Abstract
Cryptococcus neoformans is a fungal pathogen that is responsible for life-threatening disease, particularly in the context of compromised immunity. This organism makes extensive use of mannose in constructing its cell wall, glycoproteins, and glycolipids. Mannose also comprises up to two-thirds of the main cryptococcal virulence factor, a polysaccharide capsule that surrounds the cell. The glycosyltransfer reactions that generate cellular carbohydrate structures usually require activated donors such as nucleotide sugars. GDP-mannose, the mannose donor, is produced in the cytosol by the sequential actions of phosphomannose isomerase, phosphomannomutase, and GDP-mannose pyrophosphorylase. However, most mannose-containing glycoconjugates are synthesized within intracellular organelles. This topological separation necessitates a specific transport mechanism to move this key precursor across biological membranes to the appropriate site for biosynthetic reactions. We have discovered two GDP-mannose transporters in C. neoformans, in contrast to the single such protein reported previously for other fungi. Biochemical studies of each protein expressed in Saccharomyces cerevisiae show that both are functional, with similar kinetics and substrate specificities. Microarray experiments indicate that the two proteins Gmt1 and Gmt2 are transcribed with distinct patterns of expression in response to variations in growth conditions. Additionally, deletion of the GMT1 gene yields cells with small capsules and a defect in capsule induction, while deletion of GMT2 does not alter the capsule. We suggest that C. neoformans produces two GDP-mannose transporters to satisfy its enormous need for mannose utilization in glycan synthesis. Furthermore, we propose that the two proteins have distinct biological roles. This is supported by the different expression patterns of GMT1 and GMT2 in response to environmental stimuli and the dissimilar phenotypes that result when each gene is deleted.
Mannose is a key component of glycoconjugates in all eukaryotes but plays a special role in yeast, where it dominates the carbohydrate landscape. In contrast to mammals, where mannose is one of an array of moieties used to generate glycan structures, a substantially more restricted range of sugars is used in those fungi that have been examined in detail (19). The best-studied yeast, Saccharomyces cerevisiae, uses only glucose, N-acetylglucosamine, and mannose to build the vast majority of its protein- and lipid-linked carbohydrate structures, and mannose is by far the most abundant sugar in these two classes of biosynthetic products. For example, N-linked carbohydrate chains in this yeast are composed of a core of two N-acetylglucosamine and eight mannose residues, which is further extended with up to hundreds of additional mannoses (19). These mannose chains comprise the mannan that is an important component of the yeast cell wall (28). Similarly, O-linked carbohydrate chains in S. cerevisiae contain only mannose, ranging from one to five residues, and sphingolipids contain just mannose and inositol (16).
To generate the extensive array of mannosylated compounds described above, yeasts must supply appropriate mannose donor molecules to the enzymes that mediate their synthesis. The simplest donor is the nucleotide sugar GDP-mannose (GDP-Man), described by Cabib and Leloir in 1954 (11). This compound is generated in the cytosol by the sequential actions of phosphomannose isomerase, phosphomannomutase, and GDP-Man pyrophosphorylase (57). Some GDP-Man is consumed at the cytosolic face of the endoplasmic reticulum by incorporation into a dolichol phosphomannose lipid that is then flipped into that organelle and used to synthesize N-linked core oligosaccharides and glycosylphosphatidylinositol anchors (23). Much of it, however, is used within the Golgi complex for the synthesis of glycolipids and O-linked glycans (59) and for the outer-chain elaboration of core N-linked structures (14, 36). The location of these synthetic processes leads to a topological problem, which is solved in yeast by the presence of specific Golgi apparatus-localized GDP-Man transporters (1, 15, 38, 39). These proteins import the highly charged mannose precursor from the cytosol into the Golgi apparatus, operating against a concentration gradient via an antiport mechanism that exchanges GDP-Man for GMP. Notably, because mammalian cells do not perform mannosylation in the Golgi apparatus, they have no GDP-Man transporters, although these transporters have been described for plants (8, 22) and for the protozoan parasite Leishmania donovani (33).
Our studies focus on the fungal pathogen Cryptococcus neoformans. This organism causes serious disease and life-threatening meningitis, especially in the context of compromised immunity. In addition to using mannose for the synthesis of protein- and lipid-linked structures in the manner described above, C. neoformans incorporates this sugar into an extensive polysaccharide capsule that surrounds the cell wall (reviewed in reference 10). The capsule, which is required for virulence, is composed of two polysaccharides. The larger polymer, glucuronoxylomannan (GXM), has a repeating structure with monosaccharide branches of xylose and glucuronic acid linked to a mannose backbone (13). Mutants that do not synthesize GXM do not cause disease in experimental animal models (12). The second polymer, galactoxylomannan, has a galactose backbone, with side chains composed of galactose, xylose, and mannose (56); its biological function is less well understood.
We have recently found material that reacts with anti-GXM antibodies in post-Golgi exocytic vesicles in C. neoformans (60). This finding indicates that capsule components are made within secretory organelles and are then packaged into transport vesicles for release at the cell surface when those vesicles fuse with the plasma membrane. In this scenario, capsule synthesis, like the other luminal processes mentioned above, will rely on GDP-Man import via a nucleotide sugar transporter. Mannose comprises one- to two-thirds of all sugar residues in the capsule, depending on the serotype of C. neoformans, so the demands for this transport will be high.
Almost 20 years ago, Hirschberg and coworkers demonstrated GDP-Man transport in yeast by using a mixed vesicle preparation from S. cerevisiae (3). A putative GDP-Man transporter was not identified until almost 10 years later, however, when Turco, Beverley, and colleagues showed that microsomes isolated from wild-type L. donovani were able to import this compound, while cells mutated in the LPG2 gene could not. The mutant cells lost the ability to transport GDP-Man but not UDP-galactose, and the defect was corrected by the reintroduction of the gene (33). Although this initial study did not prove that Lpg2 itself was the transport protein, subsequent work showed that Lpg2 could mediate GDP-Man transport activity when expressed in mammalian cells (25), and recent reconstitution experiments proved that this protein is indeed the transporter (51). Interestingly, Lpg2 acts as a hexameric complex and is multifunctional, with the ability to transport GDP-Man, GDP-arabinose, and GDP-fucose (25). The Leishmania parasite uses GDP-Man in the synthesis of its surface coat and related molecules. lpg2 mutants lack normal forms of these molecules but still demonstrate normal N glycosylation, glycosylphosphatidylinositol anchors, glycolipids, and growth (33).
Close to the time that Lpg2 activity was described, Dean and colleagues used a permeabilized cell assay to demonstrate GDP-Man transport in S. cerevisiae and to implicate the yeast protein Vrg4 in this process (15). Those authors had previously shown that Vrg4 was required for N-linked mannosylation, secretion, and protein sorting (44); based on in vitro transport studies of mutant and wild-type strains, and the protein's homology to other known nucleotide sugar transporters, they suggested that it was the transporter itself. Subsequent work on Vrg4 defined a conserved motif required for substrate binding, showed that the protein occurred as a dimer and was localized to the Golgi apparatus, and identified regions responsible for dimerization, stability, and trafficking (1, 2, 17, 18). Unlike the case with the Leishmania Lpg2 protein, Vrg4 is essential for viability in yeast (44).
Although Vrg4 and Lpg2 have been the most thoroughly examined, several other GDP-Man transporters have been identified in the decade since these two proteins were first implicated in this process. These transporters have been identified by sequence homology and by their ability to complement a partial mutant in VRG4, vrg4-2, but they have not been characterized biochemically. In the fungal pathogen Candida albicans, the single Vrg4 homolog is Golgi apparatus localized and essential for viability, similar to S. cerevisiae Vrg4; a partial loss of function of this protein led to mannosylation defects that caused cell wall-associated phenotypes (39). In a related pathogen, Candida glabrata, deletion of the single VRG4 homolog was also lethal (38). Additional studies of Vrg4 have focused on its utility as a marker of the Golgi apparatus in fungi in both S. cerevisiae (32) and Pichia pastoris (4).
We have investigated GDP-Man transport in C. neoformans, motivated by the central role of mannose in the major cryptococcal virulence factor, the capsule, and by the fact that GDP-Man transport is a fungus-specific activity and therefore an intriguing potential drug target. We have discovered that Cryptococcus encodes and expresses two functional transporters of GDP-Man, a unique finding among fungi. Direct assays of activity in permeabilized cells show that the two proteins have similar biochemical properties. However, transcriptional analysis indicates that they are not coordinately regulated and are therefore likely to play distinct roles in cryptococcal biology. Furthermore, deletion of the two genes results in contrasting capsule phenotypes: gmt1 mutants have reduced capsule size and the ability to induce capsule, while gmt2 cells appear to have wild-type capsules.
MATERIALS AND METHODS
Strains and growth conditions.
C. neoformans strains used were serotype D strains JEC21 (MATα, from Jennifer Lodge, Saint Louis University Medical Center) and JEC43 (MATα ura5, from Joe Heitman, Duke University Medical Center), and serotype A wild-type strain H99 (MATα, from Gary Cox, Duke University Medical Center). S. cerevisiae strains used were NDY5 (MATα ura3-52 leu2-211 vrg4-2) (15) and JPY26 3d (MATα ura3-52 leu2-3,112 ade2-101 vrg4-2 dpm1) (15), provided by Neta Dean (State University of New York at Stony Brook). All cells were grown with continuous shaking at the temperatures indicated in the text in the following media: YPD (1% Bacto yeast extract, 2% peptone, 2% dextrose); YNB (6.7 g/liter Bacto yeast nitrogen base [pH 7.0]) with 2% dextrose, 2% sodium acetate, 2% sodium succinate, or 1 mM H2O2; YNB (pH 4.0) with 1 mM NaNO2; low-iron medium (LIM) (as described in reference 41) with 0.5% dextrose, 100 μM EDTA, and 100 μM bathophenanthroline disulfonic acid (BPDS); LIM lacking BPDS; Dulbecco's modified Eagle's medium (DMEM); Littman's medium (as described in reference 29) with either 0.01 or 1 mg/ml thiamine; phosphate-buffered saline (PBS); PBS with 10% fetal bovine serum (as described in reference 62); minimal medium with 2% glucose but lacking uracil (as described in reference 6); or minimal medium lacking leucine and supplemented with 0.5 M KCl.
Sequences and alignment.
The S. cerevisiae Vrg4 protein sequence (GenBank accession number NP_011290) was BLAST searched against the C. neoformans serotype D database compiled by The Institute for Genome Research (http://www.tigr.org/tdb/e2k1/cna1/). Two closely related sequences were identified and assessed using the Align X program within Vector NTI (Invitrogen, Carlsbad, CA). The first gene (TIGR accession number CNF12620 and GenBank accession number AAY85624) encoded a predicted protein that is 49% identical and 65% similar to Vrg4, which we termed GMT1 for GDP-Man transporter 1. The second gene (TIGR accession number CNG01600 and GenBank accession number AAR96298) encoded a protein with 46% identity and 63% similarity to Vrg4 and was named GMT2.
Gene cloning and expression in S. cerevisiae.
Yeast episomal plasmids containing a 2μm origin of replication, the GAP (glyceraldehyde-3-phosphate dehydrogenase) promoter, and the URA3 marker, without or with the VRG4 sequence (YEp352GAP and YEp352GAP-VRG4, respectively), were obtained from Neta Dean. We used standard PCR techniques and primers 401 to 404 in Table 1 to amplify GMT1 and GMT2 from JEC43 cDNA (prepared as described in reference 47) and incorporate flanking restriction sites (5′ EcoRI and 3′ KpnI). The resulting segments were inserted between the corresponding sites of YEp352GAP, and the constructs were transformed into NDY5 cells by electroporation for assessments of growth phenotype. Each plasmid was further modified by the insertion of LEU2 (amplified from S. cerevisiae genomic DNA using primers 405 and 406 in Table 1) at the ApaI site of each plasmid. These constructs were electroporated into JPY26 3d, and the resulting strains were used for transport assays.
TABLE 1.
Primers and probes
| Primer or probe | Description | Sequence (5′-3′)b |
|---|---|---|
| Expression | ||
| 401 | GMT1a forward primer | ATCGATGAATTCCCCACAGGAATAAC |
| 402 | GMT1 reverse primer | ATCGATGGTACCGCCTGGTTAGTCTT |
| 403 | GMT2 forward primer | GGAATTCCATGGCCTCCTACACCC |
| 404 | GMT2 reverse primer | GGGGTACCGATTACGCCTTGCTTTCTCC |
| 405 | LEU2 forward primer | ATCGATGGGCCCTAAGGCCGTTTCTGACAGAGTAAAATTCTT |
| 406 | LEU2 reverse primer | ATCGATGGGCCCACATACCTAATATTATTGCCTTATTAAAAATGG |
| Gene deletion | ||
| 407 | GMT1 forward primer | AGGCTGTCGTTGGCAGAGGTGGTGA |
| 408 | GMT1 reverse primer | GACATCTCAGCTGACGCTCTCGTCA |
| 409 | GMT2 forward primer | AGGTCTCTTGCCCCTTTTCTTCCGT |
| 410 | GMT2 reverse primer | TCCGATTCTGCTCGTCCTAGGGT |
| 411 | Geneticin forward primer | CTAGCTAGCAGCTTCTATTGTCCAGGCTG |
| 412 | Geneticin reverse primer | TTGGACCAGATGTACCCGCG |
| 413 | Nourseothricin forward primer | GCCAGTGAATTGTAATACGACTCACT |
| 414 | Nourseothricin reverse primer | GAAGATCTCTCTAGAACTAGTGGATCCC |
| Reverse transcriptase PCR | ||
| 415 | ACT1 forward primer | CGTCACAAACTGGGACGACAT |
| 416 | ACT1 reverse primer | CGACACGGAGCTCATTGTAGAA |
| 417 | ACT1 probe | FAM-AGAAGATCTGGCACCATAC-MGBNFQ |
| 418 | GMT1 forward primer | GGAATTCCATGGCCTCCTACACCC |
| 419 | GMT1 reverse primer | CACCAGCGACACCACCAA |
| 420 | GMT1 probe | FAM-CATCTCGGCCATCGC-MGBNFQ |
| 421 | GMT2 forward primer | GCCAAGAAGGAGGAGGTATGC |
| 422 | GMT2 reverse primer | CAGTAGCTCAGGATCGGCAGTA |
| 423 | GMT2 probe | FAM-CCGCGAGCACTACT MGBNFQ |
All gene sequences are from C. neoformans except for LEU2, which is from S. cerevisiae. Sequences that introduce restriction sites are italicized.
FAM, 6-carboxyfluorescein reporter dye; MGBNFQ, molecular groove binding nonfluorescent quencher.
Hygromycin sensitivity.
Cells to be tested were grown overnight in minimal medium with 2% dextrose lacking uracil and then diluted to 1 optical density at 600 nm (OD600) unit/ml and cultured for an additional 1 h. The cells were diluted to 0.1 OD600 units/ml, and 5 μl of fivefold serial dilutions were spotted onto YPD plates with or without 20 μg/ml hygromycin B (Roche Diagnostics, Indianapolis, IN). The plates were grown at 30°C for 3 days before imaging.
Transport assays using “semi-intact cells.”
“Semi-intact cells” were prepared as described previously by Dean and colleagues (15), with minor modifications. Fifty-milliliter cultures of S. cerevisiae cells carrying the indicated plasmids were grown overnight, and a portion of each culture was diluted into a 200-ml volume to achieve an OD600 reading of 1 to 2 units on the following day. Cells were collected by centrifugation (5 min at 1,800 × g at room temperature [RT]), resuspended at 50 OD600 units/ml in 100 mM Tris (pH 9.4)-10 mM dithiothreitol, and incubated at RT for 5 min. Cells were then collected as described above and resuspended at 50 OD600 units/ml in spheroplast buffer (0.75× yeast peptone, 0.7 M sorbitol, 0.5% glucose, 10 mM Tris [pH 7.5]), and the OD600 value of a 1:100 sample in water was measured. An equal volume of 20 mg/ml of Lysing Enzymes (Sigma, St. Louis, MO) in spheroplast buffer was added to the remaining cell suspension, the cells were incubated at 30°C, and the OD600 value of 1:10 dilutions of the sample was checked every 5 min. Once the cells reached 90% lysis, they were subjected to centrifugation (3 min at 1,500 × g at RT), gently resuspended at 50 OD600 units/ml in regeneration buffer (0.75× yeast peptone, 0.7 M sorbitol, 1% glucose), and incubated at 30°C for 20 min. Cells were collected as described above except that it was done at 4°C, gently resuspended at 100 OD600 units/ml in lysis buffer (0.4 M sorbitol, 0.15 M potassium acetate, 2 mM magnesium acetate, 20 mM HEPES-KOH [pH 6.8]), sedimented as described above at 4°C, and resuspended at 300 OD600 units/ml in lysis buffer. Portions (150 μl) were aliquoted into 1.5-ml microcentrifuge tubes on ice, frozen over liquid nitrogen for 1 h, and transferred to storage at −70°C.
For assays, semi-intact cells were thawed quickly and washed three times in 1 ml of ice-cold buffer H (20 mM HEPES [pH 6.8], 150 mM potassium acetate, 250 mM sorbitol, 5 mM magnesium acetate). Standard reaction mixtures included 50 nCi of GDP-[3H]mannose (GDP-[3H]Man) (20 Ci/mmol; American Radiolabeled Corporation, St. Louis, MO), 3 μM GDP-Man, and 30 μg yeast protein (determined by a protein assay; Bio-Rad, Hercules, CA) in a final volume of 25 μl buffer H. Samples were incubated at 30°C for 5 min, incubation was stopped by the addition of 500 μl of ice-cold buffer H, and samples were immediately centrifuged (15 min at 14,000 × g at 4°C). The pellets were washed gently three times with 1 ml cold buffer H, with brief centrifugation (30 s at 14,000 × g at 4°C) between each wash, and the membranes were then resuspended in 100 μl 0.1% Triton X-100 and incubated at RT for 30 min before 50-μl samples were assessed by scintillation counting.
Quantitative reverse transcriptase PCR.
RNA for reverse transcriptase PCR studies was prepared from 100-ml cultures of JEC21 cells grown at 30°C in YPD medium or LIM either overnight (for exponential phase) or for 2 days (stationary growth). Total RNA was extracted with TRIzol reagent as described previously (30), purified using an RNeasy Maxi kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions, and treated with DNase I (Invitrogen). Reverse transcriptase reactions were performed with 10 μg of total RNA, oligo(dT) primers, and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time PCRs used EZ RT-PCR Core Reagents and probes that were designed using Applied Biosystems PRIMEREXPRESS software, both obtained from Applied Biosystems. The probes and primers used (from Invitrogen) are listed as sequences 415 to 423 in Table 1. Experiments were performed using an ABI Prism 7000 instrument according to the manufacturer's instructions, with ACT1 expression serving for normalization between conditions. The amplification program consisted of 1 min at 60°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C alternating with 1 min at 60°C. Each PCR experiment included serial dilutions of known concentrations of each DNA to be analyzed, which were used to generate a standard curve for product quantitation.
RNA for microarray analysis.
Total RNA was prepared from 100-ml cultures of the indicated strain grown to an OD600 value of 1 to 2 units. Cells were sedimented (5 min at 6,000 × g at 4°C), washed with 50 ml diethyl pyrocarbonate-treated double-distilled water, and sedimented again, and the pellet was frozen in a dry ice-methanol bath and lyophilized overnight. Each dried cell pellet was vigorously vortex mixed with 3 ml of 0.5-mm glass beads (Biospec, Bartlesville, OK) for 15 min and then combined with 8 ml RT TRIzol reagent (Invitrogen) and vortex mixed for an additional 1 min. Tubes were left at RT for 5 min, mixed with 1.6 ml of chloroform, shaken gently, and incubated for 3 min at RT. The mixture was transferred to a fresh tube, covered with parafilm, and centrifuged (15 min at 7,600 × g at RT). The upper aqueous phase was removed to a fresh tube, slowly mixed with 1 volume of 70% ethanol, and applied to an RNeasy Maxi column, which was centrifuged in a swinging-bucket rotor (5 min at 3,000 × g at RT). The flowthrough fraction was reapplied to the column and eluted as described above, and the column was then washed, treated with RNase-Free DNase (QIAGEN), washed, and eluted according to the manufacturer's instructions. Eluted RNA was precipitated with a 1/10 volume of 3 M sodium acetate and 2.5 volumes of absolute ethanol on dry ice, and the pellet was recovered by centrifugation (15 min at 8,000 × g at 4°C), washed with 75% ethanol, and allowed to air dry before resuspension in formamide at 4 mg/ml and storage at −20°C. For analysis, RNA was precipitated and resuspended in diethyl pyrocarbonate-treated water, and the concentration and purity were determined from absorbance readings at 260 nm and 280 nm. Total RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) according to the manufacturer's recommendations.
We generated a reference pool of RNA by combining 67 independent RNA preparations from 27 combinations of strains and growth conditions. The cells used for RNA preparation were JEC21 grown in YPD (at 25°C, 30°C, and 37°C), YNB plus glucose (at 25°C, 30°C, 33°C, and 37°C), YNB plus acetate (at 30°C and 37°C), YNB plus succinate (at 30°C), YNB plus NaNO2 (at 25°C), YNB plus H2O2 (at 25°C), LIM with and without BPDS (at 37°C), DMEM (grown in room air or 5% CO2 at 37°C), Littman's medium (both thiamine concentrations at 37°C), and PBS (with or without fetal bovine serum at 37°C); H99 grown in YNB plus glucose (at 30°C) and JEC43, JEC43 with pCMT1/ADE2i (52), JEC43 with pADE2i (30), JEC43 with pAGS1i (46), JEC43 cmt1Δ (52), and JEC43 ags1Δ (47), all grown at 30°C in minimal medium lacking uracil. The pooled RNA samples were mixed thoroughly and aliquoted for storage at −70°C so that the same reference RNA could be used for all microarray experiments. Further details are available upon request.
Microarray construction, hybridization, and data analysis.
A total of 7,738 70-nucleotide oligonucleotide probes specific for C. neoformans serotype D were synthesized by standard methods by Illumina Corporation (San Diego, CA) and dissolved at 20 nM in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.75 M betaine. Probes were printed in duplicate, along with control oligonucleotides, on MWG Epoxy slides (MWG Biotech Inc., High Point, NC) using a locally constructed linear servo arrayer (after the DeRisi model [http://derisilabs.ucsf.edu/]). Details of probe design and sequences are available at http://genome.wustl.edu/activity/ma/cneoformans/array_spec.cgi.
To label RNA transcripts, first-strand cDNA was generated by oligo(dT)-primed reverse transcription (Superscript II; Invitrogen) using the 3DNA Array 350 kit (Genisphere, Hatfield, PA) and modified oligo(dT) primers in which a fluorophore/dendrimer-specific sequence occurs at the 5′ end of the dT primer. For cDNA synthesis, 1 μl fluorophore-specific oligo(dT) primer was added to 8 μg total RNA, and the solution was incubated at 80°C for 10 min and then cooled on ice for 2 min. Each sample was then combined with 1 μl RNase inhibitor (Superase-In; Ambion, Foster City, CA), 4 μl 5× first-strand buffer, 1 μl deoxynucleoside triphosphate mix (10 mM each dATP, dCTP, dGTP, and dTTP), 2 μl 0.1 M dithiothreitol, and 1 μl Superscript II RNase H reverse transcriptase (Invitrogen). Reverse transcription was allowed to proceed for 2 h at 42°C, at which point the reaction was terminated by the addition of 3.5 μl 0.5 M NaOH-50 mM EDTA, incubated for 15 min at 65°C, and neutralized with 5 μl 1 M Tris-HCl (pH 7). Samples to be cohybridized were then combined and concentrated using Microcon YM30 microconcentrators (Millipore, Billerica, MA) according to the manufacturer's recommendations.
For hybridization, each sample pair (∼5 μl) was mixed with 26 μl formamide-based hybridization buffer (vial 7; Genisphere), 2 μl Array 50dT blocker (Genisphere), and 19 μl RNase/DNase-free water. The primary hybridization was performed by incubating 48 μl of sample on the microarray under a supported glass coverslip (Erie Scientific, Portsmouth, NH) for 16 to 20 h at 43°C at high humidity. The slides were then gently submerged for three sequential 11-min incubations in 2× SSC with 0.2% sodium dodecyl sulfate (SDS) at 43°C, 2× SSC at RT, and 0.2× SSC at RT and dried by centrifugation. Secondary hybridization was carried out using the complementary capture reagents provided in the 3DNA Array 350 kit (Genisphere). For each reaction mixture, 2.5 μl 3DNA capture reagent with Cy3, 2.5 μl 3DNA capture reagent with Cy5, 26 μl SDS-based hybridization buffer (vial 6; Genisphere), and 21 μl RNase/DNase-free water were used. These reagents were mixed and incubated in the dark for 10 min at 80°C, followed by 15 min at 50°C. For hybridization, each slide was incubated in the dark with 48 μl of this solution under a supported glass coverslip for 3 h at 65°C at high humidity. The slides were then submerged for three sequential 11-min incubations in 2× SSC with 0.2% SDS at 65°C, 2× SSC at RT, and 0.2× SSC at RT and dried by centrifugation. To prevent fluorophore degradation, the arrays were treated with Dyesaver (Genisphere) according to the manufacturer's instructions.
For data analysis, slides were scanned on a Perkin-Elmer ScanArray Express HT scanner to detect Cy3 and Cy5 fluorescence. Laser power was kept constant for Cy3/Cy5 scans, photomultiplier tube settings were varied for each experiment to optimize signal intensity with the lowest possible background fluorescence, and a low photomultiplier tube setting scan was also performed to recover signal from saturated elements. Gridding and image analysis were performed using ScanArray v3.0 (Perkin-Elmer, Boston, MA). Each spot was defined on a pixel-by-pixel basis using a modified Mann-Whitney statistical test, and the local background was subtracted from the resulting values. The mean signal and control intensities of the on-slide duplicate spots were calculated and Lowess normalized using GeneSpring 7.2 software (Agilent, Santa Clara, CA), with 20% of the data used to calculate the Lowess fit at each point, and the signal-to-Lowess-adjusted control ratios were calculated.
Gene deletion and complementation.
To make gene deletion constructs, we PCR amplified each GMT gene and then replaced each coding sequence with a drug resistance marker. Primers listed in Table 1 were used with genomic JEC21 DNA to amplify GMT1 with 1,186 bp upstream and 590 bp downstream (primers 407 and 408) and GMT2 with 1,127 bp upstream and 1,482 bp downstream (primers 409 and 410). These products were column purified and cloned into pCR2.1-TOPO (Invitrogen). The geneticin drug resistance marker was PCR amplified from plasmid pMH12-T (from Jennifer Lodge, Saint Louis University) using primers 411 and 412 (Table 1). The forward primer introduced an NheI restriction digest site, and the reverse primer started at the SacII restriction site, truncating the 3′ untranslated region by 479 bp. This PCR product was then cloned into the NheI and SacII restriction sites of the pCR2.1-TOPO plasmid containing GMT1, replacing all of the GMT1 coding sequence except for the last 17 bp. The nourseothricin resistance marker was similarly PCR amplified using primers 413 and 414 to introduce an EcoRV restriction site at the 5′ end and an HpaI site at the 3′ end. This was cloned into the NruI and HpaI blunt restriction sites of the pCR2.1-TOPO plasmid containing GMT2, replacing all of the coding sequence except for the last 122 bp; the remaining segment begins significantly downstream of the sequence encoding the “GALNK” region and lacks an in-frame start codon. Finally, the GMT flanking sequences and drug resistance cassettes of the final constructs were amplified by PCR using primers 407 to 410, gel purified, and biolistically transformed into JEC21 cells (55). The cells were allowed to recover for 24 h on YPD medium, transferred onto YPD plates containing 100 μg/ml nourseothricin or geneticin, and incubated at 30°C for 3 to 5 days. Genomic DNA was extracted from drug-resistant transformants and screened by PCR in the presence of 5% Me2SO (35) to confirm the absence of each GMT gene and the presence of the appropriate marker in its place. DNA blotting confirmed both gene replacements and that each marker cassette was present only once in the genome, with no additional ectopic insertions. Details of these studies are available upon request.
To complement the gmt1Δ strain, GMT1 with its flanking sequences was PCR amplified from the TOPO plasmid described above by using primers 407 and 408. This DNA was then biolistically transformed into the disruption strain, together with a PCR-amplified DNA fragment containing the nourseothricin drug resistance marker (generated as described above). Cotransformation was performed as described previously (20). Transformants were identified by resistance to nourseothricin and screened for a return to geneticin sensitivity, indicating the replacement of the disruption marker with the wild-type sequence. Complementation was verified by PCR analysis and DNA blotting.
Capsule induction.
Single colonies were inoculated into 50 ml of YPD broth and grown overnight at 30°C. Cells were then washed with double-distilled water, and 2 × 106 cells/ml were inoculated into 10 ml DMEM (Sigma) in vented T-75 flasks. Flasks were incubated at 37°C in room air or in the presence of 5% CO2 for 16 h. Cells were then collected by centrifugation and resuspended in 50 μl of water, and 5 μl of this suspension was spotted onto a microscope slide with 2 μl of India ink. Cells were visualized using an Olympus BX-60 microscope (Center Valley, PA) with a ×60 objective, and random fields were photographed and printed for manual measurement of capsule radius and cell diameter of 30 to 40 cells.
RESULTS
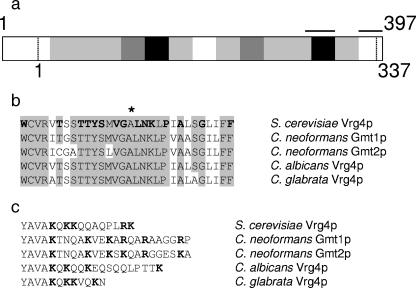
Motivated by the importance of mannose to cryptococcal biology and virulence, we sought the C. neoformans homolog of the well-characterized S. cerevisiae GDP-Man transporter Vrg4. Unexpectedly, database searches of the cryptococcal serotype D genome (http://www.tigr.org/tdb/e2k1/cna1/) (31) yielded two closely related sequences, with GenBank accession numbers AAY85624 and AAR96298. We named these sequences GMT1 and GMT2, respectively. GMT1 encodes a 397-amino-acid protein with 49% identity to Vrg4 (Fig. 1a), while GMT2 encodes a 420-amino-acid protein that is 46% identical to Vrg4 and 63% identical to the protein encoded by GMT1. Aligning by the positions of the most highly conserved sequences, the cryptococcal proteins are extended at the N terminus compared to S. cerevisiae Vrg4 (by 40 amino acids for Gmt1 and 59 amino acids for Gmt2) and contain two additional small peptide insertions (four to eight residues) in the central region of the protein as well as nine additional residues at the C terminus. A conserved “GALNK” motif that has been identified in Vrg4 as required for nucleotide sugar binding (17) is present in both of the predicted cryptococcal proteins (Fig. 1b), and the C-terminal region of each bears multiple basic residues (Fig. 1c), a feature that has been implicated in the intracellular trafficking of related proteins (2).
FIG. 1.
GDP-Man transporters from fungi. (a) The level of identity between the predicted protein sequence of C. neoformans Gmt1 and S. cerevisiae Vrg4 is indicated by the shading of each 25-amino-acid segment: 0 to 25%, white; 26 to 50%, light gray; 51 to 75%, dark gray; 76 to 100%, black. Horizontal bars indicate the regions of sequence that are expanded in b (left bar) and c (right bar). Vertical dashed lines indicate the termini of the shorter Vrg4. (b) Shown are aligned GALNK regions of GDP-Man transporters encoded by genes from S. cerevisiae (VRG4) (GenBank accession number NP_011290), C. neoformans (GMT1 and GMT2), C. albicans (GenBank accession number AAK74075), and C. glabrata (GenBank accession number AAK51897). The alignment begins with the tryptophan at position 271 of Vrg4. The originally identified components of the sequence motif (17) are indicated in boldface type in the S. cerevisiae sequence, and * indicates the amino acid mutated to aspartic acid in the S. cerevisiae vrg4-2 mutant. (c) Shown are the C-terminal hydrophilic residues that follow a conserved hydrophobic stretch (partially shown, YAVA) of each transporter. Basic residues are indicated in boldface type.
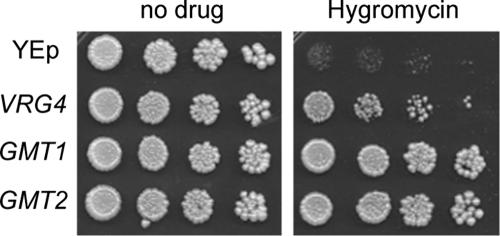
We next tested whether the putative cryptococcal GDP-Man transporters could functionally complement S. cerevisiae strain vrg4-2. This strain bears a point mutation in the GALNK motif (Fig. 1b) and is deficient in GDP-Man transport. Although these cells perform enough transport to survive, they are sensitive to the aminoglycoside hygromycin B and to osmotic stress, and they display defects in Golgi apparatus glycosylation (44). We amplified GMT1 and GMT2 from cDNA of C. neoformans and cloned each gene into a yeast episomal plasmid under the control of the GAP promoter to drive strong constitutive levels of expression. We then expressed each construct in vrg4-2 cells. As shown in Fig. 2, each cryptococcal gene complemented the vrg4-2 sensitivity to hygromycin, while plasmid alone did not. This proves that both cryptococcal proteins can function as GDP-Man transporters.
FIG. 2.
Both putative cryptococcal GDP-Man transporters complement the S. cerevisiae vrg4-2 mutant. Serial fivefold dilutions of vrg4-2 cells carrying the YEp vector alone or expressing the indicated genes were plated onto YPD medium (left) or YPD medium with hygromycin B, as described in Materials and Methods.
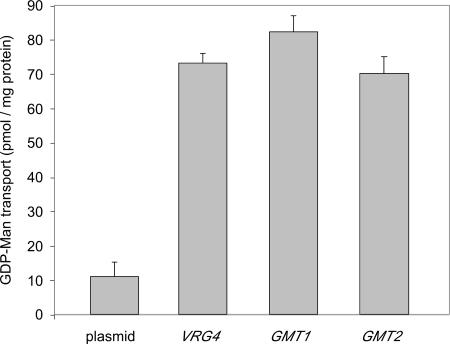
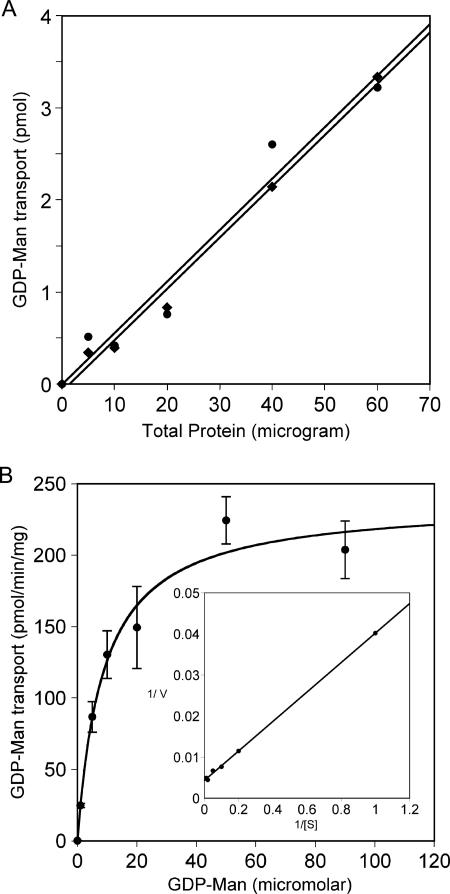
To measure the levels of activity of the two cryptococcal proteins directly, we expressed each one heterologously in S. cerevisiae and assayed GDP-[3H]Man transport in vitro, a cross-species approach that has been effective for studies of nucleotide sugar transporters from mammals and plants (7, 21, 40, 50, 54). To do this, we expressed each protein in vrg4-2 mutant cells that were also mutated in the gene encoding dolichol phosphomannose synthase (DPM1) (42, 43). This mutation serves to reduce background signal from radiolabeled mannose that becomes membrane associated independent of GDP-Man translocation into the Golgi apparatus (see above). We then measured GDP-[3H]Man transport using dpm1 vrg4-2 mutant cells with vector alone or the same cells expressing Gmt1, Gmt2, or Vrg4. As shown in Fig. 3, the two cryptococcal proteins showed levels of activity that were comparable to each other and to that of Vrg4 itself. Uptake activity in vitro was linear for the first 6 min (not shown), comparable to data from previous reports. The activity of each transporter was dependent on protein concentration (Fig. 4a), and substrate titration showed saturable uptake (shown for Gmt1 in Fig. 4b), with Km values of 8.3 μM for Gmt1 and 3.8 μM for Gmt2 (calculated from double-reciprocal plots) (shown for Gmt1 in Fig. 4b, inset). These are comparable to the Km of 6.6 μM determined for GDP-Man transport by reconstituted Leishmania Lpg2 (51) and to typical Km values for other nucleotide sugar transporters (between 1 and 10 μM) (24).
FIG. 3.
GMT1 and GMT2 expressed in S. cerevisiae enable GDP-Man transport in vitro. vrg4-2 dpm1 mutant cells expressing the indicated genes were permeabilized, washed, and used to assay GDP-[3H]Man transport (see Materials and Methods). Assays were done for 10 min at 30°C, with 15 μg of protein per reaction.
FIG. 4.
Activity of Gmt1 and Gmt2 is saturable and dependent on protein and substrate. Assays were performed for 5 min at 30°C using the indicated amounts of protein in the presence of 3 μM GDP-Man (A) or the indicated GDP-Man levels with 30 μg of protein (B). Circles, Gmt1; diamonds, Gmt2. Average and standard error values of triplicate assays are plotted. The inset of B is a double-reciprocal plot of the same data points.
The only function demonstrated for Vrg4 in S. cerevisiae is the transport of GDP-Man. However, Lpg2 from Leishmania major is multifunctional and can transport GDP-arabinose and GDP-fucose in addition to GDP-Man (less efficiently in membrane assays in vitro) (25) but at comparable levels with reconstituted protein (24). While this was a unique finding at the time that it was initially observed, in recent years, multiple substrates have been demonstrated for several additional nucleotide sugar transporters (5, 9, 37, 40, 48, 50). To assess other potential substrates of the cryptococcal proteins, we performed GDP-[3H]Man transport assays in the presence of excess amounts of the nonradiolabeled compounds indicated in Table 2. While GDP and GDP-Man substantially reduced transport activity, the inhibitory effect of GDP-fucose was not statistically significant (P = 0.90 for Gmt1 and P = 0.66 for Gmt2 by Z test) (we did not test GDP-arabinose because it is not commercially available). This indicates that the transporter either does not transport this compound or does so with extremely low efficiency.
TABLE 2.
Inhibition of GDP-[3H]Man transport
| Compound added | % Transport ± SE
|
|
|---|---|---|
| Gmt1 | Gmt2 | |
| None | 100a | 100 |
| GDPb | 12 ± 3 | 13 ± 4 |
| GDP-Man | 19 ± 5 | 19 ± 5 |
| GDP-fucose | 83 ± 13 | 94 ± 16 |
All values were normalized to assays with no added inhibitor (100%). Average values and standard errors shown were calculated from the median values of six independent experiments.
Nonradioactive compounds were present at a final concentration of 23 μM, a 230-fold molar excess over radiolabeled substrate.
The two cryptococcal transporters are similar in terms of vrg4-2 complementation and in vitro transport characteristics. However, these studies were all performed in an exogenous system, so we next investigated the expression of these genes in C. neoformans. We performed quantitative reverse transcriptase PCR studies of wild-type cryptococcal cells cultured under three growth conditions by using either a rich medium that is standard for laboratory growth or an LIM known to induce capsule production in vitro. Analysis of four independent RNA preparations for each condition showed that both genes were transcribed, although GMT1 transcripts were more abundant. The mean ratios of expression of GMT1 compared to that of GMT2 were 5.4 ± 0.3 for cells undergoing exponential growth in rich medium (YPD), 5.3 ± 1.2 for cells in stationary phase in YPD medium, and 7.9 ± 1.3 for cells undergoing exponential growth in LIM (see Materials and Methods).
To examine GMT1 and GMT2 expression in more detail, we performed microarray analysis to assess the expression of each gene under a variety of growth conditions (see Materials and Methods). The media that have been historically developed for C. neoformans culture are those that either alter capsule production or induce cell stress, so a selection of such conditions was used (see Materials and Methods). In most media tested, the expression of GMT1 exceeded that of GMT2 by severalfold, consistent with the results from the growth conditions tested by quantitative reverse transcriptase PCR. The ratio of expression of the two genes varied, ranging from 1 for samples grown in PBS to over 14 for samples grown in DMEM (data not shown).
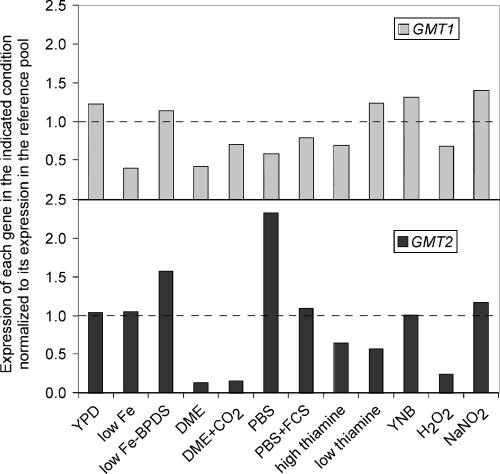
We next examined the influence of environment on the regulation of GMT1 and GMT2 independently. To do this, we assessed the expression of each gene in our set of growth conditions relative to expression of the same gene in a reference pool of mixed RNA samples (see Materials and Methods). This experiment allowed us to draw several conclusions. First, the expression of both genes changed with growth conditions (Fig. 5). Second, under the conditions tested, GMT1 expression (Fig. 5, top panel) was fairly stable, exhibiting only a 3.5-fold difference overall. In contrast, GMT2 expression (Fig. 5, lower panel) was quite variable, with a range of over 17-fold. Finally, the two genes are not coordinately regulated. For example, GMT1 expression was decreased by over 40% relative to the reference pool in PBS, while expression of GMT2 was increased severalfold. Expression of these genes reversed during growth in low-thiamine Littman's medium, where GMT1 expression was increased ∼25%, while the expression of GMT2 was reduced by almost half.
FIG. 5.
GMT1 and GMT2 are not coordinately expressed. Expression of each gene is plotted normalized to its expression in a reference pool (indicated by the dotted line). Values shown are the median values from three separate experiments using independently prepared RNA samples and performed in duplicate. The RNA samples indicated were prepared from JEC21 cells, which were grown overnight in the following media: YPD, LIM (low Fe), LIM without BPDS (low Fe − BPDS), DMEM, DMEM plus CO2, PBS, PBS plus fetal calf serum (FCS), Littman's medium with 1 μg/ml thiamine (high thiamine), Littman's medium with 0.01 μg/ml thiamine (low thiamine), YNB, YNB plus H2O2 (H2O2), or YNB plus NaNO2 (NaNO2). All cells were grown at 37°C except for YPD and YNB medium, which were grown at 30°C, and YNB medium with H2O2 or NaNO2, which was grown at 25°C. All growth was in room air, except the DMEM-plus-CO2 sample, which was grown in a 5% CO2 atmosphere. See Materials and Methods for additional details of growth media, preparation of the reference pool, and analysis.
Our expression studies suggested that Gmt1 and Gmt2 play different roles in the biology of Cryptococcus. To address this hypothesis, we replaced the gene encoding each transporter with a drug resistance marker and assessed the resulting disruption strains. Both mutants grew at the same rate as wild-type cells at 30°C (growth curves were performed as described in reference 47) (not shown). However, colonies of gmt1Δ cells appeared to be duller than those of the wild type, the gmt2Δ mutant, or the complemented gmt1Δ mutant (not shown). This feature is typical of cells with a reduced or absent capsule, a finding that intrigued us because of the extensive mannosylation of capsule polysaccharides.
To pursue the possibility that gmt1Δ cells had altered capsules, we used India ink staining and light microscopy to assess the capsule sizes of disruption and wild-type strains. Cells disrupted in GMT1 had smaller capsules than the wild type in the DMEM medium tested, with a radius of 0.67 ± 0.05 μm compared to 1.06 ± 0.06 μm (P value of <0.0001 by Student's t test). We know that cryptococcal cells modulate capsule production in response to environment both in vitro and in vivo (26, 49, 58, 62), a process that is likely to be significant for pathogenesis. We therefore probed the mutant phenotype further by testing the response of gmt1Δ cells to growth in the same medium under a 5% CO2 atmosphere, a condition that leads to the formation of large capsules (62). When grown this way, the gmt1Δ mutant cells exhibited a striking defect in capsule formation (Fig. 6), which increased only slightly, to 0.94 ± 0.05 in radius compared to 3.29 ± 0.08 for the wild type (P value of <0.0001). As shown in Fig. 6, this defect was reversed in gmt1Δ cells complemented with GMT1 (3.12 ± 0.12) (P value of 0.22 compared to the wild type). gmt2Δ capsules were also similar to those of the wild type under these inducing conditions (3.36 ± 0.08) (P value of 0.56 compared to the wild type).
FIG. 6.
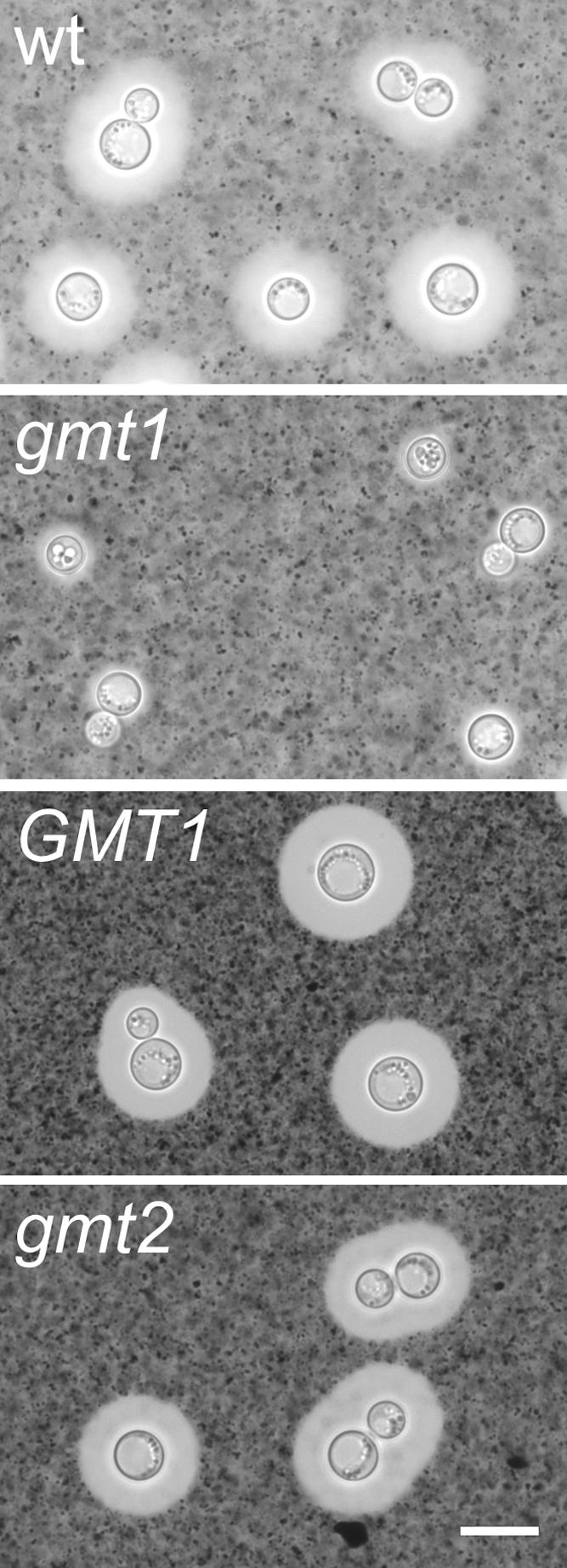
gmt1Δ cells are unable to form large capsules. Cells of the indicated strains were grown in DMEM in a 5% CO2 atmosphere and then mixed with India ink as described in Materials and Methods and examined by light microscopy. The clear halo surrounding the refractile cell wall corresponds to the space occupied by the capsule, into which the ink particles do not penetrate. wt, wild type; gmt1, gmt1Δ; GMT1, gmt1Δ complemented with GMT1; gmt2, gmt2Δ. All images are to the same scale; scale bar, 4 μm.
DISCUSSION
C. neoformans has two functional Golgi apparatus GDP-Man transporters, which bear extensive homology to Vrg4 of S. cerevisiae. One difference between the transporters from these two organisms is the presence of extended N-terminal sequences in the cryptococcal proteins (Fig. 1a). The N terminus of Vrg4 has been implicated in protein localization (18), but our complementation studies (Fig. 2) show that the cryptococcal versions function well in S. cerevisiae, suggesting that the extended sequences still allow for correct localization. In accordance with their demonstrated activity, and similar to other proteins in this class (34), the cryptococcal transporters have highly conserved GDP-Man binding domains (17). They also have several basic residues in their C-terminal tails, a feature that has been implicated in recycling to the endoplasmic reticulum (2).
The presence of two GDP-Man transporters in C. neoformans is unique compared to the other fungi studied in this regard to date: C. albicans, C. glabrata, P. pastoris, and S. cerevisiae. Beyond yeast, several examples of multiple nucleotide sugar transporters with overlapping functions have been demonstrated. These include multiple UDP-galactose transporters described for plants (7), parasites (11a), and mammals (27, 61) and a set of five Golgi apparatus-localized proteins of Arabidopsis thaliana that all exhibit GDP-Man transport activity (8, 22).
One key question is whether Gmt1 and Gmt2 play different roles in the biology and pathogenesis of C. neoformans. Our expression data suggest that they do, because they demonstrate distinct patterns of regulation in response to growth conditions, despite similar biochemical characteristics. In particular, GMT1 is modestly regulated, with only a fewfold difference in expression under the growth conditions that we tested, while the expression of GMT2 is more dramatically modulated (Fig. 5). Furthermore, under all conditions but one, the magnitude of the observed changes in expression differed between the genes, and in several cases, these changes were in opposite directions.
Several of the growth conditions that we tested yielded cells with enlarged capsules, for example, growth in DMEM under CO2 compared to growth in room air or growth in PBS with fetal calf serum as opposed to growth in PBS alone. However, we did not observe any correlation between the expression of either GMT gene and capsule size (not shown). This may be because the diverse conditions sampled here also affect the overall metabolic state of the cell and thus simultaneously alter the transport requirements for GDP-Man needed to synthesize other glycoconjugates.
Intrigued by the distinct expression patterns of two very similar genes, we deleted GMT1 and GMT2. Both deletion strains grow well in culture, but gmt1Δ cells demonstrate reduced capsule and a marked defect in capsule induction (Fig. 6), while gmt2 cells are wild type. This finding links GDP-Man transport and capsule synthesis, supporting our recent conclusion that capsule synthesis occurs inside subcellular organelles (60).
A further puzzle is how Gmt1 and Gmt2 could serve different biological functions while performing the same biochemical activity. It may be that these proteins are differentially localized, perhaps to serve distinct biosynthetic pathways. We know that localization is critical for the appropriate function of these proteins, because the S. cerevisiae protein Hvg1p, a close homolog of Vrg4 that is resident in the endoplasmic reticulum, cannot complement vrg4-2 cells unless it is engineered to reach the Golgi apparatus (18). In contrast, when the cryptococcal transporters are expressed in yeast, both are probably localized to the Golgi apparatus, since they do complement vrg4-2 cells. It is possible that the two transporters occupy different regions of the Golgi apparatus in C. neoformans. A variety of proteins have been localized to subcompartments of the Golgi apparatus in mammalian systems (45), but current techniques do not allow sufficient resolution to examine protein localization within the smaller unstacked Golgi apparatus of Cryptococcus. Another possibility is that the two transporters associate with different synthetic machinery within the cell. In mammalian systems, there is precedent for the specific association of a nucleotide sugar transporter and a glycosyltransferase, which consumes the corresponding transport substrate (53).
Ultimately, distinguishing the biological roles of the proteins encoded by GMT1 and GMT2 must await detailed biochemical and morphological examinations of the two mutant strains to examine the effects of these mutations on other mannose-requiring processes and on cryptococcal virulence. The latter studies will be particularly important, since GDP-Man transporters are present in a variety of pathogens including fungi and parasites. Because the mammalian hosts of these organisms do not have such proteins, GDP-Man transporters have been suggested as potential targets of antimicrobial chemotherapy, a direction that merits further investigation.
Acknowledgments
We thank members of the Doering Laboratory, Steve Beverley, and Sam Turco for stimulating discussions; Neta Dean for generosity in providing strains and plasmids; Michael Brent for assistance with statistics; and Morgann Reilly, Aki Yoneda, Stacey Klutts, and the reviewers for comments that improved the manuscript. We thank Barak Cohen for suggestions on the preparation of biological replicates and the RNA reference pool for microarray experiments and Seth Crosby of the Washington University Microarray Facility for microarray data analysis. C. neoformans microarrays were developed by a community effort with additional support from the Burroughs Wellcome Fund and are available from the Washington University Microarray Facility. See http://genome.wustl.edu/activity/ma/cneoformans/ for details.
This work was supported by undergraduate research awards to T.R.C. from the Howard Hughes Medical Institute and the American Society for Microbiology, support to A.A.N. from NIH T32 GM007067, and support to T.L.D. from NIH R01 GM66303.
Footnotes
Published ahead of print on 9 March 2007.
REFERENCES
- 1.Abe, M., H. Hashimoto, and K. Yoda. 1999. Molecular characterization of Vig4/Vrg4 GDP-mannose transporter of the yeast Saccharomyces cerevisiae. FEBS Lett. 458:309-312. [DOI] [PubMed] [Google Scholar]
- 2.Abe, M., Y. Noda, H. Adachi, and K. Yoda. 2004. Localization of GDP-mannose transporter in the Golgi requires retrieval to the endoplasmic reticulum depending on its cytoplasmic tail and coatomer. J. Cell Sci. 117:5687-5696. [DOI] [PubMed] [Google Scholar]
- 3.Abeijon, C., P. Orlean, P. W. Robbins, and C. B. Hirschberg. 1989. Topography of glycosylation in yeast: characterization of GDP-mannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc. Natl. Acad. Sci. USA 86:6935-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arakawa, K., M. Abe, Y. Noda, H. Adachi, and K. Yoda. 2006. Molecular cloning and characterization of a Pichia pastoris ortholog of the yeast Golgi GDP-mannose transporter gene. J Gen. Appl. Microbiol. 52:137-145. [DOI] [PubMed] [Google Scholar]
- 5.Ashikov, A., F. Routier, J. Fuhlrott, Y. Helmus, M. Wild, R. Gerardy-Schahn, and H. Bakker. 2005. The human solute carrier gene SLC35B4 encodes a bifunctional nucleotide sugar transporter with specificity for UDP-xylose and UDP-N-acetylglucosamine. J. Biol. Chem. 280:27230-27235. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, K. Struhl, L. M. Albright, D. M. Coen, and A. Varki (ed.). 2006. Current protocols in molecular biology. John Wiley & Sons, Inc., Hoboken, NJ.
- 7.Bakker, H., F. Routier, S. Oelmann, W. Jordi, A. Lommen, R. Gerardy-Schahn, and D. Bosch. 2005. Molecular cloning of two Arabidopsis UDP-galactose transporters by complementation of a deficient Chinese hamster ovary cell line. Glycobiology 15:193-201. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin, T. C., M. G. Handford, M. I. Yuseff, A. Orellana, and P. Dupree. 2001. Identification and characterization of GONST1, a Golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13:2283-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berninsone, P., H.-Y. Hwang, I. Zemtseva, H. R. Horvitz, and C. B. Hirschberg. 2001. SQV-7, a protein involved in Caenorhabditis elegans epithelial invagination and early embryogenesis, transports UDP-glucuronic acid, UDP-N-acetylgalactosamine, and UDP-galactose. Proc. Natl. Acad. Sci. USA 98:3738-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabib, E., and L. F. Leloir. 1954. Guanosine diphosphate mannose. J. Biol. Chem. 206:779-790. [PubMed] [Google Scholar]
- 11a.Capul, A. A., T. Barron, D. E. Dobson, S. J. Turco, and S. M. Beverley. 8 March 2007, posting date. Two functionally divergent UDP-gal nucleotide-sugar transporters participate in phosphoglycan synthesis in Leishmania major. J. Biol. Chem. doi: 10.1074/jbc.M610869200. [DOI] [PMC free article] [PubMed]
- 12.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherniak, R., H. Valafar, L. C. Morris, and F. Valafar. 1998. Cryptococcus neoformans chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, N. 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426:309-322. [DOI] [PubMed] [Google Scholar]
- 15.Dean, N., Y. B. Zhang, and J. B. Poster. 1997. The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 272:31908-31914. [DOI] [PubMed] [Google Scholar]
- 16.Dickson, R. C., and R. L. Lester. 1999. Yeast sphingolipids. Biochim. Biophys. Acta 1426:347-357. [DOI] [PubMed] [Google Scholar]
- 17.Gao, X.-D., A. Nishikawa, and N. Dean. 2001. Identification of a conserved motif in the yeast Golgi GDP-mannose transporter required for binding to nucleotide sugar. J. Biol. Chem. 276:4424-4432. [DOI] [PubMed] [Google Scholar]
- 18.Gao, X. D., and N. Dean. 2000. Distinct protein domains of the yeast Golgi GDP-mannose transporter mediate oligomer assembly and export from the endoplasmic reticulum. J. Biol. Chem. 275:17718-17727. [DOI] [PubMed] [Google Scholar]
- 19.Gemmill, T. R., and R. B. Trimble. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426:227-238. [DOI] [PubMed] [Google Scholar]
- 20.Goins, C., K. Gerik, and J. K. Lodge. 2006. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet. Biol. 43:531-534. [DOI] [PubMed] [Google Scholar]
- 21.Guillen, E., C. Abeijon, and C. B. Hirschberg. 1998. Mammalian Golgi apparatus UDP-N-acetylglucosamine transporter: molecular cloning by phenotypic correction of a yeast mutant. Proc. Natl. Acad. Sci. USA 95:7888-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handford, M. G., F. Sicilia, F. Brandizzi, J. H. Chung, and P. Dupree. 2004. Arabidopsis thaliana expresses multiple Golgi-localised nucleotide-sugar transporters related to GONST1. Mol. Genet. Genomics 272:397-410. [DOI] [PubMed] [Google Scholar]
- 23.Helenius, J., and M. Aebi. 2002. Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell Dev. Biol. 13:171-178. [DOI] [PubMed] [Google Scholar]
- 24.Hirschberg, C. B., P. W. Robbins, and C. Abeijon. 1998. Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67:49-69. [DOI] [PubMed] [Google Scholar]
- 25.Hong, K., D. Ma, S. M. Beverley, and S. J. Turco. 2000. The Leishmania GDP-mannose transporter is an autonomous, multi-specific, hexameric complex of LPG2 subunits. Biochemistry 39:2013-2022. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson, E. S., M. J. Tingler, and P. L. Quynn. 1989. Effect of hypertonic solutes upon the polysaccharide capsule in Cryptococcus neoformans. Mycoses 32:14-23. [DOI] [PubMed] [Google Scholar]
- 27.Kabuss, R., A. Ashikov, S. Oelmann, R. Gerardy-Schahn, and H. Bakker. 2005. Endoplasmic reticulum retention of the large splice variant of the UDP-galactose transporter is caused by a dilysine motif. Glycobiology 15:905-911. [DOI] [PubMed] [Google Scholar]
- 28.Klis, F. M., A. Boorsma, and P. W. De Groot. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185-202. [DOI] [PubMed] [Google Scholar]
- 29.Littman, M. L. 1958. Capsule synthesis by Cryptococcus neoformans. Trans. Acad. Sci. 20:623-648. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H., T. R. Cottrell, L. M. Pierini, W. E. Goldman, and T. L. Doering. 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160:463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Losev, E., C. A. Reinke, J. Jellen, D. E. Strongin, B. J. Bevis, and B. S. Glick. 2006. Golgi maturation visualized in living yeast. Nature 441:1002-1006. [DOI] [PubMed] [Google Scholar]
- 33.Ma, D., D. G. Russell, S. M. Beverley, and S. J. Turco. 1997. Golgi GDP-mannose uptake requires Leishmania LPG2. J. Biol. Chem. 272:3799-3805. [PubMed] [Google Scholar]
- 34.Martinez-Duncker, I., R. Mollicone, P. Codogno, and R. Oriol. 2003. The nucleotide-sugar transporter family: a phylogenetic approach. Biochimie 85:245-260. [DOI] [PubMed] [Google Scholar]
- 35.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 36.Munro, S. 2001. What can yeast tell us about N-linked glycosylation in the Golgi apparatus? FEBS Lett. 498:223-227. [DOI] [PubMed] [Google Scholar]
- 37.Muraoka, M., M. Kawakita, and N. Ishida. 2001. Molecular characterization of human UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 495:87-93. [DOI] [PubMed] [Google Scholar]
- 38.Nishikawa, A., B. Mendez, Y. Jigami, and N. Dean. 2002. Identification of a Candida glabrata homologue of the S. cerevisiae VRG4 gene, encoding the Golgi GDP-mannose transporter. Yeast 19:691-698. [DOI] [PubMed] [Google Scholar]
- 39.Nishikawa, A., J. B. Poster, Y. Jigami, and N. Dean. 2002. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J. Bacteriol. 184:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norambuena, L., L. Marchant, P. Berninsone, C. B. Hirschberg, H. Silva, and A. Orellana. 2002. Transport of UDP-galactose in plants. Identification and functional characterization of AtUTr1, an Arabidopsis thaliana UDP-galactose/UDP-glucose transporter. J. Biol. Chem. 277:32923-32929. [DOI] [PubMed] [Google Scholar]
- 41.Nyhus, K. J., and E. S. Jacobson. 1999. Genetic and physiologic characterization of ferric/cupric reductase constitutive mutants of Cryptococcus neoformans. Infect. Immun. 67:2357-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orlean, P. 1990. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:5796-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orlean, P., C. Albright, and P. W. Robbins. 1988. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 263:17499-17507. [PubMed] [Google Scholar]
- 44.Poster, J. B., and N. Dean. 1996. The yeast VRG4 gene is required for normal Golgi functions and defines a new family of related genes. J. Biol. Chem. 271:3837-3845. [DOI] [PubMed] [Google Scholar]
- 45.Puthenveedu, M. A., and A. D. Linstedt. 2005. Subcompartmentalizing the Golgi apparatus. Curr. Opin. Cell Biol. 17:369-375. [DOI] [PubMed] [Google Scholar]
- 46.Reese, A. J., and T. L. Doering. 2003. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50:1401-1409. [DOI] [PubMed] [Google Scholar]
- 47.Reese, A. J., A. Yoneda, J. A. Breger, A. Beauvais, H. Liu, C. L. Griffith, I. Bose, M.-J. Kim, C. Skau, S. Yang, J. A. Sefko, M. Osumi, J. P. Latge, E. Mylonakis, and T. L. Doering. 2007. Loss of cell wall alpha-1,3-glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 63:1385-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyes, F., L. Marchant, L. Norambuena, R. Nilo, H. Silva, and A. Orellana. 2006. AtUTr1, a UDP-glucose/UDP-galactose transporter from Arabidopsis thaliana, is located in the endoplasmic reticulum and up-regulated by the unfolded protein response. J. Biol. Chem. 281:9145-9151. [DOI] [PubMed] [Google Scholar]
- 49.Rivera, J., M. Feldmesser, M. Cammer, and A. Casadevall. 1998. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect. Immun. 66:5027-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segawa, H., M. Kawakita, and N. Ishida. 2002. Human and Drosophila UDP-galactose transporters transport UDP-N-acetylgalactosamine in addition to UDP-galactose. Eur. J. Biochem. 269:128-138. [DOI] [PubMed] [Google Scholar]
- 51.Segawa, H., R. P. Soares, M. Kawakita, S. M. Beverley, and S. J. Turco. 2005. Reconstitution of GDP-mannose transport activity with purified Leishmania LPG2 protein in liposomes. J. Biol. Chem. 280:2028-2035. [DOI] [PubMed] [Google Scholar]
- 52.Sommer, U., H. Liu, and T. L. Doering. 2003. An alpha-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 278:47724-47730. [DOI] [PubMed] [Google Scholar]
- 53.Sprong, H., S. Degroote, T. Nilsson, M. Kawakita, N. Ishida, P. van der Sluijs, and G. van Meer. 2003. Association of the Golgi UDP-galactose transporter with UDP-galactose:ceramide galactosyltransferase allows UDP-galactose import in the endoplasmic reticulum. Mol. Biol. Cell 14:3482-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun-Wada, G. H., S. Yoshioka, N. Ishida, and M. Kawakita. 1998. Functional expression of the human UDP-galactose transporters in the yeast Saccharomyces cerevisiae. J. Biochem. (Tokyo) 123:912-917. [DOI] [PubMed] [Google Scholar]
- 55.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaishnav, V. V., B. E. Bacon, M. O. O'Neill, and R. Cherniak. 1998. Structural characterization of the galactoxylomannan of Cryptococcus neoformans CAP67. Carbohydr. Res. 306:315-330. [DOI] [PubMed] [Google Scholar]
- 57.Varki, A., R. Cummings, J. Esko, H. Freeze, G. Hart, and J. Marth (ed.). 1999. Essentials of glycobiology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 58.Vartivarian, S. E., E. J. Anaissie, R. E. Cowart, H. A. Sprigg, M. J. Tingler, and E. S. Jacobson. 1993. Regulation of cryptococcal polysaccharide by iron. J. Infect. Dis. 167:186-190. [DOI] [PubMed] [Google Scholar]
- 59.Willer, T., M. C. Valero, W. Tanner, J. Cruces, and S. Strahl. 2003. O-Mannosyl glycans: from yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 13:621-630. [DOI] [PubMed] [Google Scholar]
- 60.Yoneda, A., and T. L. Doering. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 17:5131-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshioka, S., G. H. Sun-Wada, N. Ishida, and M. Kawakita. 1997. Expression of the human UDP-galactose transporter in the Golgi membranes of murine Had-1 cells that lack the endogenous transporter. J. Biochem. (Tokyo) 122:691-695. [DOI] [PubMed] [Google Scholar]
- 62.Zaragoza, O., and A. Casadevall. 2004. Experimental modulation of capsule size in Cryptococcus neoformans. Biol. Proced. Online 6:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]