taylor. 1998. Evolutionary relationships in Aspergillus section Fumigati inferred from partial β-tubulin and hydrophobin DNA sequences. Mycologia 90:831-845. [Google Scholar]
- 15.Glass, L., and C. Staben. 1997. Neurospora mating type symbol mt revised to mat. Fungal Genet. Newsl. 44:64. [Google Scholar]
- 16.Grosschedl, R., K. Giese, and J. Pagel. 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 3:94-100. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, P., A. Kumar, N. Lan, N. Echols, M. Snyder, and M. Gerstein. 2002. A small reservoir of disabled ORFs in the yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol. 316:409-419. [DOI] [PubMed] [Google Scholar]
- 18.Holtzman, D. A., S. Yang, and D. G. Drubin. 1993. Synthetic-lethal interactions identity two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122:635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhls, K., E. Lieckfeldt, G. J. Samuels, W. Kovacs, W. Meyer, O. Petrini, W. Gams, T. Börner, and C. P. Kubicek. 1996. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina. Proc. Natl. Acad. Sci. USA 93:7755-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igic, B., L. Bohs, and J. R. Kohn. 2006. Ancient polymorphism reveals unidirectional breeding system shifts. Proc. Natl. Acad. Sci. USA 103:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inderbitzin, P., J. Harkness, B. G. Turgeon, and M. L. Berbee. 2005. Lateral transfer of mating system in Stemphylium. Proc. Natl. Acad. Sci. USA 102:11390-11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LoBuglio, K. F., and J. W. Taylor. 1993. Molecular phylogeny of Talaromyces and Penicillium species in subgenus Biverticillium, p. 115-119. In D. R. Reyolds and J. W. Taylor (ed.), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. C.A.B. International, Surrey, United Kingdom.
- 24.Lonial, S., L. Williams, G. Carrum, M. Ostrowski, and P. McCarthy, Jr. 1997. Neosartorya fischeri: an invasive fungal pathogen in an allogeneic bone marrow transplant patient. Bone Marrow Transplant. 19:753-755. [DOI] [PubMed] [Google Scholar]
- 25.Maddison, W., and D. Maddison. 2005. MacClade: analysis of phylogeny and character evolution, version 4.01. Sinauer, Sunderland, MA. [DOI] [PubMed]
- 26.Malloch, D., and R. F. Cain. 1972. The Trichomataceae: Ascomycetes with Aspergillus, Paecilomyces, and Penicillium imperfect states. Can. J. Bot. 50:2613-2628. [Google Scholar]
- 27.Metzenberg, R. L., and N. L. Glass. 1990. Mating type and mating strategies in Neurospora. Bioessays 12:53-59. [DOI] [PubMed] [Google Scholar]
- 28.Murtagh, G. J., P. S. Dyer, and P. D. Crittenden. 2000. Sex and the single lichen. Nature 404:564. [DOI] [PubMed] [Google Scholar]
- 29.Nauta, M. J., and R. F. Hoekstra. 1992. Evolution of reproductive systems in filamentous ascomycetes. I. Evolution of mating types. Heredity 68:405-410. [DOI] [PubMed] [Google Scholar]
- 30.Nauta, M. J., and R. F. Hoekstra. 1992. Evolution of reproductive systems in filamentous ascomycetes. II. Evolution of hermaphroditism and other reproductive strategies. Heredity 68:537-546. [DOI] [PubMed] [Google Scholar]
- 31.Nierman, W., A. Pain, M. J. Anderson, J. Wortman, J. Arroya, M. Berriman, K. Abe, D. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latgé, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 32.Paoletti, M., C. Rydholm, E. U. Schwier, M. J. Anderson, G. Szakacs, F. Lutzoni, J. P. Debeaupuis, J. P. Latgé, D. W. Denning, and P. S. Dyer. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242-1248. [DOI] [PubMed] [Google Scholar]
- 33.Peterson, S. W. 2000. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis, p. 323-356. In R. A. Samson and J. I. Pitt (ed.), Classification of Penicillium and Aspergillus: integration of modern taxonomic methods. Harwood Publishers, Reading, United Kingdom.
- 34.Pöggeler, S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42:153-160. [DOI] [PubMed] [Google Scholar]
- 35.Raeder, U. A., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 36.Raper, K. B., and D. I. Fennell. 1965. The genus Aspergillus, p. 1-686. Williams & Wilkins, Baltimore, MD.
- 37.Rehner, S. A., and G. J. Samuels. 1995. Molecular systematics of the Hypocreales: a teleomorph gene phylogeny and the status of their anamorphs. Can. J. Bot. 73:S816-823. [Google Scholar]
- 38.Ribar, B., T. Izumi, and S. Mitra. 2004. The major role of human AP-endonuclease homolog APN1 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res. 32:115-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scherrer, S., U. Zippler, and R. Honegger. 2005. Characterisation of the mating-type locus in the genus Xanthoria (lichen-forming ascomycetes, Lecanoromycetes). Fungal Genet. Biol. 42:976-988. [DOI] [PubMed] [Google Scholar]
- 40.Turgeon, B. G., and O. C. Yoder. 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 31:1-5. [DOI] [PubMed] [Google Scholar]
- 41.Varga, J. 2003. Mating-type gene homologues in Aspergillus fumigatus. Microbiology 149:816-819. [DOI] [PubMed] [Google Scholar]
- 42.Varga, J., Z. Vida, B. Tóth, F. Debets, and Y. Horie. 2000. Phylogenetic analysis of newly described Neosartorya species. Antonie Leeuwenhoek 77:235-239. [DOI] [PubMed] [Google Scholar]
- 43.Wirsel, S., B. Horwitz, K. Yamaguchi, O. C. Yoder, and B. G. Turgeon. 1998. Single mating type-specific genes and their 3′ UTRs control mating and fertility in Cochliobolus heterostrophus. Mol. Gen. Genet. 259:272-281. [DOI] [PubMed] [Google Scholar]
- 44.Yun, S.-H., T. Arie, I. Kaneko, O. C. Yoder, and B. G. Turgeon. 2000. Molecular organization of mating-type loci in heterothallic, homothallic and asexual Gibberella and Fusarium species. Fungal Genet. Biol. 31:7-20. [DOI] [PubMed] [Google Scholar]
- 45.Yun, S.-H., M. L. Berbee, O. C. Yoder, and B. G. Turgeon. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA 96:5592-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zolan, M. E., and P. J. Pukkila. 1986. A rapid, high yield mini-prep method for isolation of total genomic DNA. Mol. Cell. Biol. 6:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]