Abstract
Metarhizium anisopliae is a fungus of considerable metabolic and ecological versatility, being a potent insect pathogen that can also colonize plant roots. The mechanistic details of these interactions are unresolved. We provide evidence that M. anisopliae adheres to insects and plants using two different proteins, MAD1 and MAD2, that are differentially induced in insect hemolymph and plant root exudates, respectively, and produce regional localization of adhesive conidial surfaces. Expression of Mad1 in Saccharomyces cerevisiae allowed this yeast to adhere to insect cuticle. Expression of Mad2 caused yeast cells to adhere to a plant surface. Our study demonstrated that as well as allowing adhesion to insects, MAD1 at the surface of M. anisopliae conidia or blastospores is required to orientate the cytoskeleton and stimulate the expression of genes involved in the cell cycle. Consequently, the disruption of Mad1 in M. anisopliae delayed germination, suppressed blastospore formation, and greatly reduced virulence to caterpillars. The disruption of Mad2 blocked the adhesion of M. anisopliae to plant epidermis but had no effects on fungal differentiation and entomopathogenicity. Thus, regulation, localization, and specificity control the functional distinction between Mad1 and Mad2 and enable M. anisopliae cells to adapt their adhesive properties to different habitats.
As judged by species number, the ascomycetes are the most successful fungi, comprising some 75% of known species (30). They include many pathogens of plants and animals, and ascomycetes are also the principal inhabitants of soils containing solid resources such as plant and animal remains. Many economically important pathogenic fungi have the ability to be free living in soil and elsewhere, but there is limited knowledge of the range of ecological niches that they can occupy in the absence of a host (16). For example, the commercially important biocontrol agent Metarhizium anisopliae is a ubiquitous pathogen of insects and a well-established model organism for the study of insect-microbe interactions (2, 26). However, only recently was it reported to colonize the rhizosphere (the layer of soil influenced by root metabolism) and adhere to plant root surfaces (15). M. anisopliae is believed to exert a considerable influence within this ecological niche by repelling and killing soil insects (15).
A major goal of evolutionary and ecological genetics is to identify genes that adapt organisms to their ecological niches and to understand the interaction between their products and the environment. However, genes directly involved in ecological attributes are hard to identify (4). Most forms of fungal adaptation at the molecular level, such as circadian rhythms (20), have focused on the traditional model systems used for genetic analysis such as yeasts and Neurospora crassa. However, many ecologically important lifestyles such as pathogenicity are not represented by these models. Acquiring a detailed understanding of each species in every ecosystem is unrealistic, but M. anisopliae provides a genetically tractable model to address important questions regarding the ecological genetics and molecular mechanisms of two different interspecific interactions, particularly what genes make a fungus an entomopathogen and what genes make a fungus competent to colonize plant roots. In addition, strains of M. anisopliae have an extremely wide distribution within vastly different climates (arctic to true desert) and ecological settings. Hence, many important environmental adaptations made by fungi may be displayed by strains of M. anisopliae. Finally, M. anisopliae produces many different cell types for developmental studies including conidia, hyphae, appressoria (prepenetration swellings produced by many plant and insect pathogens), unicellular blastospores (budding yeast-like form), and multicellular hyphal bodies of various sizes and shapes that differentiate from blastospores in insect hemolymph (24).
Current ecological knowledge of the conditions in which M. anisopliae lives has allowed us to consider gene function in laboratory conditions that model these environments in order to identify genes underlying ecologically relevant traits. Our previous expressed-sequence-tag (EST) analysis identified two very-high-frequency contigs, CN808227 (4.08% of all clones in a cDNA library of M. anisopliae genes expressed in insect hemolymph) and CN809626 (8.9% of clones in a cDNA library of M. anisopliae genes expressed in root exudate), that had unknown functions (31). This study greatly extends our understanding of how M. anisopliae interacts with insects and plants by establishing that CN808227 (designated Mad1 for Metarhizium adhesin-like protein 1) and CN809626 (designated Mad2) are responsible for the capacity to anchor to insect and plant surfaces that enables M. anisopliae to effectively persist and colonize these different environments.
MATERIALS AND METHODS
Gene cloning and knockout.
Full DNA sequences of Mad1 (based on the sequence of CN808227) and Mad2 (CN809626) were obtained by primer walking (DNA Walking Speedup kit; Seegene). Strains lacking either Mad1 or Mad2 were constructed using plasmid pGPS3Bar as described previously (32). The 5′ region of the target gene Mad1 was amplified using primers Mad1-1U (GACTAGTTTCGTTTCCTTCGACTACCATT [position 75]) and Mad1-1L (GACTAGTGTGGTCTCCTTTCCAGGTGTAG [position 1269]). The 3′ region was amplified with Mad1-2U (ATTTGCGGCCGCCCTCTACCGTTTACACGACCTC [position 1410]) and Mad1-2L (ATTTGCGGCCGCAACTGGTTGTTGAGCATGTGTC [position 2678]). The two PCR products were digested with SpeI and NotI enzymes, respectively (underlined), and then inserted into the SpeI and NotI sites of pGPS3Bar using two steps of subcloning to generate the disruption cassette pBarMad1. Similarly, primers Mad2-1U (CCTGGATCCAGGAGAAGCCCGAAGTGAC) and Mad2-1L (CCTGGATCCGGCATGGTAGCAGGAACAGT) and Mad2-2U (ATTTGCGGCCGCCTCGGGTCCCGGTACTGT) and Mad2-2L (ATTTGCGGCCGCGCTTGATGGCAGCAGACC) were used to amplify the 5′ and 3′ regions of Mad2, respectively. The PCR products were digested with BamHI and NotI (underlined) and then inserted into pGPS3Bar to generate pBarMad2. Both constructs were linearized before being used to transform the protoplasts of M. anisopliae. Putative transformants were verified by PCR and reverse transcription (RT)-PCR. At least three verified transformants of Mad1 or Mad2 were used for parallel functional analysis to compare them with the wild type (WT).
Adherence assays.
Suspensions of WT, ΔMad1, and ΔMad2 conidia were prepared in 0.05% Tween 20 at a concentration of 2 × 107 conidia/ml, washed twice with water, and resuspended in water for use. Locust hind wings, fly wings, and epidermis peeled from pieces of onion or celery (1 by 0.5 cm) were sterilized in 37% H2O2 for 5 min, washed twice in water, immersed in spore suspensions for 20 s, and placed on 0.7% water agar. After incubation for 8 h (to induce spore swelling and initiation of germination), the number of conidia in five objective fields was counted under a light microscope before and after washing out the less adherent conidia in 0.05% Tween 20 for 30 s. Adherence by the WT and both mutants on the different materials was estimated using the average number of conidia per objective field after washing compared to the average count before washing (always >300 conidia per treatment). Experiments were done in triplicate, and each experiment was repeated at least twice.
Yeast transformation and adherence assays.
To confirm the binding properties of MAD1 and MAD2, the full cDNAs of Mad1 and Mad2 were amplified from corresponding EST clones (31). PCR was conducted using Pfu Ultra polymerase (Stratagene) and primers Mad1U (CCGGGATCCCATTGCTGTCTTCGTCGTCT) and Mad1L (ATTTGCGGCCGCCATACCCGCTTAGCACAACA) and Mad2U (CCGGGATCCTCCAACCGCTCTCACCTTAT) and Mad2L (ATTTGCGGCCGCATATGCTGTGCGGTCAACAC) by introducing BamHI and NotI restriction sites (underlined). The digested product was inserted into BamHI/NotI-digested pYes2 (Invitrogen) under the control of a Gal1 promoter to generate pYes2Mad1 and pYes2Mad2, respectively. Yeast transformation (using Saccharomyces cerevisiae strain INVSc1 [Invitrogen]) and protein induction were performed according to the manufacturer's instructions. Briefly, after 6 h of incubation in galactose medium, potentially adherent yeast clones were screened by placing locust hind wings and peeled onion skin epidermis (see above) into the induction medium and incubating them for up to 10 h on a gyratory shaker (150 rpm). The control experiments were conducted by incubating wings or onion skins with untransformed yeasts or yeast cells transformed with unmodified pYes2 plasmid. After incubation, the substrates were washed for 30 s in 0.05% Tween 20 to assay for cell binding abilities as described above.
Insect bioassays.
Virulence was assayed using conidia of the WT and mutant strains against newly emerged fifth-instar Manduca sexta larvae (28). Conidia were applied topically by immersing larvae for 20 s in an aqueous suspension containing 2 ×107 conidia/ml. Each treatment had three replicates with 10 insects each, and the experiments were repeated twice. Mortality was recorded every 12 h. Additional infected insects were bled for microscopic observation of fungal development within the insect hemocoel.
RT-PCR.
To determine the inductive effects of different growth conditions, 36-h Sabouraud dextrose broth (SDB) (Difco) cultures were harvested and washed, and equal amounts (0.2 g [wet weight]) of mycelia were inoculated into 10 ml of minimal medium (10 g/liter glucose, 6 g/liter NaNO3, 0.52 g/liter KCl, 0.52 g/liter MgSO4·7H2O, 0.25 g/liter KH2PO4), SDB, sterile water, 0.1% bean root exudate, 1% locust cuticle, or M. sexta hemolymph for 6 h. The root exudates, locust cuticle, and M. sexta hemolymph were prepared as described previously (31). The RNA was extracted using a QIAGEN RNeasy Plant Mini kit and treated with DNase I, and 1 μg was converted into single-stranded cDNA using an anchored oligo(dT) primer (ABgene).
To monitor the influence of deleting Mad1 or Mad2 on gene expression, primers for various genes were designed (data not shown) and used for RT-PCR analyses of WT and mutants cultured in hemolymph or root exudate for 6 h.
IIF assay.
Predicted antigenic sequences from the N-terminal domains of MAD1 (CESNFGKRGDIQGR [85 to 98 amino acids {aa}]) and MAD2 (CDDNDNDEWHYVHP [80 to 93 aa]) were synthesized commercially and used for raising antibodies in New England white rabbits (GeneScript). To localize MAD proteins, WT conidia harvested from 20-day-old potato dextrose agar (Difco) plates were grown in SDB or 1% bean root exudates for 0 to 12 h, fixed in 3.7% formaldehyde, and used for indirect immunofluorescence (IIF) analysis using previously described protocols (32). Control samples were incubated without either the primary or secondary antibody. To explore the potential interactions between MAD1 and septin(s), an antibody against Aspergillus nidulans septin B (AspB) (33) was used to localize septin distribution on WT and mutant hyphal bodies.
Fluorescent staining.
Cell actin distribution was visualized with fluorescein isothiocyanate (FITC)-labeled phalloidin (Sigma) at a final concentration of 0.05 mg/ml in phosphate-buffered saline plus 1% dimethyl sulfoxide. Cell walls and nuclei were stained with calcofluor white and DAPI (4′,6′-diamidino-2-phenylindole) (Sigma), respectively.
Nucleotide sequence accession numbers.
Data reported here have been deposited in the GenBank database under the following accession numbers: DQ338437 for Mad1 mRNA, DQ338438 for Mad2 mRNA, and DQ338439 for genomic DNA.
RESULTS
Molecular characteristics of MAD1 and MAD2.
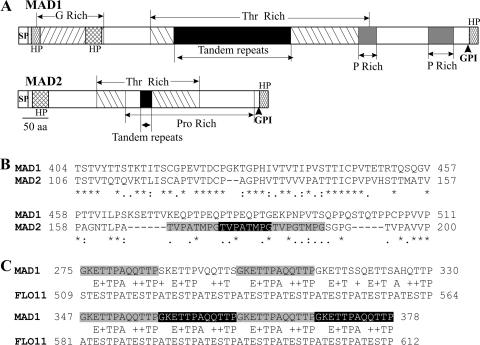
The full open reading frame of Mad1 cDNA encodes a protein of 717 aa (74.6 kDa, with a predicted pI of 6.1). MAD2 comprises 306 aa (30.5 kDa, with a pI of 6.0). They share a three-domain structure similar to that of the Candida albicans cell wall ALS (agglutinin-like sequence) proteins (14, 27) that form rapid and extremely stable H-bond-dependent associations with host proteins and peptides (9). Consistent with their being cell wall proteins, both MAD1 and MAD2 possess a hydrophobic signal peptide and a predicted glycosylphosphatidylinositol cell wall anchor site (5) at their N- and C-terminal ends, respectively. Also, similar to ALS adhesins (14, 29) (Fig. 1A), the MAD1 and MAD2 N-terminal regions (domain A) downstream of the signal peptides are predicted to be highly hydrophobic, and the middle regions (domain B) contain Thr-rich tandem repeats. Sequence similarities between MAD1 and MAD2 exist largely within this middle region (Fig. 1B). In MAD1, it contains six tandem repeats comprised of 12 residues, GKETTPAQQTTP, while in MAD2, it has three repeats of TVPATMPG (Fig. 1B and C). The tandem repeat region of MAD1 also shows limited sequence similarities to C. albicans FLO11 (Fig. 1C and data not shown), which is required for yeast pseudohypha formation (21). The working model for ALS proteins is that the tandem repeats are heavily glycosylated to produce a rigid elongated structure that holds the adhesive N-terminal domain at the cell surface (14, 25). Presumably, the smaller number of repeats in MAD2 means that the distance between the cell surface and the N-terminal ligand binding region will be shorter than in MAD1.
FIG. 1.
Structural features of MAD1 and MAD2. (A) Schematic structure of MAD1 and MAD2 showing glycine-, threonine-, and proline-rich regions. SP, signal peptide; GPI, glycosylphosphatidylinositol anchor site; HP, hydrophobic region. (B) ClustalX alignment of the conserved region in MAD1 and MAD2 (* indicates consensus sites). The MAD2 tandem repeats are shadowed. (C) Conservation between tandem repeat regions (shadowed) in MAD1 and C. albicans FLO11.
Heterologous expression of M. anisopliae genes in a nonadherent strain of S. cerevisiae.
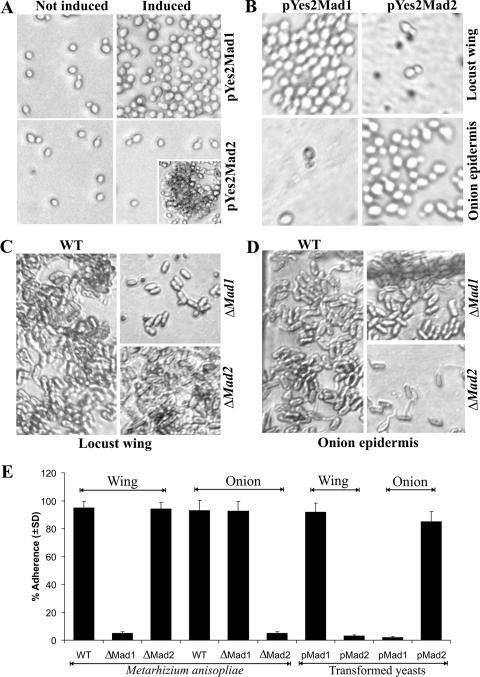
Mad1 and Mad2 genes were expressed individually in yeast cells to determine if they conferred adhesive properties. Cells were transformed with plasmid pYes2 containing either Mad1 or Mad2 cDNA under the control of the tightly regulated Gal1 promoter. After 10 h of induction with 2% galactose, the cells expressing MAD1 adhered to plastic so that they did not wash off. In contrast, the MAD2-expressing cells adhered only to each other (a process called flocculation in yeasts) (Fig. 2A). In contrast to Candida adhesin INT1 (1), heterologous expressions of MAD1 or MAD2 in S. cerevisiae did not result in the yeast producing filamentous growth.
FIG. 2.
Adherence assays. (A) Binding of yeast cells to plastic in spite of washing with 0.05% Tween 20. Yeast cells were transformed with pYes2Mad1 or pYes2Mad2 and incubated for 10 h in 2% raffinose (noninducing) or galactose (inducing) medium. In contrast to noninduced yeast cells, cells expressing MAD1 (induced) became adherent to the plastic surface so that they did not wash off. In contrast, MAD2-expressing cells (induced) were readily washed off plates but adhered to each other in the absence of washing (inset). (B) Binding of yeast cells to locust wing cuticle and onion epidermis. Yeast cells expressing MAD1, but not MAD2, adhere to locust cuticle, while only yeast cells expressing MAD2 adhere to onion epidermis. (C) Binding of M. anisopliae conidia incubated with locust wing cuticle for 8 h. M. anisopliae WT and ΔMad2 conidia adhere to locust cuticle, but deleting Mad1 results in a loss of adherence. (D) Binding of M. anisopliae conidia incubated with onion epidermis for 8 h. Deletion of Mad2 but not Mad1 leads to the loss of conidial adherence to onion skin. (E) Bar graph quantifying adherence of M. anisopliae conidia and yeast cells to locust wing cuticle and onion epidermis. Values represent means ± standard deviations.
Given that Mad1 and Mad2 are highly expressed in insect hemolymph and bean root exudates, respectively (31), and M. anisopliae is an insect pathogen that also adheres to plant surfaces (15), additional adherence assays were conducted using locust wings and epidermal tissue peeled from onions. Yeast cells expressing MAD1 adhered to locust cuticle (92.2% ± 6.1% adherence) but not to onion epidermis, while the cells expressing MAD2 adhered to onion epidermis (85.5% ± 7.3%) but not to locust cuticle (Fig. 2B and E). Untransformed yeast cells and cells transformed with unmodified pYes2 were nonadherent to either wing or onion surfaces.
Disruption of Mad1 and Mad2 blocks adhesion of M. anisopliae to insect and plant surfaces.
To determine the contributions of MAD1 and MAD2 to conidial adherence, we used homologous replacement to obtain mutants of M. anisopliae that were disrupted in each gene. Adhesion assays showed that >90% of the parental WT conidia adhered to either locust cuticle or onion epidermis and could not be washed off with 0.05% Tween 20. Only 5.3% ± 1.2% of the ΔMad1 conidia adhered to locust cuticle, but ΔMad2 conidia were as adherent as the WT (Fig. 2C and E). Conversely, ΔMad2 conidia showed very little adherence (4.8% ± 1.3%) to onion epidermis, while ΔMad1 conidia were as adherent as the WT (Fig. 2D and E). Very similar results were obtained when locust and onion surfaces were replaced with fly wings and celery epidermis (data not shown). Thus, MAD1 and MAD2 have unique (i.e., nonsubstitutable) functions and are principally responsible for the ability of M. anisopliae to adhere to insect hosts and plant surfaces, respectively.
Expression and localization of MAD1 and MAD2.
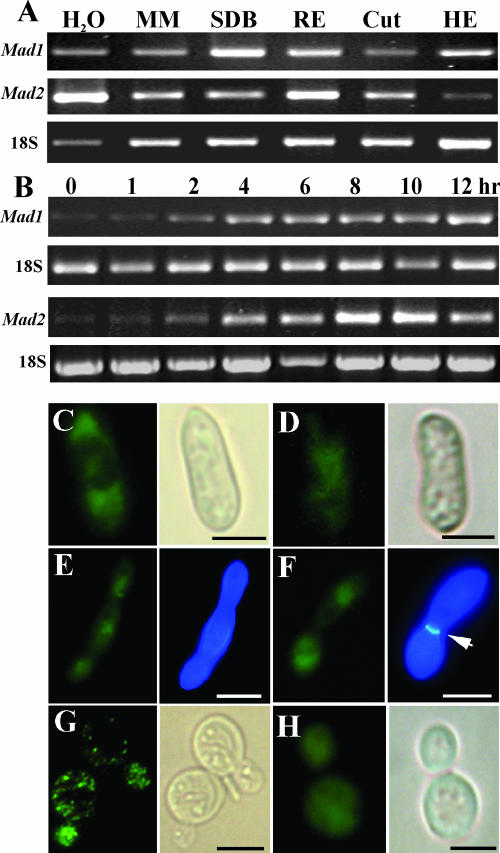
RT-PCR analyses were performed to measure the expression of Mad1 and Mad2 in different media. Consistent with previous EST and microarray analyses (31), Mad1 and Mad2 were expressed at various levels in all tested media. However, the Mad1 gene was up-regulated in nutrient-rich media such as SDB and cell-free insect (M. sexta) hemolymph, while the Mad2 gene was up-regulated in bean root exudates and water (Fig. 3A). Time course studies showed that Mad1 and Mad2 transcripts are present at low levels in freshly collected conidia from 20-day-old potato dextrose agar plates but that transcripts started to accumulate within 2 h of incubation in SDB or root exudates (Fig. 3B).
FIG. 3.
Expression and localization of MAD1 and MAD2. (A) RT-PCR analysis of Mad1 and Mad2 expression by WT M. anisopliae transferred from SDB cultures to water, minimum medium (MM), 1% bean root exudate (RE), 1% Manduca larval cuticle (Cut), or cell-free hemolymph (HE) for 6 h. (B) RT-PCR time course analysis of Mad1 and Mad2 expression. Conidia were incubated in SDB (for Mad1 induction) or bean root exudate (for Mad2 induction) for up to 12 h as indicated. (C) IIF with anti-MAD1 localizing MAD1 at the poles of a conidium incubated in SDB for 8 h. (D) IIF with anti-MAD2 of a conidium incubated in bean root exudates (8 h) demonstrating that MAD2 is more centrally localized than MAD1. (E and F) IIF with anti-MAD1 demonstrating MAD1 production on unicellular (E) and multicellular (F) hyphal bodies harvested from infected M. sexta caterpillars. The panels on the right of E and F show the same cells stained with calcofluor white. The arrow points to a septum. (G and H) MAD1 and MAD2 production by transgenic yeast incubated with galactose for 10 h demonstrating a patchy distribution of MAD1 (G) and an even distribution of MAD2 (H) on yeast cells. The panels on the right of C, D, G, and H show bright-field microscopy of the same cells. Scale bar, 5 μm.
For MAD1 and MAD2 to function as adhesins, they must be located on the cell surface. This was verified by using an IIF assay with antibodies raised against predicted antigenic amino acid sequences from the N-terminal regions of MAD1 (85 to 98 aa) or MAD2 (80 to 93 aa). Neither MAD1 nor MAD2 could be detected on freshly collected conidia, suggesting an absence of surface antigens, as antibodies do not penetrate the cell (32). Before germination, conidia absorb water and swell. MAD1 was not detected in spores that had not swelled, irrespective of the time point. However, coincident with swelling (6 to 8 h for most spores in SDB), MAD1 localized at the ends of the bar-shaped M. anisopliae conidia (Fig. 3C). One hundred percent of conidia germinating in SDB or successfully adhering to plastic or cuticle surfaces showed polar localization of MAD1 (Fig. 3C). Spores germinated in minimum medium or root exudates take longer (>12 h) to produce detectable MAD1, but it was similarly localized. In contrast, MAD2 was preferentially localized in the middle of conidia when germinated in bean root exudates for 8 h (Fig. 3D). Presumably, this asymmetry results in the regional localization of the adhesive surfaces related to the roles played by MAD1 and MAD2. The distribution patterns of MAD1 in ΔMad2 mutant cells or MAD2 in ΔMad1 cells were identical to those of WT cells. No immunofluorescence was observed when the ΔMad1 and ΔMad2 mutants were tested with antibodies to MAD1 and MAD2, respectively.
We also investigated the localization of adhesins on hyphal bodies harvested from insects infected with WT M. anisopliae. Consistent with RT-PCR analysis (Fig. 3A), MAD1 but not MAD2 was detected on the surfaces of hyphal bodies. MAD1 was produced irrespective of whether the hyphal bodies were unicellular (blastospores) or multicellular (Fig. 3E and F).
A punctuate distribution of MAD1 was detected on the surface of yeast cells expressing MAD1 (Fig. 3G), suggesting that MAD1 was heterogeneously and randomly distributed within the cell wall structure, while MAD2 expressed by yeast cells was distributed more uniformly on the surface of cells (Fig. 3H).
Linkage of adhesion, germination, blastospore formation, and virulence of M. anisopliae to MAD1.
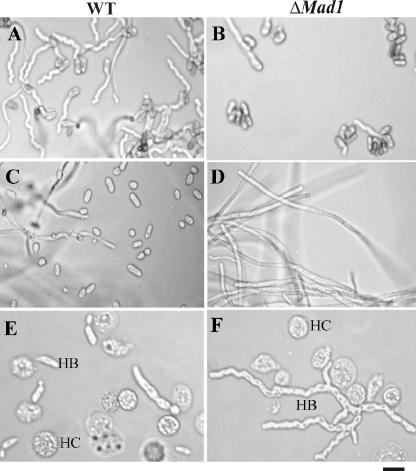
In addition to adhesion, MAD1, but not MAD2, affects conidial germination, blastospore formation, and in insecta hyphal body differentiation (Fig. 4). Thus, 14 h postinoculation in SDB, 78.7% ± 2.3% of WT conidia and only 18.9% ± 3.6% of ΔMad1 conidia had germinated (Fig. 4A and B). During 3 days of culturing in isolated M. sexta hemolymphs, the WT produced large numbers of blastospores (Fig. 4C), while the ΔMad1 mutant grew as long hyphae and produced very few blastospores (Fig. 4D). Likewise, the WT strain infecting fifth-instar M. sexta larvae produced short, variably shaped hyphal bodies composed of one to three cells (Fig. 4E). In contrast, 73.5% ± 4.32% of ΔMad1 hyphal bodies were long (typically composed of more than five cells) and branched (Fig. 4F). However, the number of WT hyphal bodies in hemolymph (3.45 × 106 ± 1.5 × 106 cell per ml) was >20-fold greater than the number produced by ΔMad1 (1.5 × 105 ± 1.1 × 105 cell per ml) (n = 20 insects). Similar results were obtained using independently acquired ΔMad1 mutants (data not shown).
FIG. 4.
Deletion of Mad1 affects conidial germination, blastospore formation, and hyphal body differentiation. (A and B) Conidial germination. Fourteen hours postinoculation in SDB medium, germination levels were higher for the WT (A) than for ΔMad1 (B). (C and D) Blastospore formation. When cultured for 3 days in cell-free insect hemolymph, the WT budded off blastospores (C), while ΔMad1 grew as hyphae (D). (E and F) Hyphal body differentiation. M. sexta larvae were injected with conidia and bled at 10-h intervals. At 50 h, the WT (E) had formed variably shaped hyphal bodies with one to three cells, but the ΔMad1 mutant (F) had produced long and branched chains of cells (more than five cells). HB, hyphal body; HC, insect hemocytes. Scale bar, 10 μm.
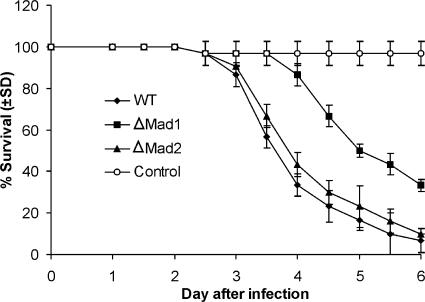
To test whether deleting MAD1 or MAD2 influences fungal virulence, we bioassayed newly emerged fifth-instar M. sexta caterpillars (Fig. 5). The LT50 values (time needed for the pathogen to kill 50% of caterpillars) showed that the ΔMad1 mutant takes a significantly longer time (5.35 ± 0.22 days) to kill insects than the WT does (3.92 ± 0.36 days) (t = 24.76; P = 0.00081). In contrast, the difference between ΔMad2 (4.11 ± 0.18 days) and the WT (3.92 ± 0.36 days) is not significant (t = 2.92; P = 0.12), showing that only MAD1 is a virulence factor.
FIG. 5.
Kinetics of insect survivorship in bioassays. Shown is the survival of Manduca larvae following topical application with suspensions of 2 × 107 conidia/ml of WT, ΔMad1, or ΔMad2 strains (control insects were dipped in water). LT50 values were 3.92 ± 0.36 days for the WT, 5.35 ± 0.22 days for ΔMad1, and 4.11 ± 0.18 days for ΔMad2. The difference between the WT and ΔMad1 is significant (t = 24.76; P = 0.00081). The difference between the WT and ΔMad2 is not significant (t = 2.92; P = 0.12).
MAD1 is required for normal cytoskeleton organization and cell division.
Since we showed that disrupting the Mad1 gene delays spore germination and results in large multicellular hyphal bodies (Fig. 4), experiments were performed to further explore the effects of MAD1 on the cell cycle. The initiation of spore germination in fungi is followed by actin polarization (alignment of actin to the poles of the conidia) and mitosis (18). Six hours postinoculation in SDB, actin polymerization is evident in both WT (Fig. 6A) and ΔMad1 conidia (Fig. 6B). However, it took 10 h for 90% of ΔMad1 spores to have polarized actin compared to 6 h for WT spores. The polymerized actin in ΔMad1 conidia was scattered in clumps throughout the cytoplasm 6 h postinoculation (Fig. 6B). These results suggest that ΔMad1 interferes with the organization of the cytoskeleton that is not due to a failure in assembly but is rather due to deficient interactions with proteins that regulate its distribution. This could be a direct interaction between MAD1 and actin or indirect via a regulator such as septin.
FIG. 6.
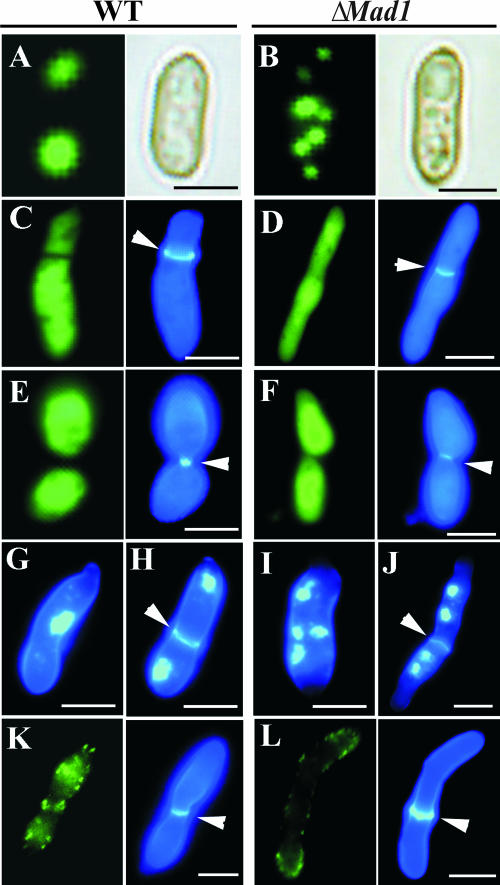
MAD1 is involved in cytoskeletal organization and cell division. (A) FITC-phalloidin fluorescent staining of a WT conidium incubated in SDB for 6 h showed polymerized actin aligning at the poles (the panel on the right shows bright-field microscopy of the same cell). (B) Actin in conidia of ΔMad1 also polymerizes but is patchily distributed in the cytoplasm. (C to F) Double-staining hyphal bodies with FITC-phalloidin (the panels on the left of C to E) and calcofluor (the panels on the right of C to E). Note that where septa form, the actin cytoskeleton is more clearly delineated between daughter cells in WT M. anisopliae (C and E) than in the ΔMad1 mutant (D and F). (G and H) Double-staining cells of WT hyphal bodies harvested from infected insects with DAPI and calcofluor white showed that they are uninucleate whether comprised of one (G) or more (H) cells. In contrast, ΔMad1 hyphal bodies are usually multinucleate, even when septa are formed (I and J). (K and L) Immunofluorescent staining of hyphal bodies with an Aspergillus anti-septin B antibody shows the formation of a septin ring at the septum of a WT cell (K), which is lacking in ΔMad1 cells (L) (the panels on the right show the same cells stained with calcofluor). The septa are indicated by arrows. Scale bar, 5 μm.
Septins play crucial roles in cell division/compartmentalization (3). For example, budding yeasts show a narrow mother-bud neck (the “septin hourglass” shape). Mutants deficient in septins have very wide necks, providing a rapid diagnostic for septin defects (10). WT M. anisopliae budding hyphal bodies in hemolymph exhibit an hourglass shape almost indistinguishable from that of yeasts during septum formation (Fig. 6E). However, <10% of dividing ΔMad1 cells form an hourglass shape (Fig. 6F). Even where septa had formed, the actin cytoskeleton was usually less clearly delineated between daughter cells in ΔMad1 than in the WT (Fig. 6C to F). Consistent with MAD1 being involved in the separation of cells after mitosis, 85.3% ± 9.8% of ΔMad1 hyphal bodies were multinucleate, whereas WT cells were invariably uninucleate (Fig. 6G to J). These results suggest that MAD1 is involved in achieving the “septin hourglass” shape and subsequent separation of cells.
The M. anisopliae EST clone with GenBank accession number AJ274373 is similar (E = 3 × 10−34) to Aspergillus nidulans septin B (AspB). It is more-than-twofold up-regulated by WT cells in hemolymph (31). To label this septin, we performed IIF assays using a polyclonal antibody against AspB (33). Fluorescent signals detected using the antibody demonstrated that septin rings are formed at the septa of WT but not ΔMad1 hyphal bodies (Fig. 6K and L), confirming a role for MAD1 in septin organization. The dividing ΔMad1 cell shown in Fig. 6L has an unusually well-staining septum for this mutation, suggesting that septin B is a contributing factor but not a prerequisite for differentiation of the cell wall. The involvement of MAD1 in cytoskeletal organization and cell division was confirmed by testing additional MAD1 mutants (data not shown).
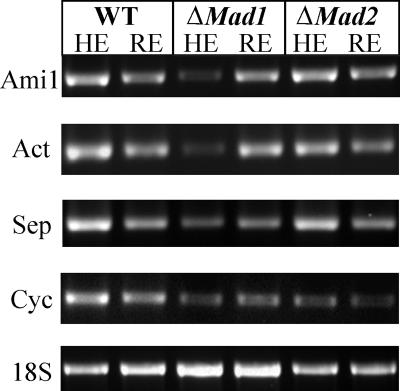
To test whether MAD1 affects gene expression, we performed RT-PCR analyses of selected genes involved in cell division (Fig. 7). Deletion of Mad1, but not Mad2, down-regulated conserved Metarhizium homologs of an actin filament organization factor (CN809636) as well as septin B (GenBank accession number AJ274373) (Fig. 7). Interestingly, although microscopy studies did not implicate MAD1 as being important for nuclear division, Ami1 (accession number AJ274250), involved in nuclear migration, and a G2/mitosis-specific cyclin B (CN808863) were also down-regulated. However, diverse genes involved in carbohydrate metabolism (e.g., glucosidases) (accession number AJ272748 and CN807981), proteolysis (e.g., subtilisin Pr1A [CN808958] and ubiquitin-specific proteinase [accession number AJ273678]), and the synthesis of secondary toxic metabolites (e.g., phenazine biosynthesis-like protein [accession number AJ273180] and polyketide synthase [accession number AJ273296]) did not show transcriptional differences between WT and MAD deletion mutants (data not shown).
FIG. 7.
RT-PCR analysis of selected genes involved in cell division. Thirty-six-hour SDB cultures of the WT, ΔMad1, or ΔMad2 were transferred into M. sexta hemolymph (HE) or 1% (wt/vol) root exudate (RE) for 6 h. The mycelia were harvested for RNA extraction, and 1 μg RNA was converted into cDNA for RT-PCR analysis. The selected genes include Metarhizium homologs of Ami1 (GenBank accession number AJ274250) involved in nuclear migration, an actin filament organization factor (Act) (CN809636), septin B (Sep) (accession number AJ274373), and cyclin B (Cyc) (CN808863).
DISCUSSION
Hundreds of fungal species are pathogenic to insects or can colonize plant roots. The ubiquitous soil fungus M. anisopliae, however, is remarkably versatile in being able to do both. This suggests that it must have evolved mechanisms to adhere to a variety of biological surfaces to initiate and maintain pathogenic and mutualistic interactions. Our study demonstrates that MAD1 and MAD2 are chiefly responsible for conidia of M. anisopliae adhering to insect and plant surfaces, respectively. Disruption of these genes produced approximately 90% reduction in adherence, suggesting that M. anisopliae conidia possess little if any redundancy of adhesion molecules for the ligands present on plant and insect surfaces. We also constructed S. cerevisiae strains that express individual MAD proteins to determine if that confers the ability to adhere to insect or plant surfaces. The same method of heterologous expression has been used to characterize adhesins of Candida spp. (8) and has the advantage of guarding against any effect of additional adhesins in M. anisopliae on adherence. Transformation of Saccharomyces with Mad1 produced a strain capable of adhering to a plastic surface as well as insect cuticle. By contrast, the expression of Mad2 causes yeast cells on plastic to aggregate (flocculation) due to cell-cell adhesion. Thus, although Mad2 has little sequence similarity with the Saccharomyces flocculin gene family, it resembles them in promoting cell-cell adhesion.
It is not surprising that the deletion of Mad1 significantly reduced the virulence of the mutant strain, as it contributes to many of the characteristics that are associated with the pathogenicity of M. anisopliae. Attachment and adherence to the host surface are the key initial steps for colonization. Passive hydrophobic interactions mediated by the hydrophobins are responsible for the initial adherence of conidia to insect cuticle (13). As the spores swell preceding germination, the hydrophobin layer disintegrates, and new cell wall materials are laid down (34). This is coincident with the appearance of MAD1, suggesting that its role in adhesion is to replace the hydrophobins with tighter, more specific interactions. However, disrupting Mad1, but not Mad2, produced phenotypes that extended beyond adhesion to changes in germination and morphology. The formation of small hyphal bodies is a virulence strategy that facilitates the rapid multiplication and dispersal of infectious propagules within the insect body (17, 32). It is likely, therefore, that the reduced number of large and therefore presumably less mobile hyphal bodies produced by ΔMad1 would contribute to diminished virulence.
Like MAD1, Candida adhesins Int1 (8) and Als1 (6) and S. cerevisiae Flo11 (12) have also been reported to be involved in both adhesion and morphogenesis. Als1 and Flo11 may promote filamentation by mediating the effects of transcription factors (6, 12). However, the intermediate components involved in these pathways have not been determined. In this study, we demonstrated that knocking out MAD1 down-regulated various genes, including a septin, involved in regulating the cytoskeleton and the cell cycle. A simple hypothesis to account for our findings would be that adhesion itself triggers gene expression. However, these changes were observed in hyphal bodies freely circulating in the hemolymph, suggesting that adhesins have more effects than previously realized. Consistent with the transcriptional changes, MAD1 influences the organization of the cytoskeleton. A delay in actin polarization correlates with the delay in ΔMad1 spore germination. Likewise, dysfunctions of actin and septin in ΔMad1 hyphal bodies could cause failures in cell division/compartmentalization. It is known that fungal cell compartmentalization is septin dependent (22), and interactions between actin and septins occur in yeast (23) and A. nidulans (33). In yeasts, septins function in bud site selection, recruitment of proteins involved in budding, proper cell wall disposition, and cytokinesis (10, 19). The intracellular domain of C. albicans adhesin Int1 is believed to interact directly with septins Cdc11 and Cdc12 (7, 11). Colocalization of MAD1 and septin B was not observed in WT hyphal bodies in this study. Because MAD1 is localized outside the cell, it follows that the link with morphogenesis is likely to involve transmembrane signaling/interactions via its C-terminal tail, although we have not proven that this is the case.
The involvement of adhesin MAD1 in cytokinesis clearly merits further investigations. Attachment to insect surfaces and subsequent infection processes involve a variety of cellular structures and complicated patterns of gene expression and likely therefore require several different signaling pathways to accomplish them. The Mad genes themselves are differentially regulated by physiologically relevant conditions such as growth medium changes, morphological form, and stage of growth. Thus, Mad1 transcripts are induced during spore swelling and during growth in hemolymphs, two key stages of host invasion. There is overlap in regulation, as Mad1 and Mad2 were produced at various levels in all media tested, albeit sometimes at low levels. Further studies are needed to determine the effects of the dosage of MAD proteins for the full expression of the adhesive phenotype. It is possible that low-level constitutive production of MAD proteins may provide sensors that trigger decision processes in response to different subsets of environmental conditions such as insect and plant ligands. It will be interesting, therefore, to determine how Mad1 and Mad2 regulate, or are regulated by, other pathways involved in ecological adaptation. The results of this study could also be relevant to innovating new methods in agriculture. For example, the ability of MAD1 and MAD2 to cause yeast cells to differentially adhere to insect or plant surfaces suggests they could be used to target otherwise nonadherent pathogenic or symbiotic organisms to beneficial insects or pests.
Acknowledgments
This work was supported by NSF grant MCB-0542904.
We also highly appreciate Michelle Momany's gift of the septin antibody.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Asleson, C. M., E. S. Bensen, C. A. Gale, A. S. Melms, C. Kurischko, and J. Berman. 2001. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol. Cell. Biol. 21:1272-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanford, S., B. H. Chan, N. Jenkins, D. Sim, R. J. Turner, A. F. Read, and M. B. Thomas. 2005. Fungal pathogen reduces potential for malaria transmission. Science 308:1638-1641. [DOI] [PubMed] [Google Scholar]
- 3.Douglas, L. M., F. J. Alvarez, C. McCreary, and J. B. Konopka. 2005. Septin function in yeast model systems and pathogenic fungi. Eukaryot. Cell 4:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duda, T. F., Jr., and S. R. Palumb. 1999. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. USA 96:6820-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenhaber, B., G. Schneider, M. Wildpaner, and F. Eisenhaber. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. J. Mol. Biol. 337:243-253. [DOI] [PubMed] [Google Scholar]
- 6.Fu, Y., A. S. Ibrahim, D. C. Sheppard, Y. C. Chen, S. W. French, J. E. Cutler, S. G. Filler, and J. E. Edwards, Jr. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61-72. [DOI] [PubMed] [Google Scholar]
- 7.Gale, C., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gale, C. A., C. M. Bendel, M. McClellan, M. Hauser, J. M. Becker, J. Berman, and M. K. Hostetter. 1998. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science 279:1355-1358. [DOI] [PubMed] [Google Scholar]
- 9.Gaur, N. K., and S. A. Klotz. 2004. Accessibility of the peptide backbone of protein ligands is a key specificity determinant in Candida albicans SRS adherence. Microbiology 150:277-284. [DOI] [PubMed] [Google Scholar]
- 10.Gladfelter, A. S., L. Kozubowski, T. R. Zyla, and D. J. Lew. 2005. Interplay between septin organization, cell cycle and cell shape in yeast. J. Cell Sci. 118:1617-1628. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Novo, A., L. Labrador, A. Jimenez, M. Sanchez-Perez, and J. Jimenez. 2006. Role of the septin Cdc10 in the virulence of Candida albicans. Microbiol. Immunol. 50:499-511. [DOI] [PubMed] [Google Scholar]
- 12.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holder, D. J., and N. O. Keyhani. 2005. Adhesion of the entomopathogenic fungus Beauveria (Cordyceps) bassiana to substrata. Appl. Environ. Microbiol. 71:5260-52066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176-180. [DOI] [PubMed] [Google Scholar]
- 15.Hu, G., and R. J. St. Leger. 2002. Field studies using a recombinant mycoinsecticide (Metarhizium anisopliae) reveal that it is rhizosphere competent. Appl. Environ. Microbiol. 68:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerry, B. R. 2000. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant-parasitic nematodes. Annu. Rev. Phytopathol. 38:423-441. [DOI] [PubMed] [Google Scholar]
- 17.Kershaw, M. J., E. R. Moorhouse, R. Bateman, S. E. Reynolds, and A. K. Charnley. 1999. The role of destruxins in the pathogenicity of Metarhizium anisopliae for three species of insect. J. Invertebr. Pathol. 74:213-223. [DOI] [PubMed] [Google Scholar]
- 18.Kono, K., R. Matsunaga, A. Hirata, G. Suzuki, M. Abe, and Y. Ohya. 2005. Involvement of actin and polarisome in morphological change during spore germination of Saccharomyces cerevisiae. Yeast 22:129-139. [DOI] [PubMed] [Google Scholar]
- 19.Kozubowski, L., J. R. Larson, and K. Tatchell. 2005. Role of the septin ring in the asymmetric localization of proteins at the mother-bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell 16:3455-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakin-Thomas, P. L., and S. Brody. 2004. Circadian rhythms in microorganisms: new complexities. Annu. Rev. Microbiol. 558:489-519. [DOI] [PubMed] [Google Scholar]
- 21.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luedeke, C., S. B. Frei, I. Sbalzarini, H. Schwarz, A. Spang, and Y. Barral. 2005. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 169:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norden, C., D. Liakopoulos, and Y. Barral. 2004. Dissection of septin actin interactions using actin overexpression in Saccharomyces cerevisiae. Mol. Microbiol. 53:469-483. [DOI] [PubMed] [Google Scholar]
- 24.Prasertphon, S., and Y. Tanada. 1968. The formation and circulation, in Galleria, of hyphal bodies of entomophthoraceous fungi. J. Invertebr. Pathol. 11:260-280. [Google Scholar]
- 25.Rauceo, J. M., R. de Armond, H. Otoo, P. C. Kahn, S. A. Klotz, N. K. Gaur, and P. N. Lipke. 2006. Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot. Cell 5:1664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts, D. W., and R. J. St. Leger. 2004. Metarhizium spp., cosmopolitan insect-pathogenic fungi: mycological aspects. Adv. Appl. Microbiol. 54:1-70. [DOI] [PubMed] [Google Scholar]
- 27.Sheppard, D. C., M. R. Yeaman, W. H. Welch, Q. T. Phan, Y. Fu, A. S. Ibrahim, S. G. Filler, M. Zhang, A. J. Waring, and J. E. Edwards, Jr. 2004. Functional and structural diversity in the Als protein family of Candida albicans. J. Biol. Chem. 279:30480-30489. [DOI] [PubMed] [Google Scholar]
- 28.St. Leger, R. J., L. Joshi, M. J. Bidochka, and D. W. Roberts. 1996. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. USA 93:6349-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, J. W. 1995. Molecular phylogenetic classification of fungi. Arch. Med. Res. 6:307-314. [PubMed] [Google Scholar]
- 31.Wang, C., G. Hu, and R. J. St. Leger. 2005. Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manduca sexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Genet. Biol. 42:704-718. [DOI] [PubMed] [Google Scholar]
- 32.Wang, C., and R. J. St. Leger. 2006. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc. Natl. Acad. Sci. USA 103:6647-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westfall, P. J., and M. Momany. 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wosten, H. A. 2001. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55:625-646. [DOI] [PubMed] [Google Scholar]