Abstract
Synchronous primary breast cancer describes the occurrence of multiple tumors affecting one or both breasts at initial diagnosis. This provides a unique opportunity to identify tissue-specific genomic markers that characterize each tumor while controlling for the individual genetic background of a patient. The aim of this study was to examine the genomic alterations and degree of similarity between synchronous cancers. Using metaphase comparative genomic hybridization and array comparative genomic hybridization (aCGH), the genomic alterations of 23 synchronous breast cancers from 10 patients were examined at both chromosomal and gene levels. Synchronous breast cancers, when compared to their matched counterparts, were found to have a common core set of genetic alterations, with additional unique changes present in each. They also frequently exhibited features distinct from the more usual solitary primary breast cancers. The most frequent genomic alterations included chromosomal gains of 1q, 3p, 4q, and 8q, and losses of 11q, 12q, 16q, and 17p. aCGH identified copy number amplification in regions that are present in all 23 tumor samples, including 1p31.3-1p32.3 harboring JAK1. Our findings suggest that synchronous primary breast cancers represent a unique type of breast cancer and, at least in some instances, one tumor may give rise to the other.
Keywords: Synchronous breast cancer, microdissection, comparative genomic hybridization (CGH), array CGH (aCGH), metaphase CGH (mCGH)
Introduction
Although many genetic perturbations have been detected in a proportion of breast cancer cases, common events underlying the majority of breast cancer cases have been much harder to find, due partly to differences in genetic background between individuals. Studying synchronous tumors from the same patient can overcome such confounding factors by allowing investigators to identify genetic markers specific to individual tumors while controlling for both tissue-specific and patient-specific genetic background factors.
The prevalence of synchronous primary breast carcinoma is approximately 1% to 11% [1,2], although the incidence of these cases may be underreported because clinical staging is based on the single largest tumor. Genetic events underlying the occurrence of synchronous breast tumors are poorly understood. For example, it is not clear whether multiple synchronous tumors develop through intramammary spread of a primary tumor or they are pathogenetically independent primary tumors that develop simultaneously. Cytogenetic [3–7] and clonal analysis studies [8–12] have not provided a consistent genomic assessment of synchronous tumors, and it remains inconclusive whether this group of breast cancers represents a synchronous presentation of genetically unique tumors or reflects molecular divergence in clonal progression after one tumor has spread to a second site.
Conventional metaphase comparative genomic hybridization (mCGH) [13] is a valuable tool used to obtain cytogenetic signatures of tumors with a resolution of > 20 Mb [14] and, in this study, to rapidly compare overall patterns of imbalance within synchronous tumor subgroups. In contrast, array comparative genomic hybridization (aCGH) permits a more detailed analysis with refined resolution at the level of genes and expressed sequence tags (ESTs) [15]. We have previously demonstrated the fidelity and applicability of single-cell comparative genomic hybridization (SCOMP) of whole genomic amplification using minute amounts of microdissected formalin-fixed paraffin-embedded (FFPE) materials for aCGH studies [16]. The combination of tissue microdissection and whole genome amplification allows for an in-depth high-throughput genomic analysis of well-defined cell populations. In this study, genetic aberrations and the degree of genomic association between 23 synchronous breast cancers from 10 patients were investigated using mCHG and aCGH to define alterations at chromosomal and locus-specific/gene levels.
Materials and Methods
Sample Collection
Samples from 23 synchronous tumors were collected from 10 patients at the University Health Network and Mount Sinai Hospital (Toronto, Ontario, Canada), with approval from the Research Ethics Boards at each institution. All 10 patients had no family history of breast cancer. Cases from nine patients were unilateral, and one patient presented with synchronous tumors in both breasts (bilateral). All tumors exhibited no apparent physical connection by standard histopathological analysis. The distance between tumors is given in Table 1. FFPE tumor sections from each case were evaluated by a pathologist (S.J.D.), and representative tumor blocks were selected.
Table 1.
Histologic Features of Synchronous Breast Tumors.
| Cases | Type | Laterality | Tumor Size (cm) | Tumor Distance (cm) | Histologic Grade |
Estrogen Receptor | Progesterone Receptor | HER2 | |||
| Grade | T | P | M | ||||||||
| P1T1 | IDC (invasive micropapillary) | R | 1.3 | 1.3 | II | T2 | P3 | M1 | + | + | ND |
| P1T2 | IDC (NOS) | R | 0.8 | 1.3 | II | T2 | P3 | M1 | + | + | ND |
| P1T3 | IDC (NOS) | L | 0.7 | 0.75 | II | T3 | P2 | M1 | + | + | ND |
| P1T4 | IDC (NOS) | L | 0.7 | 0.75 | II | T3 | P2 | M1 | + | + | ND |
| P2T1 | IDC | L | 0.25 | ND | I | T1 | P2 | M1 | + | + | ND |
| P2T2 | IDC | L | 1.7 | ND | II | T2 | P2 | M1 | + | - | ND |
| P3T1 | IDC | L | 1.6 | ND | II | T3 | P2 | M1 | + | + | - |
| P3T2 | IDC | L | 2.0 | ND | II | T3 | P2–3 | M1 | + | + | - |
| P4T1 | IDC (tubular) | R | 0.8 | 0.4 | I | T1 | P2 | M1 | + | + | ND |
| P4T2 | IDC (tubular) | R | 1.6 | 0.4 | I | T1 | P2 | M1 | + | + | ND |
| P5T1 | IDC (NOS) | R | 2.3 | 1.8 | II | T2 | P2 | M3 | - | - | - |
| P5T2 | IDC (NOS) | R | 1.0 | 1.8 | II | T3 | P2 | M1 | - | - | - |
| P6T1 | IDC (NOS) | L | 2.7 | 3.0 | III | T3 | P3 | M3 | - | - | - |
| P6T2 | IDC (NOS) | L | 0.4 | 3.0 | I | T1 | P2 | M1 | + | - | - |
| P6T3 | IDC (NOS) | L | 0.5 | 1.7 | II | T3 | P2 | M1 | - | ||
| P7T1 | IDC (NOS) | L | 1.5 | 1.5 | II | T2 | P3 | M1 | + | + | - |
| P7T2 | IDC (NOS) | L | 1.1 | 1.5 | I | T1 | P3 | M1 | + | + | - |
| P8T1 | IDC (NOS) | R | 6.7 | 3.2 | III | T3 | P3 | M3 | - | - | - |
| P8T2 | IDC (NOS) | R | 2.5 | 3.2 | III | T3 | P3 | M3 | - | - | - |
| P9T1 | IDC | R | 1.5 | 5.0 | II | N2/3 | ND | ||||
| P9T2 | IDC | R | 1.0 | 5.0 | II | N2/3 | ND | ||||
| P10T1 | ND | ND | ND | ND | ND | ||||||
| P10T2 | ND | ND | ND | ND | ND | ND | |||||
Tumors are tabulated based on type, laterality, size, relative distance between synchronous tumors, histology, nuclear grading, and hormone receptor status (+, positive; -, negative). Histologic grade is composed of three parameters each scored 1–3: tubules, nuclear pleomorphism, and mitotic rate. Scores are added to give an overall grade (grade I = 3–5; grade II = 6 or 7; grade III = 8 or 9). Twenty-three samples from 10 patients were used for the study. From the 10 patient cases, 9 cases were unilateral synchronous tumors and 1 case was bilateral. All cases were invasive ductal carcinomas. ND = no data; NOS = not otherwise specified.
Tissue Microdissection and DNA Extraction
For each case, 20 to 40 unstained 5-mm-thick sections and one H&E-stained reference section were cut from a representative tumor block. FFPE tissue sections were treated with xylene to remove paraffin and subsequently dehydrated in an ethanol series. The slides were stained briefly with hematoxylin. With the use of the reference H&E slide for orientation, invasive carcinoma was microdissected manually from FFPE tissue sections using 18-gauge needles and a stereomicroscope. Small tumor regions that could not be microdissected manually were isolated using the PixCell II Laser Capture Microdissection System (Arcturus, Mountain View, CA). DNA was extracted with the use of 3-day proteinase K digestion and the Qiagen DNA Mini Kit (Qiagen, Inc., Mississauga, Ontario, Canada) according to the manufacturer's instructions. The presence of amplifiable DNA was confirmed by polymerase chain reaction (PCR) for a housekeeping gene (asparagine synthetase). The forward and reverse primers of 5′-ACATTGAAGCACTCCGCGAC-3′ and 5′-CACATTGTCATAGAGGGCG-3′ (Qiagen) were used to amplify a DNA fragment of 160 bp.
Whole Genomic Amplification
Three of 23 samples produced sufficient DNA for direct aCGH study. SCOMP was performed on the remaining 20 samples, as DNA quantities were limited. We have previously demonstrated the fidelity of the SCOMP method of whole genomic amplification in CGH studies using the MCF-7 breast cancer cell line [16]. Test (tumor) DNA (100 ng) and reference DNA (uninvolved pool of lymph nodes) were subjected to MseI (New England Biolabs, Pickering, Ontario, Canada) digestion in three separate reactions. For each reaction, adaptor formation and ligation were carried out according to Stoecklein et al. [17].
mCGH
Lymphocyte metaphase spreads were prepared using standard methods [18]. For the labeling of DNA, 2 µl of primary SCOMP product of both test (tumor) and reference DNA (lymph node) was simultaneously PCR-amplified and labeled with biotin-16-dUTP (Roche Diagnostics, Laval, Canada) and digoxigenin-11-dUTP (Roche), respectively. Subsequently, 8 µg of biotin-labeled test DNA was combined with 8 µg of digoxigenin-labeled reference DNA and precipitated with 100 µg of unlabeled human CotI DNA (Invitrogen, Burlington, Ontario, Canada) and 100 µg of sonicated salmon sperm DNA (Invitrogen) to suppress the hybridization of repetitive sequences. Normal male lymphocyte metaphase slides were treated with pepsin at 37°C for 5 minutes, washed with 1× phosphate-buffered saline and 2× sodium saline citrate (SSC), dehydrated in an ethanol series, a with 70% formamide/2× SSC at 70°C for 2 minutes. Probes were denatured and hybridized to denatured metaphase slides. Following a 48-hour hybridization, posthybridization washes and antibody detection were carried out as described by Speicher et al. [19]. Metaphase spreads were captured using the Vysis Quips SmartCapture imaging system, and image analysis was performed using the Quips CGH/Karyotyper and Interpreter software (Vysis, Downers Grove, IL). Final CGH profiles were analyzed at 95% confidence intervals. The cutoff values for chromosomal gains and losses were 1.2 and 0.8, respectively.
aCGH
For all 23 samples, 1 µg of test DNA and 1 µg of FFPEDNA extracted from a pool of lymph node tissues (reference) were labeled by random priming using Cy3-dUTP (Amersham Biosciences, Piscataway, NJ) and Cy5-dUTP (Amersham) fluorescent nucleotides, respectively, in triplicate reactions. Purified labeled products were pooled and hybridized onto Human 19K cDNA single-spot arrays (Clinical Genomics Center; http://www.microarrays.ca/) in duplicate experiments. The 19K cDNA arrays contained 19,000 cDNA and ESTs. Images were captured using a GenePix 4000A scanner (Axon Instruments, Union City, CA) and analyzed using the GenePix Pro 3.0 software (Axon Instruments).
Data Analysis
Analysis of microarray data was performed using Normalise Suite software (Normalise Suite, Toronto, Ontario, Canada) [20] (available as free download at http://www.utoronto.ca/cancyto/) to graphically illustrate regions of gains or losses along chromosomes. Array data were loaded in GPR format and matched to the appropriate “gene list” file (corresponding to Human 19K cDNA arrays used in the experiment). Duplicate normalized files were combined together into one “project file.” Biomathematical analysis of the data was carried out using Eisen clustering software (available at http://rana.lbl.gov/EisenSoftware.htm) to calculate distance matrices and to plot a hierarchical clustering map. The significance analysis of microarrays (SAM) method was used to determine statistically significant recurrent amplified and deleted regions associated with synchronous tumors. A log2 ratio threshold of ± 0.25 was used to define all copy number amplifications and deletions. This threshold value is within the acceptable range for the number of false positives. mCGH profiles were compiled using the Progenetix software (Progenetix, Stanford, CA) [21] (available at http://www.progenetix.net) to produce a frequency graph of regions of chromosomal imbalances. Constitutive heterochromatic regions were excluded from all analyses.
Real-Time Quantitative PCR (Q-PCR)
Real-Time Q-PCR was conducted using the 2× Quantitect SYBR Green PCR kit (Qiagen) and the ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) to validate array data. Oligonucleotide primers were designed using Primer Express (version 1.5; PE Applied Biosystems, Foster City, CA). Quantitative reactions for the target gene JAK1 on 1p32.3-p31.3 and a reference gene γ-interferon (IFNG) on 12q24.1 were performed in 50-µl volumes and in separate tubes. For each sample, reactions were performed in duplicate for both the reference gene and the target gene. A threshold of 1.5 was used for copy number amplification. The following forward and reverse primers (Qiagen) were used: JAK1: 5′-TCCCTGATAACAGCACATGCA-3′ and 5′-ACCTTCCCAAAGTGGCCC-3′; IFNG: 5′-GCCTCCCTAACCTGATTGGT-3′ and 5′-CAATCCCTGACTCGCTCTC-3′.
Results
Clinical Characteristics
The histopathological characteristics of tumors were tabulated based on type, relative distance between synchronous tumors, histology, grade, and hormone receptor status. Twenty-three samples from 10 patients were used for the study. From the 10 patient cases, 9 were unilateral synchronous tumors and 1 was bilateral. All cases were invasive duct carcinomas (Table 1).
mCGH from SCOMP-Amplified Tumor DNA
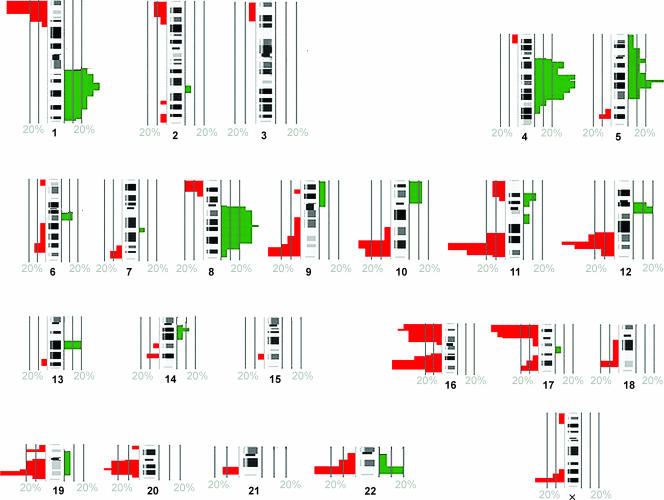
Reproducible profiles were obtained from 15 of 23 samples revealing numerous chromosomal imbalances of synchronous breast cancer samples (Table 2). Across all tumors, the most frequent chromosomal gains were observed on 1q (80%), 3p (70%), 4q (60%), and 8q (60%). Regions exhibiting marked loss were 11q (70%), 12q (60%), 16q (60%), 17p (60%), 16 p (50%), and 19q (50%) (Figure 1).
Table 2.
A List of Chromosomal Alterations in Cases of Synchronous Breast Cancer.
| Tumor Cases | Gain | Deletion |
| P1T1 | 5p11.2-23, 8q | 8p21-22, 9q13-qter, 19q13.3, 22q |
| P1T2 | 3p12-q12, 4q13, 4q22-28, 6q11-14, 8q11.2-24.1 | 1p33-pter, 1q34, 10q25, 12q24.1, 12q24.3, 16q12.2-24.1, 19q, 20q11.2-13.2, 22q13 |
| P1T3 | 1q12, 2p31.3, 3p12-q12, 4q12, 5q21.1, 9q12, 12q12, 12q13.2 | 11p13-15, 11q21-24, 16q12.2-24.1, 17p |
| P1T4 | 1q12, 3p12, 4q22-31.2, 5q15, 12q13.2, 13q21 | 11p14-15.3, 11q21-24, 16q21-23, 17p, 19q13.3, 22q13.1 |
| P2T1 | 1q12-43, 5q15, 11p11.2-14, 11q13 | 1p35-pter, 3p24-pter, 6q16-24, 16p12-pter, 16q22-23, 17p, 17q11.1-12, 18q12-22, 22q13 |
| P3T1 | 1q12, 1q22-31.3, 3p12, 4q11-28, 5p13-q23, 19q11-13.3, 22q13 | 8p23, 11q23-25, 12q22-24.2, 16p12-pter, 16q22-23, 17p11.2-12, 17q24 |
| P3T2 | 1q12, 3p12, 4q11-12, 4q22-25, 5q11.2, 6q12, 8q21.2, 22q13 | 12q24.1, 16p12-pter |
| P5T2 | 1q41-pter, 2q31, 4q24 | 1p33-pter, 2q33-pter, 5q34, 9q33-34, 10q25-qter, 14q23, 16p12-pter, 17p, 17q25, 18q22, 19q13.11-13.3, 22q12-13, Xq27 |
| P6T1 | 1q12, 8q12-23, 11p13-14 | 1p35-pter, 5q33.1-34, 6q25, 7q33-36, 11q23-24, 12q24.2, 13q32, 14q31, 16p13.2-pter, 16q22-23, 17p12-pter, 19q13.2-13.3 |
| P6T2 | 4q13-26, 5q14-21, 8q11.2-23, 13q21, 14q12, 17q21 | 1p35-pter, 2p21-pter, 6q24-25, 7q35-36, 8p22-pter, 11q21-24, 12q23-qter, 16p, 16q21-23, 17p, 18q22, 19q |
| P7T2 | 1q11-12, 3p12, 4q13-28, 5q15, 13q21 | 1p36-pter, 6q24-qter, 8p22-pter, 9q34, 11q23, 16p12-pter, 17p, Xq27 |
| P8T1 | 1q, 3p12, 6q12, 7q22, 8q11.2-24.11, 9p, 10p, 12q12 | 2q36-qter, 9q31-34, 10q23-qter, 11q23, 12q22-24.2, 16p13.2, 17q23-25 |
| P8T2 | 1q, 10p | 10q25-qter, 11q23-25, 12q22-24.2, Xq25-27 |
| P10T1 | 1p13-q32, 4q13-32, 6q12-14, 9q12 | 2p23-ter, 4p15.3-pter, 9p22, 10q25-qter, 11q23, 12q24.1, 14q31, 16q13.1-qter, 16q21-23, 20p13, 20q13.1, Xq27 |
| P10T2 | 1q12, 1q25, 3q11.2, 5q11.1-23, 9q12, 12q13.2, 22q | 1p35-pter, 6p24-pter, 8p22-pter, 9q34, 12q24.1-24.2, 13q33, 15q25, 16q22-23, 17p13, 17q25, 19q13.3, Xp22.1-pter |
Common changes between paired samples are italicized.
Figure 1.
Frequency graph of aberrations in 15 cases of synchronous breast cancer obtained by mCGH. The green marks on the right side of the chromosome ideograms indicate the frequency of chromosomal gains, and the red marks on the left show the frequency of losses. The cutoff values for chromosomal gain and losses were 1.2 and 08, respectively. The graph was created with the Progenetix software [21].
The CGH findings between synchronous tumors (Table 2) revealed at least three common regions of chromosomal gains and losses (Table 2, italicized). Interestingly, when the imbalance pattern was compared within synchronous tumors from each patient, regions of chromosomal loss were more concordant than regions of gain.
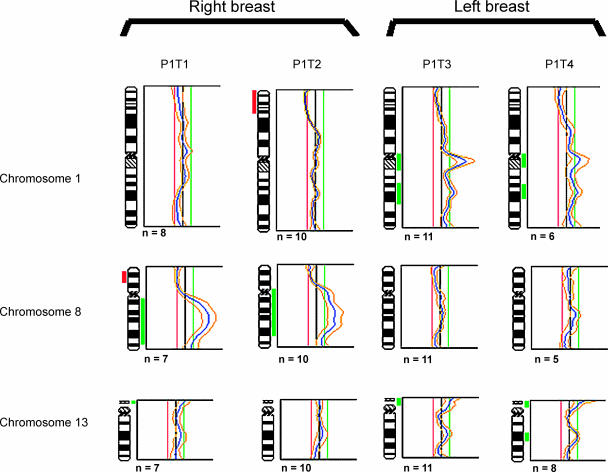
Patient 1, the only case of bilateral breast cancer, presented with four tumors: two in the right breast (P1T1 and P1T2) and two in the left breast (P1T3 and P1T4). The paired tumors P1T1 and P1T2 possessed common changes, namely, the gains of 8q and the losses of regions on 19q and 22q, with tumor P1T2 displaying far more regions of chromosomal gains and losses. Tumors P1T3 and P1T4 from the left breast showed greater similarities in overall genomic gains and losses, with tumor P1T3 showing six additional regions of chromosomal gain and with tumor P1T4 showing two regions of additional chromosomal gain. Regions of deletion were similar in the two tumors, with tumor P1T4 showing additional losses on 19q and 22q. However, a comparison between tumors from the right breast and those from the left breast showed marked dissimilarities (Table 2 and Figure 2). Overall comparisons using chromosomes 1, 8, and 13 for these four tumors showed a general similarity in the profiles of tumor pairs arising in the same breast, but a different pattern of imbalance was apparent when profiles from opposite breasts were compared for these chromosomes.
Figure 2.
Comparison of mCGH profiles of synchronous tumors in patient 1 with four synchronous breast tumors. Tumors P1T1 and P1T2 show no gain on 1q, whereas tumors 3 and 4 are similar in showing gains on chromosome 1q. Tumors P1T1 and P1T2 have high gains of 8q, whereas tumors P1T3 and P1T4 show normal profiles for chromosome 8. Chromosome 13 in tumor 4 shows a low-level gain in the q region. Although tumors P1T1, P1T2, and P1T3 have not been (marked to have) scored as having this chromosomal gain, this region in these cases has “spikes” just below the threshold, suggesting a possible gradual increase in chromosomal instability in this region. For each profile, the black vertical line on the middle represents a ratio of 1.0; the red line on the left represents a ratio of 0.8; and the green line on the right represents a ratio of 1.2. Alterations to the right represent chromosomal gains, and alterations to the left are chromosomal losses. Gains and deletions are marked next to chromosome ideograms as green and red bars, respectively.
aCGH
aCGH analysis of 23 samples using the Human 19K cDNA array revealed copy number changes comparable to findings by mCGH analysis and can be accessed from http://www.uhnresearch.ca/labs/done/publications.htm.
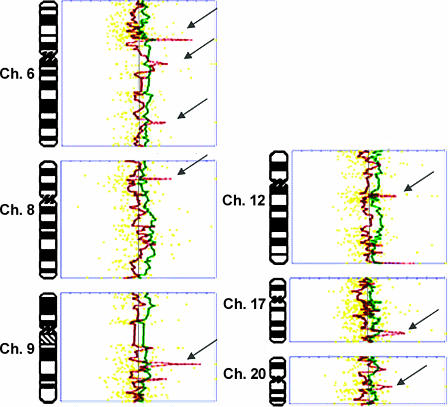
The increased resolution revealed regions of focal gains and loci of amplification at 1q, 3p, 4q, and 8q. Moreover, the profiles of each tumor could be compared to their matched synchronous counterpart, and the extent of genomic imbalance could be determined with greater mapping precision (Figure 3).
Figure 3.
Comparison of aCGH profiles of synchronous tumors from patient 9. Profiles of P9T1 (green) and P9T2 (red) were generated by Normalise Suite software and superimposed to facilitate comparison. Note the dissimilar regions in chromosomes 6, 8, 9, 12, 17, and 20 (arrows). For each profile, the vertical line on the middle represents a ratio of 1.0. By convention, alterations to the right are copy number amplifications, and alterations to the left are copy number deletions. Thresholds are marked on the graph (top) and indicate 3 SD from the mean. Yellow dotted lines are data points.
The comparative values of known breast cancer-related genes that have their corresponding sequences represented on the array were investigated. Array features corresponding to TP53 (17p13.1), IGF2 receptor (11p15.5), IL-6 (7p21), ERBB2-interacting protein (5p14.3-q12.3), IFNα receptor 1 (21q22), MYC (8q24.21), MAP3K12 (12q13), TIMP2 (17q25.3), TIMP3 (22q12.1-q13.2), IFNGR2 (21q22), CCND1 (11q13.1), and ATM (11q22.3) exhibit normal copy number values in all samples. The increased mapping resolution of aCGH revealed structural conformation of amplicons in greater detail compared to mCGH data, as shown by the mCGH profiles of the 17p13 and 17q25 regions, detecting a net chromosomal gain of the region. The aCGH data of the corresponding region included TP53 (17p13.1) and TIMP2 (17q25.3), respectively, and revealed normal copy number values for both genes. However, array features mapping telomerically and centromerically to these genes exhibited low-level amplification of nonannotated genes (supplementary data available at http://www/uhnresearch.ca/labs/done/publications.htm). These changes were collectively detectable as chromosomal gains in 17p13 and 17q25 regions by mCGH. Thus, as expected, aCGH was able to reveal the complexity of amplicon copy number at a resolution higher than that in mCGH.
Using SAM, the genes on the array that exhibited significant copy number alterations and were consistently amplified or deleted in all 23 samples were identified. The analysis produced a list of 189 ESTs and genes, 159 copy number amplifications, and 30 deletions, the majority of which were nonannotated. (This list is available at http://www.uhnresearch.ca/labs/done/publications.htm.) The exception was the 1p31.3-1p32.3 region harboring JAK1. Varying levels of JAK1 amplification were detected in all samples. The copy number amplification of JAK1 in all 23 samples was confirmed by Q-PCR, with copy numbers ranging from a 1.5-fold to a 6.3-fold increase compared to JAK1 levels in the reference sample (a pool of lymph nodes). All alterations obtained from SAM were categorized based on type (copy number amplification or deletion) and further grouped based on the chromosomal arm where the alteration occurred. The frequency graph of aCGH data is shown in Table 3.
Table 3.
Frequency of Copy Number Amplifications and Deletions Present in All 23 Synchronous Tumors Samples from aCGH Data.
| Copy Number Amplifications | Copy Number Deletions |
| 1p (39.1%), 1q (52.2%), 2q (21.7%), 3p (4.4%), 4q (30.4%), 5q (4.3%), 6p (4.3%), 6q (8.7%), 7p (17.4%), 7q (21.7%), 8q (21.7%), 9p (8.7%), 9q (17.4%), 10q (8.7%), 11p (21.8%), 11q (21.8%), 12p (4.4%), 12q (21.8%), 13q (8.7%), 14q (13.0%), 16p (4.4%), 17q (4.4%), 18p (8.7%), 18q (8.7%), 20q (4.4%), 21q (8.7%), 22q (4.4%), Xp (17.4%) | 1p (39.1%), 1q (53.2%), 2q (21.8%), 3p (4.4%), 4q (30.4%), 5p (4.4%), 5q (4.4.%), 6p (4.4%), 6q (8.7%), 7p (17.4%), 7p (17.4%), 7q (17.4%), 8q (13.0%), 10q (8.7%), 11p (30.4%), 11q (13.0%), 12p (8.7%), 17q (13.0%), 18p (13.0%), 18q (8.7%), Xq (26.1%) |
The SAM method was used to determine statistically significant recurrent amplified and deleted regions associated with synchronous tumors. Copy number alterations were categorized based on type (amplification or deletion) and further grouped based on the chromosomal arm where the alterations are present.
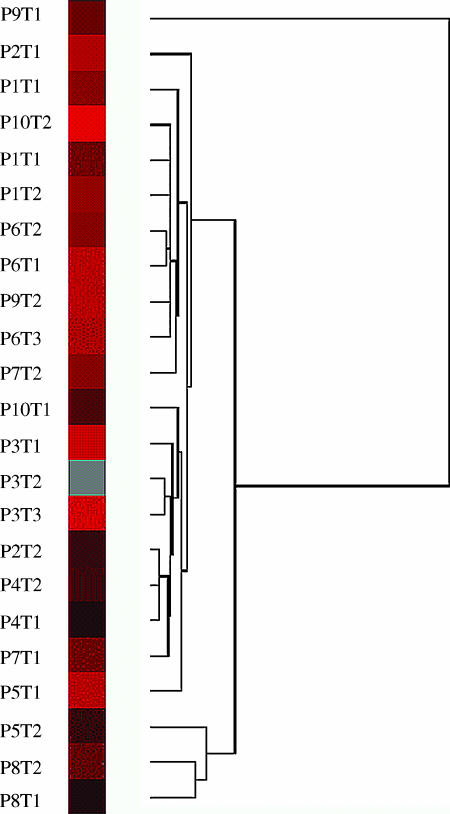
Eisen cluster analysis of the aCGH data was performed to identify the degree of similarities between matched synchronous tumors (Figure 4). The analysis clustered patients 6 and 8 with the greatest degree of similarity between their respective tumors. Tumors P1T3 and P1T4 (left breast) from patient 1 also clustered closely but separated from P1T1 and P1T2 (right breast). These patterns were consistent with the groupings determined by similar patterns of chromosomal change identified by mCGH analysis. All other tumors showed a pattern of imbalance clustered away from these tumors.
Figure 4.
Cluster analysis of array data showing the relative relationship between synchronous breast cancer tumors. Tumor samples with a high degree of similarity are connected to the tree by very short branches. Tumors with decreased similarity are joined by increasingly longer branches. Tumors in patient 8 (P8T1 and P8T2) and patient 3 (P3T1 and P3T2) show the greatest degree of similarity to their synchronous counterparts. In patient 6, only tumors P6T1 and P6T2 are clustered closely, whereas P6T3 is only distantly related. Tumors P1T3 and P1T4 (left breast) from patient 1 also clustered closely but separated from P1T1 and P1T2 (right breast). Note that not all tumors are matched to their synchronous counterparts. The clustering map was created with Eisen cluster.
To compare the profiles of synchronous tumors in a given patient, the log2 values of array data were plotted on chromosomes 1 to X using Normalise Suite. The profiles were superimposed to facilitate comparison and to determine regions with different imbalance levels. An example is shown in Figure 3.
Discussion
The identification of genomic alterations associated with the development of breast cancer has been hampered by the large variation in the genetic changes found between tumors. Part of this variation may be patient-specific. The study of multiple tumors in the same patient, therefore, is of great value as it allows direct genomic comparison of (synchronous) tumors while excluding confounding genetic variations, such as copy number polymorphisms (i.e., variation in the number of copies of a sequence within the DNA), that exist between individuals. To date, the relationship between synchronous tumors remains unclear. The histologic comparison of synchronous tumors in the same patient often reveals different morphologic features. The limited number of genetic analyses of this presentation of breast cancer shows variations in results and is inconclusive as to whether or not synchronous tumors share common molecular events or indeed represent truly distinct entities. A clonal analysis based on restriction fragment length polymorphism of the X-chromosome phosphoglycerokinase (PGK) gene [8] revealed that the same X-chromosome was inactivated in all 10 tumors from three patients. This finding was taken to suggest that the multiple tumors were clonally related. However, a somewhat different conclusion was arrived at by Shibata et al. [10], who demonstrated that a subset of synchronous cases had discordant allelic patterns and inferred that synchronous tumors in those cases arose independently. The clonality of multiple tumors was also examined by loss-of-heterozygosity studies on 16q in 60 cases of multiple tumors [9], on 5 chromosomal arms (11p/q, 13q, and 7p,q) in 8 cases [22], and on 7 chromosome arms (3p, 11p, 11q, 13q, 14q, 17p, and 17q) in 26 tumors [11]. All three studies concluded that monoclonality could be established only in some cases and that the remaining proportion of tumors could be distinct primaries. An immunohistochemical study based on p53 and c-erbB2 expression was in agreement with this finding and reported that only a fraction of multiple tumors were clonally related [23], whereas a more recent immunohistochemical study on bilateral breast cancer reported dissimilar patterns of p53 and osteopontin expression [24]. Cytogenetic studies and karyotypic comparison of multiple tumors indicated that only a proportion of synchronous breast tumors were distinct primaries at the chromosomal level and that the differences in those cases may be reflective of their independent origin [3–7,25]. The inconsistency in these reports could be due to the limitations of the techniques used, which examine only a few parameters and do not give an overall assessment of genomic anomalies. Although classic karyotyping studies of breast cancer samples provide a genomewide evaluation of the samples, they have a greater risk of introducing in vitro selection biases.
In this study, we have: utilized microdissection and genome amplification techniques for the specific isolation of cells from each tumor; used standard mCGH to identify gross chromosomal aberrations; and performed aCGH to improve the resolution of an imbalance map and to identify loci/gene-specific regions of copy number changes. The resolution of mCGH is known to be 20 Mb, making the detection of smaller regions much more difficult; thus, the limitation in resolution can be compensated for by aCGH. Conversely, mCGH can provide a rapid means of determining overall patterns of imbalance for comparative purposes, such as within our various synchronous tumor subsets. Therefore, the aim of this study was to objectively examine the degree of genomic similarity between synchronous tumors and to identify regions of recurrent alteration at higher resolution by aCGH.
The mCGH analysis of synchronous breast tumor samples revealed a number of chromosomal aberrations, which are in part consistent with genomic signatures of breast carcinomas (http://www.progenetix.de/~pgscripts/progenetix/LC50/ideogram.html). The results were compared to a breast cancer CGH database, a map of chromosomal alterations in 686 cases of breast cancers of different types [21]. They can be categorized into two groups: chromosomal aberrations known to be associated with breast cancer such as gains of 1q and 8q and loss of 17p, and those that are not commonly implicated in breast cancer. The chromosomal gain of the 4q region, for instance, has not been reported to be frequently involved in breast cancer [21]. Patterns of 4q gain are similar in the majority of our cases and include the region 4q22-4q26. The chromosomal gain of this region obtained by mCGH was supported by array data showing a cluster of mostly nonannotated genes in the 4q region with high copy number values. The function of the majority of these genes is unknown. To date, putative oncogenes in 4q that may have biologic roles that are important in breast cancer development and/or progression have not yet been reported; future studies may thus reveal novel oncogenes in this region. The additional frequent chromosomal alterations in our synchronous cases were the losses of 16q and 16p. The latter is more frequently reported as amplified in breast cancer, whereas 16q, at times, is associated with loss [21]. It has been reported that the loss of genomic material from 16q is present in low-grade to intermediate-grade ductal carcinoma in situ [26] and high-grade ductal carcinoma in situ [27].
The comparison of mCGH profiles revealed that all synchronous tumors had unique features and that paired tumors in a given patient, although similar, were not identical. The results were compared to those obtained from unifocal tumors (data not shown) and validated that the alterations are not the consequence of experimental artifacts. The genomic profiles of the majority of tumors (11 of 15) were shown to be distantly related to their matched synchronous pair. This is an interesting observation because it might be expected that the profiles of multiple tumors in a given patient, having grown in a common milieu, would be more similar to one another than to tumors of different patients. Our finding demonstrates that the genetic pathology of synchronous tumors of the same patient goes beyond the boundaries of patient specificity and that the profiles of synchronous tumors within the same microenvironment may be different. Some of the subtle differences in imbalance level may be related to the scale of an alteration. For example, comparisons between the four tumors from patient 1 showed minor differences in 13q levels of chromosomal gain. It can be postulated that this subtle increase in gain may be a function of differential levels of chromosomal instability and/or polysomy for this region in all four synchronous tumors. It may well be that tumor 4 was the first tumor to develop in this patient and, therefore, the incongruity in the CGH profile of 13q may reflect temporal differences in the progression of breast cancer in this patient.
Hierarchical clustering of aCGH data was performed to reveal the relative relationship between tumors. Interestingly, again, the analysis did not group together all paired tumors in a given patient. In fact, with the exception of P8T1/T2 and P6T1/T2, all other tumors were only distantly related to their matched synchronous tumor and showed similarities to tumors in different hosts. P8T1 and P8T2 were grouped closely together, and yet the distance between these unilateral tumors was 3.7 cm—more than the distance between the other tumors with distantly related profiles. The relationship between synchronous tumors in a given patient, therefore, does not seem to be a function of the physical distance between them in the host microenvironment.
Amplification of the 1p31.3-p32.3 region was found in all synchronous breast cancer samples by high-resolution aCGH. A candidate gene of interest in this region and on the array included the JAK1 gene. JAK1 has been reported to be involved in various biologic pathways, including epidermal growth factor signaling [28], IFN [29], and IL-6 [30] pathways, all of which have been shown be associated with breast cancer [31–34]. To our knowledge, this is the first time that copy number amplification of JAK1 has been reported in breast cancer. The family of JAK tyrosine kinases (JAK 1–3), through the activation of the STAT signaling pathway, demonstrates a dual role: apoptosis and transcription of negative regulators of cell cycle such as p21, or cell proliferation through PI3K and MAP kinase activation [35]. In the MDA-MB-468 breast cancer cell line, JAK1-mediated activation of STAT, a downstream target of JAK, could induce apoptosis [36]. Another study examined the expression pattern of tyrosine kinases in 13 breast cancer cell lines and 2 normal immortalized breast epithelial cell lines, and demonstrated high levels of tyrosine kinases, including HER2/neu and JAK1, in breast cancer cells [36]. In the quest to identify transcriptional targets of BRCA1 involved in breast cancer, one study examined BRCA1 effects on expression in a generated epithelial cell line (derived from human embryonic kidney) with inducible BRCA1 expression. Microarray analysis revealed a group of highly overexpressed genes, including JAK1, STAT, CCND1, and MYC [37]. The association of JAK1 and BRCA1 has also been reported in prostate cancer. Immunoprecipitation studies showed that BRCA1 interacts with JAK1 in prostate cancer and blocks the activation of the JAK-STAT signaling pathway [38]. Because BRCA1 is a tumor suppressor that is also involved in familial breast cancer, a potential relation between BRCA1 and JAK1 was postulated. The role of JAK1 in other malignancies suggests a possible role of this gene in the pathology of several tumor types [38].
The mechanism(s) underlying the pathogenesis of synchronous primary breast cancers is not well understood. Several different possibilities exist. Genetically similar tumors arising in a single individual may suggest a “field effect” whereby genetic aberrations affect a region or a “field” of the breast, but each tumor develops different additional genetic aberrations [39]. Multiple tumors in a given patient may also have a metastatic relationship and, therefore, similarities between tumors can merely be the consequence of a metastatic event from a common primary site. If one tumor metastasizes within the breast, presumably each will continue to accumulate genetic alterations independently after separation, so the time since separation would affect the similarity of genetic profiles. The genetic differences we see in synchronous tumors in a given patient may also be due to tumor heterogeneity. Indeed, studies have shown that breast cancers exhibit high levels of tumor heterogeneity [40–42]. Alternatively, it may be indicative of independent primary tumors that undergo different tumorigenic mechanisms and that are present within the same microenvironment merely by coincidence. Genomic findings show that, although some paired tumors shared similarities in imbalance profiles, none of the matched synchronous tumors had identical profiles. This result is suggestive of a model for the occurrence of synchronous breast cancer that could involve genomic divergence away from a “related” common progenitor. Such a mechanism of origin would be consistent with current ideas concerning tumor-initiating cells [43]. This finding is also in agreement with previously reported clonal studies of bilateral breast cancers based on TP53 mutation [44] and phenotypic characteristics [45] and studies on metastatic breast lesions [44] where no relation between molecular genetics and histopathological parameters could be established. According to the tumor progression model [46], metastases derived from monoclonal tumors are expected to contain the core genetic features of the primary tumor. It may be, however, that such a simple linear model may be inadequate to explain the genetic progression of a primary breast cancer (Figure 5). Our data suggest that both models may be occurring. Cluster analysis suggests a linear model for patients 1, 6, and 8 because paired tumors appear to be more closely related to each other. The findings of the bilateral case of patient 1 are interesting as there is close relatedness for the right (P1T1 and P1T2) and left (P1T3 and P1T4) tumors, but little overall similarity when the tumor groups from the right and left breasts are compared. Thus, it would appear that a linear model may hold for genomically related synchronous tumors within the same breast, but a model involving early genomic divergence is more likely to explain the CGH dissimilarity of the right and left breast tumors. For the remaining cases, the situation is unclear. The factors influencing the observed genomic changes are numerous and include the effects of the microenvironment, the level of genomic heterogeneity, and the karyotypic characteristics associated with disease progression.
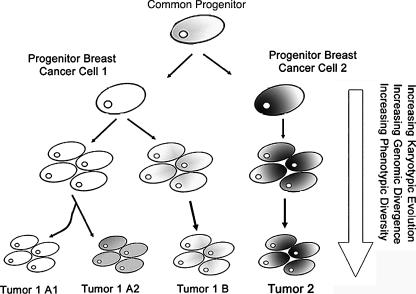
Figure 5.
Shown is a schematic model used to explain the genomic relationships of synchronous breast cancers arising from a common progenitor. In the first model, the progenitor breast cancer cell 1 (left) contains the “core” genomic information, and genomic divergence during tumor growth leads to synchronous tumors possessing related genomic signatures (shown as a different gray tone). The expansion of clones from this common progenitor, through either selection of the microenvironment or karyotypic viability, results in clones that will accumulate unique genomic changes. The second model implicates the presence of a progenitor breast cancer cell 2 shown in gray, also possessing the “core” genomic information for breast cancers. Collectively, it can be seen that tumors (1 and 2) can evolve independently from the progenitor cell with no common lineage, but may possess common changes.
In conclusion, our CGH data revealed that all synchronous tumors, despite their similar features, have unique genomic characteristics and that no two synchronous tumors in a given patient are identical. This bears potential therapeutic implications in designing more tailored clinical treatments for patients with synchronous breast cancer.
Acknowledgements
We thank F. O'Malley for her assistance in obtaining some of the samples.
Footnotes
This work was made possible by grants (to S.J.D.) from the Cancer Research Society, Inc., and the Canadian Breast Cancer Research Alliance.
References
- 1.Tinnemans JG, Wobbes T, van der Sluis RF, Lubbers EJ, de Boer HH. Multicentricity in nonpalpable breast carcinoma and its implications for treatment. Am J Surg. 1986;151(3):334–338. doi: 10.1016/0002-9610(86)90462-9. [DOI] [PubMed] [Google Scholar]
- 2.Kollias J, Pinder SE, Denley HE, Ellis IO, Wencyk P, Bell JA, Elston CW, Blamey RW. Phenotypic similarities in bilateral breast cancer. Breast Cancer Res Treat. 2004;85(3):255–261. doi: 10.1023/B:BREA.0000025421.00599.b7. [DOI] [PubMed] [Google Scholar]
- 3.Teixeira MR, Pandis N, Bardi G, Andersen JA, Mandahl N, Mitelman F, Heim S. Cytogenetic analysis of multifocal breast carcinomas: detection of karyotypically unrelated clones as well as clonal similarities between tumour foci. Br J Cancer. 1994;70(5):922–927. doi: 10.1038/bjc.1994.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandis N, Teixeira MR, Gerdes AM, Limon J, Bardi G, Andersen JA, Idvall I, Mandahl N, Mitelman F, Heim S. Chromosome abnormalities in bilateral breast carcinomas. Cytogenetic evaluation of the clonal origin of multiple primary tumors. Cancer. 1995;76(2):250–258. doi: 10.1002/1097-0142(19950715)76:2<250::aid-cncr2820760215>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 5.Teixeira MR, Pandis N, Bardi G, Andersen JA, Heim S. Karyotypic comparisons of multiple tumorous and macroscopically normal surrounding tissue samples from patients with breast cancer. Cancer Res. 1996;56(4):855–859. [PubMed] [Google Scholar]
- 6.Teixeira MR, Pandis N, Bardi G, Andersen JA, Bohler PJ, Qvist H, Heim S. Discrimination between multicentric and multifocal breast carcinoma by cytogenetic investigation of macroscopically distinct ipsilateral lesions. Genes Chromosomes Cancer. 1997;18(3):170–174. doi: 10.1002/(sici)1098-2264(199703)18:3<170::aid-gcc3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 7.Teixeira MR, Ribeiro FR, Torres L, Pandis N, Andersen JA, Lothe RA, Heim S. Assessment of clonal relationships in ipsilateral and bilateral multiple breast carcinomas by comparative genomic hybridisation and hierarchical clustering analysis. Br J Cancer. 2004;91(4):775–782. doi: 10.1038/sj.bjc.6602021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noguchi S, Aihara T, Koyama H, Motomura K, Inaji H, Imaoka S. Discrimination between multicentric and multifocal carcinomas of the breast through clonal analysis. Cancer. 1994;74(3):872–877. doi: 10.1002/1097-0142(19940801)74:3<872::aid-cncr2820740313>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 9.Tsuda H, Hirohashi S. Identification of multiple breast cancers of multicentric origin by histological observations and distribution of allele loss on chromosome 16q. Cancer Res. 1995;55(15):3395–3398. [PubMed] [Google Scholar]
- 10.Shibata A, Tsai YC, Press MF, Henderson BE, Jones PA, Ross RK. Clonal analysis of bilateral breast cancer. Clin Cancer Res. 1996;2(4):743–748. [PubMed] [Google Scholar]
- 11.Tse GM, Kung FY, Chan AB, Law BK, Chang AR, Lo KW. Clonal analysis of bilateral mammary carcinomas by clinical evaluation and partial allelotyping. Am J Clin Pathol. 2003;120(2):168–174. doi: 10.1309/6YEP-MCHA-CPG2-BD15. [DOI] [PubMed] [Google Scholar]
- 12.Unger MA, Rishi M, Clemmer VB, Hartman JL, Keiper EA, Greshock JD, Chodosh LA, Liebman MN, Weber BL. Characterization of adjacent breast tumors using oligonucleotide microarrays. Breast Cancer Res. 2001;3(5):336–341. doi: 10.1186/bcr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258(5083):818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 14.Albertson DG. Profiling breast cancer by array CGH. Breast Cancer Res Treat. 2003;78(3):289–298. doi: 10.1023/a:1023025506386. [DOI] [PubMed] [Google Scholar]
- 15.Bashyam M, Ryan B, Young K, Wang P, Hernandez-Boussard T, Karikari C, Tibshirani R, Maitra A, Pollack JR. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazani AA, Arneson NC, Warren K, Done SJ. Limited tissue fixation times and whole genomic amplification do not impact array CGH profiles. J Clin Pathol. 2006;59(3):311–315. doi: 10.1136/jcp.2005.029777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoecklein NH, Erbersdobler A, Schmidt-Kittler O, Diebold J, Schardt JA, Izbicki JR, Klein CA. SCOMP is superior to degenerated oligonucleotide primed-polymerase chain reaction for global amplification of minute amounts of DNA from microdissected archival tissue samples. Am J Pathol. 2002;161(1):43–51. doi: 10.1016/S0002-9440(10)64155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dracopoli NC. Current Protocols in Human Genetics. New York, NY: John Wiley and Sons Inc.; 2000. [Google Scholar]
- 19.Speicher MR, du Manoir S, Schrock E, Holtgreve-Grez H, Schoell B, Lengauer C, Cremer T, Ried T. Molecular cytogenetic analysis of formalin-fixed, paraffin-embedded solid tumors by comparative genomic hybridization after universal DNA-amplification. Hum Mol Genet. 1993;2(11):1907–1914. doi: 10.1093/hmg/2.11.1907. [DOI] [PubMed] [Google Scholar]
- 20.Beheshti B, Braude I, Marrano P, Thorner P, Zielenska M, Squire JA. Chromosomal localization of DNA amplifications in neuroblastoma tumors using cDNA microarray comparative genomic hybridization. Neoplasia. 2003;5(1):53–62. doi: 10.1016/s1476-5586(03)80017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baudis M, Cleary ML. Progenetix.net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17(12):1228–1229. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- 22.Chunder N, Roy A, Roychoudhury S, Panda CK. Molecular study of clonality in multifocal and bilateral breast tumors. Pathol Res Pract. 2004;200(10):735–741. doi: 10.1016/j.prp.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Dawson PJ, Baekey PA, Clark RA. Mechanisms of multifocal breast cancer: an immunocytochemical study. Hum Pathol. 1995;26(9):965–969. doi: 10.1016/0046-8177(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 24.Tuck AB, O'Malley FP, Singhal H, Tonkin KS, Harris JF, Bautista D, Chambers AF. Osteopontin and p53 expression are associated with tumor progression in a case of synchronous, bilateral, invasive mammary carcinomas. Arch Pathol Lab Med. 1997;121(6):578–584. [PubMed] [Google Scholar]
- 25.Agelopoulos K, Tidow N, Korsching E, Voss R, Hinrichs B, Brandt B, Boecker W, Buerger H. Molecular cytogenetic investigations of synchronous bilateral breast cancer. J Clin Pathol. 2003;56(9):660–665. doi: 10.1136/jcp.56.9.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James LA, Mitchell EL, Menasce L, Varley JM. Comparative genomic hybridisation of ductal carcinoma in situ of the breast: identification of regions of DNA amplification and deletion in common with invasive breast carcinoma. Oncogene. 1997;14(9):1059–1065. doi: 10.1038/sj.onc.1200923. [DOI] [PubMed] [Google Scholar]
- 27.Buerger H, Otterbach F, Simon R, Poremba C, Diallo R, Decker T, Riethdorf L, Brinkschmidt C, Dockhorn-Dworniczak B, Boecker W. Comparative genomic hybridization of ductal carcinoma in situ of the breast—evidence of multiple genetic pathways. J Pathol. 1999;187(4):396–402. doi: 10.1002/(SICI)1096-9896(199903)187:4<396::AID-PATH286>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M, Honjo M, Takahashi M, Takahashi T, Hirai H, et al. Growth hormone-induced tyrosine phosphorylation of EGF receptor as an essential element leading to MAP kinase activation and gene expression. Endocr J Suppl. 1998;45:S27–S31. doi: 10.1507/endocrj.45.suppl_s27. [DOI] [PubMed] [Google Scholar]
- 29.Caraglia M, Marra M, Pelaia G, Maselli R, Caputi M, Marsico SA, Abbruzzese A. Alpha-interferon and its effects on signal transduction pathways. J Cell Physiol. 2005;202(2):323–335. doi: 10.1002/jcp.20137. [DOI] [PubMed] [Google Scholar]
- 30.Horvath CM. The JAK-STAT pathway stimulated by interleukin 6. Sci STKE. 2004;2004(260):tr9. doi: 10.1126/stke.2602004tr9. [DOI] [PubMed] [Google Scholar]
- 31.Connett JM, Hunt SR, Hickerson SM, Wu SJ, Doherty GM. Localization of IFN-gamma-activated Stat1 and IFN regulatory factors 1 and 2 in breast cancer cells. J Interferon Cytokine Res. 2003;23(11):621–630. doi: 10.1089/107999003322558755. [DOI] [PubMed] [Google Scholar]
- 32.Kozlowski L, Zakrzewska I, Tokajuk P, Wojtukiewicz MZ. Concentration of interleukin-6 (IL-6), interleukin-8 (IL-8) and interleukin-10 (IL-10) in blood serum of breast cancer patients. Rocz Akad Med Bialymst. 2003;48:82–84. [PubMed] [Google Scholar]
- 33.Lemmon MA. The EGF receptor family as therapeutic targets in breast cancer. Breast Dis. 2003;18:33–43. doi: 10.3233/bd-2003-18105. [DOI] [PubMed] [Google Scholar]
- 34.Saha A, Bairwa NK, Ranjan A, Gupta V, Bamezai R. Two novel somatic mutations in the human interleukin 6 promoter region in a patient with sporadic breast cancer. Eur J Immunogenet. 2003;30(6):397–400. doi: 10.1111/j.1365-2370.2003.00423.x. [DOI] [PubMed] [Google Scholar]
- 35.Chin YE, Kitagawa M, Kuida K, Flavell RA, Fu XY. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol Cell Biol. 1997;17(9):5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meric F, Lee WP, Sahin A, Zhang H, Kung HJ, Hung MC. Expression profile of tyrosine kinases in breast cancer. Clin Cancer Res. 2002;8(2):361–367. [PubMed] [Google Scholar]
- 37.Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, Warrington JA, King MC. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci USA. 2002;99(11):7560–7565. doi: 10.1073/pnas.062181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao B, Shen X, Kunos G, Meng Q, Goldberg ID, Rosen EM, Fan S. Constitutive activation of JAK-STAT3 signaling by BRCA1 in human prostate cancer cells. FEBS Lett. 2001;488(3):179–184. doi: 10.1016/s0014-5793(00)02430-3. [DOI] [PubMed] [Google Scholar]
- 39.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274(5295):2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Rosen JM. Stem/progenitor cells in mouse mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2005;10(1):17–24. doi: 10.1007/s10911-005-2537-2. [DOI] [PubMed] [Google Scholar]
- 41.Chang JC, Hilsenbeck SG, Fuqua SA. The promise of microarrays in the management and treatment of breast cancer. Breast Cancer Res. 2005;7(3):100–104. doi: 10.1186/bcr1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuqua SA, Chang JC, Hilsenbeck SG. Genomic approaches to understanding and treating breast cancer. Breast Dis. 2004;19:35–46. doi: 10.3233/bd-2004-19106. [DOI] [PubMed] [Google Scholar]
- 43.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. (discussion, 1895–1896) [DOI] [PubMed] [Google Scholar]
- 44.Janschek E, Kandioler-Eckersberger D, Ludwig C, Kappel S, Wolf B, Taucher S, Rudas M, Gnant M, Jakesz R. Contralateral breast cancer: molecular differentiation between metastasis and second primary cancer. Breast Cancer Res Treat. 2001;67(1):1–8. doi: 10.1023/a:1010661514306. [DOI] [PubMed] [Google Scholar]
- 45.Schlechter BL, Yang Q, Larson PS, Golubeva A, Blanchard RA, de las Morenas A, Rosenberg CL. Quantitative DNA fingerprinting may distinguish new primary breast cancer from disease recurrence. J Clin Oncol. 2004;22(10):1830–1838. doi: 10.1200/JCO.2004.05.123. [DOI] [PubMed] [Google Scholar]
- 46.Simpson PT, Reis-Filho JS, Gale T, Lakhani SR. Molecular evolution of breast cancer. J Pathol. 2005;205(2):248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]