Abstract
Background: Severe asthma has been associated with severe exacerbations, lower lung function and greater tissue inflammation. Previous studies have suggested that mutations in interleukin-4 receptor α (IL4Rα) are associated with lower lung function, higher IgE, and a gain in receptor function. However, an effect on exacerbations and tissue inflammation has not been shown.
Hypothesis: Allelic substitutions in IL4Rα are associated with asthma exacerbations, lower lung function, and tissue inflammation, in particular to mast cells and IgE.
Methods: Two well-characterized cohorts of subjects with severe asthma were analyzed for five single nucleotide polymorphisms (SNPs) in IL4Rα. These polymorphisms were compared with the history of severe asthma exacerbations and lung function. In the primary (National Jewish) cohort, these polymorphisms were also compared with endobronchial tissue inflammatory cells and local IgE.
Results: In both cohorts, the presence of the minor alleles at E375A and Q551R, which were more common in African Americans, was associated with a history of severe exacerbations and lower lung function. In the National Jewish cohort, the C allele at E375A was associated with higher tissue mast cells and higher levels of IgE bound to mast cells. The significance for most of these associations remained when whites (the larger racial subgroup) were analyzed separately.
Conclusions: SNPs in IL4Rα, which are more common in African Americans, are associated with severe asthma exacerbations, lower lung function, and increased mast cell–related tissue inflammation. Further studies of the impact of these mutations in African Americans and on receptor function are indicated.
Keywords: asthma, genetics, IL4Rα, exacerbations, mast cells, IgE
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Mutations in IL4Rα have previously been associated with asthma, airflow limitation, and receptor function.
What This Study Adds to the Field
Mutations in the IL4Rα gene are associated with asthma exacerbations and immunopathologic changes in the airways.
One of the major distinguishing characteristics of severe asthma is the presence of severe, and often near-fatal asthma exacerbations (1). Although multiple factors, including those related to baseline airway obstruction, socioeconomic status, treatment, and psychologic factors, have been proposed to contribute to these exacerbations, it remains unclear whether underlying pathobiologic or genetic variations (or their interactions) predispose certain individuals to more life-threatening forms of asthma.
Numerous genes have been associated with asthma and related phenotypes, including those related to Th2 pathways (interleukin [IL]-4, IL-13, IL-4Rα, as well as other immunomodulatory and structure-related genes (2–4). Single nucleotide polymorphisms (SNPs) in both IL-4 and IL4Rα have been associated with severity of asthma as measured by baseline FEV1 (2, 3). However, of these genes, only the C-589T SNP in IL-4 has been associated with severe exacerbations (5).
Both IL-4 and IL-13 signal through the common IL-4Rα molecule, either in combination with the common γ chain (IL-4) or in combination with IL-13Rα (both IL-4 and IL-13) (6). The IL4Rα gene is located on chromosome 16, in an area associated with asthma susceptibility in some studies (7). Many SNPs have been identified in the IL4Rα gene, with the majority present in exon 12, one identified in the promoter, and one coding for amino acid changes in the extracellular portion of the receptor (8). The polymorphisms in exon 12 code for amino acid changes in the signaling portion of the receptor, making this a region of interest for its potential contribution to both cellular and pathobiologic changes (6, 9). SNPs in this region have previously been related to both lower FEV1 and higher IgE levels (3, 10, 11).
We hypothesized based on the association of polymorphisms in the gene for the IL-4Rα with lower FEV1 levels, and the indication that these SNPs alter the function of the receptor, that IL4Rα polymorphisms would be associated with severe exacerbations of asthma, namely intensive care unit (ICU) hospitalizations or near-fatal events, in addition to increased airflow limitation. We further proposed that these SNPs would associate with enhanced cellular inflammation in endobronchial biopsies, particularly as increases in mast cells and their level of IgE expression, which might contribute biologically to development of severe exacerbations. To address these hypotheses, questionnaire, lung function, and biopsy data from subjects with severe asthma studied at National Jewish Medical and Research Center (NJ) were characterized by IL4Rα genotype. The results from this cohort were replicated in a larger dataset (the Severe Asthma Research Program [SARP] cohort), where all data except endobronchial biopsy data were also available. Some of the data were previously reported in abstract form (12).
METHODS
Subjects
Primary cohort.
Adult subjects with asthma (18–60 yr old) recruited to the NJ from 1997 to 2005 were enrolled in various investigational protocols that involved pulmonary function testing, questionnaires, IgE levels, and bronchoscopy with endobronchial biopsy. Asthma was diagnosed on the basis of clinical history and a greater than 12% bronchodilator response to albuterol or a methacholine provocation concentration (PC20) of 8 mg/ml or less. Subjects with asthma were classified by severity using previously published criteria, including the necessity for oral corticosteroids (CS) for 50% or more of the previous year (13, 14). Because the majority of these subjects had been recruited before the implementation of the multicenter SARP, these subjects were analyzed separately (see below).
Replication cohort.
The SARP has recruited over 500 patients with asthma of varying severity from eight separate sites (Cleveland Clinic/Emory University/University of Virginia, Harvard/Brigham and Women's Hospital, Imperial College [UK], National Jewish Medical and Research Center/University of Colorado, University of Pittsburgh, Wake Forest University, Washington University, University of Wisconsin) (1). Asthma was defined as in the primary (NJ) cohort. The American Thoracic Society workshop definition of refractory asthma was used to characterize the subjects as “severe” and “not severe,” when the criteria for refractory asthma were not met. Only subjects from the seven non-NJ sites were included in this analysis (see above). All studies in both cohorts were approved by the local institutional review boards, and all subjects gave informed consent.
Evaluation of Subjects
All subjects specifically answered the questions, “Have you ever spent the night in the intensive care unit due to an asthma attack” and “Have you ever been intubated/mechanically ventilated due to an asthma attack?” In the SARP cohort, subjects also responded to similar questions regarding hospitalization and ED visits. In addition, subjects in both cohorts performed spirometry before and after up to 8 puffs (90 μg/puff) of albuterol. The baseline FEV1 testing required a 4- to 6-hour withholding of short-acting bronchodilators and a 10- to 12-hour hold on long-acting bronchodilators. Hankonsen predicted values (with race correction) were used to obtain “percent predicted” values. For safety reasons, methacholine challenge was only performed on the smaller subset of subjects whose FEV1 was greater than 55% predicted; therefore, bronchial hyperresponsiveness data were not analyzed due to the small sample size. NJ/SARP subjects had total serum IgE levels measured and blood drawn for DNA isolation and genetic analysis.
In the NJ cohort, bronchoscopy with endobronchial biopsy was specifically evaluated. Biopsy tissue sections were immunostained for IgE (PharMingen International, San Diego, CA), mast cell tryptase, CD3 (both from Dako, Carpinteria, CA), and eosinophil major basic protein (clone BMK-13; Accurate Chemical and Scientific Corp., Westbury, NY) using peroxidase-based Vectastain Elite ABC detection system (Vector Laboratories, Burlingame, CA) and amino-ethylcarbazole as a chromogen as previously described (14). IgE+ mast cells (only available on a subset) were reported as both total number of IgE+ cells per millimeter squared and percentage of total tryptase-positive mast cells.
Genotyping
All subjects were genotyped for five common SNPs in IL4Rα at the Center for Human Genomics at Wake Forest University Health Sciences. Genotyping was performed using the MassArray genotyping system (Sequenom, Inc., San Diego, CA) as previously described (10, 15). Four SNPs in exon 12, which code for amino acid changes in the intracellular signaling portion of the receptor (E375A, S411L, S478P, Q551R), and one SNP coding for an amino acid substitution in the extracellular portion of the receptor (Ile50) were identified and analyzed.
To evaluate the accuracy of genotyping, all polymorphisms were assessed to determine if the observed genotype frequencies reflected the measured allele frequencies with Hardy-Weinberg equilibrium using chi-square tests. To determine whether the presence of certain genotypes was more likely to be seen in association with SNPs, pair-wise marker linkage disequilibrium (LD) was estimated using Lewontin's D′ statistic and r2 using Haploview (16).
Statistics
All continuous data were analyzed for normality. Data not normally distributed (IgE, tissue cell counts) were log transformed. Categorical data were compared using chi-square testing for multiple groups, whereas analysis of variance was used for continuous variables. When three groups (two homozygote groups and one heterozygote group) were compared, only the overall p value was generated to determine whether an overall difference in the three groups existed. For presentation, all transformed variables were reconverted to the linear scale, with standard errors unequal after conversion back to the linear scale. Hence, both positive and negative standard errors are shown. p values of less than 0.05 were considered significant. Haplotyping analysis was performed using Haplo.SCORE (17) for both qualitative and quantitative traits.
RESULTS
Subjects
A total of 175 subjects were genotyped from NJ, including 140 patients with asthma and 35 normal subjects (NJ cohort) (Table 1). Nearly 70% of the subjects with asthma in this cohort had severe asthma as defined previously (13, 14). Fifty-two percent of the subjects with asthma were on daily oral CS and 80% were receiving inhaled CS. Slightly less than 10% of the NJ cohort were African American. A total of 179 subjects with severe and 244 subjects with milder asthma were analyzed from the replication SARP cohort. The SARP cohort (with the NJ subjects excluded) was more racially diverse than the NJ cohort (29% African American), and only 42% of the cohort subjects were categorized as severe.
TABLE 1.
DEMOGRAPHICS FOR SUBJECTS WITH ASTHMA IN THE NATIONAL JEWISH AND SEVERE ASTHMA RESEARCH PROGRAM COHORTS
| NJ Cohort* (n = 140) | SARP Cohort (n = 423) | |
|---|---|---|
| Age ± SD, yr | 41 ± 12 | 36 ± 14 |
| Sex, %, M/F | 44/56 | 34/65 |
| Race, %, white/African American/other | 84/10/6 | 64/29/7 |
| FEV1, % predicted ± SEM | 60 ± 2 | 76 ± 1 |
| Atopy, % | 82 | 82 |
| Severe, % | 74 | 42 |
Definition of abbreviations: F = female; M = male; NJ = National Jewish Medical and Research Center; SARP = Severe Asthma Research Program.
In the NJ cohort, an additional 35 normal subjects were included for inflammatory cell comparisons. Furthermore, the NJ severe cohort was recruited using higher oral corticosteroid dosing standards.
IL4Rα Polymorphisms
All five SNPs were in Hardy-Weinberg equilibrium when stratified by race. Within each racial group, there were no differences in allele frequencies between the two cohorts. Allele frequencies were evaluated in whites and African Americans for E375A, S411L, and Q551R. Significant differences in allele frequencies occurred between whites and African Americans, such that the minor alleles for E375A and Q551R in whites were the more frequent alleles in the African Americans (in both cohorts). When stratified by race, the allele frequency for C at E375A was 0.55 for African Americans and 0.09 for whites, whereas the allele frequency for G at Q551R was 0.68 in African Americans and 0.20 in whites. In contrast, the rare S411L SNP was three times more common in whites than in African Americans in the replicate (larger) SARP cohort (p = 0.01). Three of the five SNPs (E375A, S478P, Q551R) were in strong LD in both cohorts (Figures E1a–E1d in the online supplement). The strongest LD was seen in whites and was between S478P and Q551R (D′ = 0.89 and 0.96, r2 = 0.54 and 0.49 for NJ and SARP, respectively). The LD between these two SNPs in African Americans was much less (D′ = 0.8, r2 = 0.27 in SARP only), whereas in this population, the strongest LD was observed between E375A and Q551R (D′ = 1, r2 = 0.59) (Figures E1c and E1d). This was similar to the values in whites in the NJ cohort (D′ = 1, r2 = 0.53) and the SARP replication cohort (D′ = 0.98, r2 = 0.41). The less common allele at S411L was rare (< 5%). It was not in LD with E375A but was in LD with S478P and Q551R in whites (D′ = 1 for both, r2 from 0.17 to 0.30). The low-frequency T allele for S411L only occurred in association with the low-frequency alleles at S478P and Q551R. There was no LD for S411L with any SNP in African Americans. The A to G change, which codes for the Ile50 amino acid switch, was not in LD with the other SNPs.
IL4Rα Allele Distribution across Subject Groups
The allele frequencies were similar across the NJ and SARP cohorts (0.31/0.37 for C at E375A and 0.44/0.55 for G at Q551R, 0.50/0.50 for A at Ile50). There were no differences in allele frequencies between subjects with asthma of different severities in either cohort. The low numbers of normal subjects in the NJ cohort limited comparisons to subjects with asthma.
IL4Rα Polymorphisms and Asthma Exacerbations
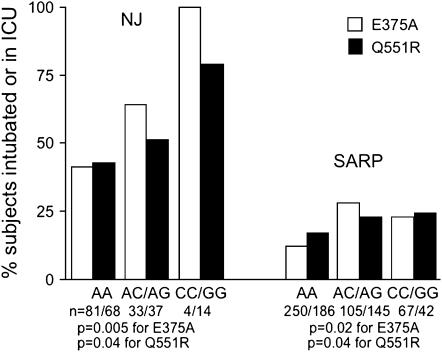
In the NJ asthma cohort, both the C allele at E375A and the G allele at Q551R were associated with a history of an asthma exacerbation requiring an ICU stay and/or mechanical ventilation (Figure 1). These relationships were replicated in the SARP population, with slightly higher p values (0.02–0.09) for history of ICU/mechanical ventilation exacerbations (Figure 1), perhaps on the basis of the less severe nature of this cohort compared with the NJ cohort (9% of the SARP cohort vs. 25% of the NJ cohort had been intubated). In addition, the history of hospitalizations and/or ED visits (only collected in the SARP cohort) was also significantly associated with those alleles (p value < 0.001 and 0.003 for E375A and Q551R, respectively). No other single SNP was associated with asthma exacerbations.
Figure 1.
The history of intensive care unit (ICU) stay or intubations was significantly greater in subjects with the C allele at E375A and the G allele at Q551R for both the National Jewish Medical and Research Center (NJ) and Severe Asthma Research Program (SARP) cohorts.
IL4Rα Polymorphisms and Pulmonary Function
In the predominantly severe (70%) NJ cohort, no associations of IL4Rα SNPs with prebronchodilator %predicted FEV1, FEV1/FVC, or reversibility were observed, although %predicted FEV1 was numerically lower in individuals with the C or G alleles at E375A and Q551R (Tables 2 and 3). In the larger replication SARP cohort, particularly in the two-thirds who did not have severe asthma, there were associations with %predicted FEV1 and reversibility (Table 2). In the replicate cohort, homozygotes for the C allele at E375A had significantly greater maximal reversibility than observed in the heterozygotes or the AA homozygotes (p = 0.02), with a similar trend observed for Q551R (p = 0.08). When pulmonary function parameters in the not-severe subgroup of the SARP cohort were evaluated by IL4Rα genotype, significant associations were observed for baseline FEV1 (p = 0.01 and 0.006 for the C allele at E375A and for the G allele at Q551R, respectively; Table 2). These were the same alleles that were significant for bronchodilator response and for severe exacerbations in both cohorts. However, the associations with maximal reversibility were no longer significant (p = 0.25 and 0.36). No other single SNPs were associated with lung function.
TABLE 2.
PULMONARY FUNCTION (ALL SUBJECTS)
| National Jewish Medical and Research Center
|
SARP (all)
|
SARP (not severe)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | FEV1 | FEV1/FVC | BD Response | SNP | FEV1 | FEV1/FVC | BD Response | SNP | FEV1 | FEV1/FVC | BD Response |
| E375A | E375A | E375A | |||||||||
| AA, n = 86 | 61 ± 3 | 0.63 ± 0.01 | 23 ± 2 | AA, n = 245 | 75 ± 1 | 0.70 ± 0.01 | 16 ± 1 | AA, n = 140 | 85 ± 1 | 0.75 ± 0.01 | 13 ± 1 |
| AC, n = 35 | 59 ± 3 | 0.62 ± 0.01 | 18 ± 3 | AC, n = 104 | 73 ± 2 | 0.71 ± 0.01 | 16 ± 1 | AC, n = 62 | 78 ± 2 | 0.72 ± 0.01 | 15 ± 2 |
| CC, n = 5 | 48 ± 6 | 0.56 ± 0.01 | 31 ± 11 | CC, n = 42 | 74 ± 3 | 0.71 ± 0.02 | 24 ± 5 | CC, n = 25 | 79 ± 4 | 0.74 ± 0.03 | 17 ± 3 |
| p = | NS | NS | NS | NS | NS | 0.02 | 0.01 | 0.16 | NS | ||
| Q55IR | Q551R | Q551R | |||||||||
| AA, n = 72 | 60 ± 3 | 0.62 ± 0.02 | 24 ± 2 | AA, n = 182 | 76 ± 2 | 0.71 ± 0.01 | 16 ± 1 | AA, n = 104 | 87 ± 2 | 0.76 ± 0.01 | 13 ± 1 |
| AG, n = 40 | 62 ± 3 | 0.63 ± 0.02 | 18 ± 3 | AG, n = 143 | 73 ± 2 | 0.70 ± 0.01 | 15 ± 1 | AG, n = 86 | 80 ± 2 | 0.73 ± 0.01 | 14 ± 1 |
| GG, n = 15 | 55 ± 4 | 0.61 ± 0.02 | 24 ± 5 | GG, n = 68 | 74 ± 3 | 0.72 ± 0.02 | 21 ± 4 | GG, n = 38 | 79 ± 3 | 0.74 ± 0.02 | 17 ± 3 |
| p = | NS | NS | NS | NS | NS | 0.08 | 0.006 | 0.19 | NS | ||
Definition of abbreviations: BD = bronchodilator; NS = not significant; SARP = Severe Asthma Research Program; SNP = single nucleotide polymorphism.
All data are mean ± SEM.
TABLE 3.
PULMONARY FUNCTION—WHITES IN SEVERE ASTHMA RESEARCH PROGRAM
| SNP | FEV1 | FEV1/FVC | BD Response | SNP | FEV1 | FEV1/FVC | BD Response |
|---|---|---|---|---|---|---|---|
| E375A | E375A | ||||||
| AA n = 202 | 76 ± 2 | 0.70 ± 0.01 | 16 ± 1 | AA, n = 121 | 86 ± 1 | 0.75 ± 0.01 | 12 ± 1 |
| AC n = 32 | 71 ± 3 | 0.71 ± 0.02 | 15 ± 2 | AC, n = 23 | 81 ± 3 | 0.73 ± 0.02 | 11 ± 1 |
| CC n = 2 | 51 ± 10 | 0.55 ± 0.04 | 28 | CC, n = 1 | 41 | 0.50 | — |
| p = | 0.12 | 0.16 | NS | 0.004 | 0.02 | NS | |
| Q55IR | Q551R | ||||||
| AA n = 158 | 77 ± 2 | 0.71 ± 0.01 | 16 ± 1 | AA, n = 95 | 87 ± 1 | 0.76 ± 0.01 | 13 ± 1 |
| AG n = 79 | 80 ± 2 | 0.69 ± 0.01 | 15 ± 2 | AG, n = 46 | 83 ± 2 | 0.74 ± 0.01 | 11 ± 1 |
| GG n = 8 | 65 ± 6 | 0.70 ± 0.04 | 12 ± 6 | GG, n = 5 | 66 ± 8 | 0.72 ± 0.06 | 6 ± 1 |
| p = | 0.05 | 0.32 | NS | 0.005 | NS | NS |
Definition of abbreviations: BD = bronchodilator; NS = not significant; SNP = single nucleotide polymorphism.
All data are mean ± SEM.
IL4Rα Polymorphisms and Tissue Inflammation
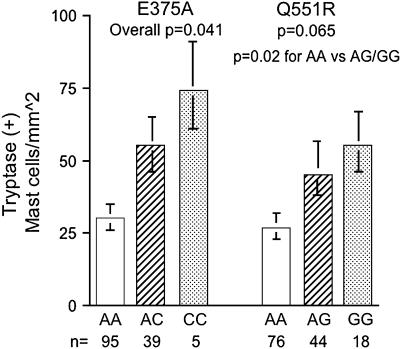
Because tissue inflammation has been associated with asthma, exacerbations of asthma, and its overall severity, the relationship between tissue inflammatory cells and IL4Rα polymorphisms was evaluated in the subgroup (140/175 total subjects) of the NJ cohort in whom endobronchial biopsies had been performed. Only those cells typically associated with allergic inflammation (lymphocytes, eosinophils, and mast cells) were analyzed. Normal subjects (n = 29) were included in this analysis because tissue cell counts (unlike asthma exacerbations and degree of airflow limitation) are not specific to asthma. There was no relationship between tissue eosinophils or CD3+ lymphocytes and IL4Rα polymorphisms (p > 0.29 for all comparisons). However, there were significant associations between tryptase-positive mast cells and IL4Rα polymorphisms, in a manner consistent with the “risk” alleles for exacerbations and lung function (Figure 2). The associations for E375A and mast cells were significant when analyzed across the homozygotes and heterozygotes for the C allele, whereas the association with Q551R was not significant (p = 0.06) across the three groups, but was significant when the combined GA/GG genotypes were compared with the AA genotype (p = 0.02). There were no differences in inhaled or oral CS use by genotype (p = 0.15 for inhaled CS to p = 0.6 for oral CS).
Figure 2.
Tryptase-positive (total) mast cell numbers in endobronchial biopsies are significantly higher in the presence of the C allele at E375A and the G allele at Q551R in the NJ cohort.
IL4Rα Polymorphisms and Systemic/Local IgE
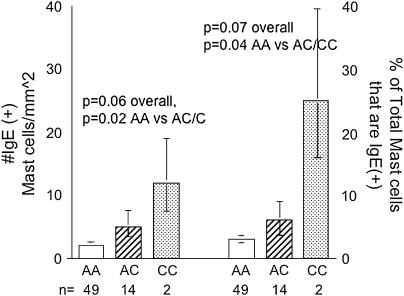
Serum IgE levels were available for less than 50% of the NJ cohort. No differences in IgE levels by genotype were observed in this small subset. Despite previous reports, in the SARP cohort, there were no significant associations between age- and sex-adjusted serum IgE levels and IL4Rα polymorphisms. However, in the NJ cohort, one-third of the subjects also had tissue IgE measured (both as the number of mast cells positive for IgE and the percentage of mast cells positive for IgE). There were higher numbers of IgE+ mast cells and a higher percentage of mast cells positive for IgE in the presence of the C allele at E375A. These were both significant when the homozygotes and heterozygotes for the C allele of the E375A SNP were combined, again consistent with the pattern observed for exacerbations, lung function, and tissue mast cells (Figure 3). In contrast, there was no relationship of the Q551R SNP with IgE+ mast cells.
Figure 3.
Endobronchial tissue IgE+ mast cells, as well as the percentage of total (tryptase positive) mast cells that are IgE positive, are significantly greater in subjects with the C allele at E375A.
Population Stratification
In the NJ cohort, removing the small numbers of African Americans had only a small effect on the significance of the association with the E375A SNP. Severe exacerbations decreased in significance to p = 0.07. Significance was maintained for mast cell counts in tissue (p = 0.03) and a trend was maintained for IgE+ cells in tissue, despite a nearly 25% loss of subjects in that analysis (p = 0.11). However, the significance for the Q551R associations was lost when only evaluating whites. In the replicate SARP cohort, the tests for gene-by-race interaction were not significant; therefore, separate analyses by racial group were not necessary but performed for comparison purposes. The significant associations with severe exacerbations were not observed when the African-American and white subgroups were studied separately (p values, all > 0.25), but a marginal relationship for ED exacerbations remained in the SARP whites (again, for E375A) (p = 0.08). In contrast to exacerbations, better differentiation by genotype was seen when analyzing whites (only) in SARP for %predicted FEV1 (p = 0.12 for E375A and p = 0.048 for Q551R) (Table 3). These associations became stronger when separately evaluating the nonsevere white cohort (p = 0.004 and p = 0.005, respectively) (Table 3).
Haplotype Analysis
Haplotype analysis was performed for all SNPs as well as by using a two-SNP sliding window, which in general did not add insight into the single SNP results, with no p values less than 0.01 (analyzed separately for NJ whites, SARP whites, SARP African Americans). In contrast to other studies, haplotypes that included Ile50 did not better differentiate groups than the individual SNPs. The few nominally significant p values (between 0.01 and 0.05) reflected the single SNP results with the “at risk” haplotypes containing the C allele at E375 or the G allele at Q551.
DISCUSSION
The results from these two cohorts confirm data from other studies, which have reported that common polymorphisms in IL4Rα are associated with asthma outcomes, such as FEV1. However, this study expands the understanding of the importance of the variation in this gene by also identifying a relationship between SNPs coding for amino acid substitutions in the intracellular signaling portion of the receptor and severe exacerbations of asthma. This study further identified a possible inflammatory mechanism for this effect, specifically an increase in mast cells and their IgE binding in airway tissue. Finally, there are distinct differences in allele frequencies at all coding SNPs in exon 12 between African Americans and whites that have the potential to contribute to disparities in asthma severity/health care utilization in these two racial groups (18).
The primary and replicating databases used in this study contain large numbers of the most extensively phenotyped subjects with severe and milder asthma studied to date (1, 19). This phenotyping enabled us to associate relatively small numbers of subjects with asthma with specific asthma outcomes, such as health care utilization, lung function, and inflammatory processes. More general asthma phenotypes, such as distinctions by level of severity and total serum IgE levels, were not associated with the IL4Rα SNPs that were studied. Because severe asthma, in particular, is recognized to consist of those individuals with both frequent/severe exacerbations and those with more persistent but severe airway obstruction (often without severe exacerbations), a specific association of genotype with one particular phenotype could be predicted. In contrast, those with milder asthma may be a slightly more homogeneous population, less confounded by treatment modalities, comorbid conditions, and phenotypes. This may explain why, in the SARP cohort, in whom the majority had mild or moderate asthma, a stronger association of IL4Rα SNPs to %predicted FEV1 was present for both E375A and Q551R than was observed in the full cohort. This relationship was even stronger when analyzing whites separately. Interestingly, even in this milder subgroup, there remained a marginal relationship to higher intensity health care utilization (p = 0.08 for hospitalizations/ED visits in the nonsevere white subgroup for the C allele at E375A).
Th2 inflammatory pathways have long been associated with asthma, including severe asthma. IL-4Rα is likely the most important receptor involved in these pathways. IL4Rα is known to be a highly polymorphic gene, yet only a few SNPs in this gene have been evaluated to date (8). Of these, the SNPs in exon 12, which encode for amino acid changes in the intracellular signaling portion of the receptor, have received the most attention. These SNPs are of interest both from their persistent significance in genetic association studies of asthma and allergy and because of their potential to alter receptor function. Published studies have suggested the less common alleles at E375A, S478P, or Q551R are associated with lower lung function, higher IgE levels, and an in vitro gain of function of the receptor (5, 10, 11, 20, 21). In contrast to our study, where the evaluation of haplotypes consisting of the Ile50 (in the extracellular portion of the receptor) and exon 12 SNPs did not add to the significance of our findings, others have suggested that change in function of the receptor requires the presence of two specific amino acid changes in these two regions of the gene (20–22). Interestingly, one study has suggested that these IL4Rα polymorphisms contribute to a loss of function of the receptor, perhaps through a lower level of activation of STAT-6 (9). Certainly, interactions between other genes in the Th2 pathway (including IL-4, IL-13, STAT-6, and others) and IL4Rα could also contribute to clinical, physiologic, and immunologic phenotypes (4, 10). Future studies in larger numbers of subjects will need to address gene-by-gene interactions in these cohorts.
This is the first genetic study of subjects with asthma to evaluate the association of genotypes with tissue inflammatory findings. In subjects with the higher risk IL4Rα polymorphisms, there was an increase in tissue mast cell numbers and a more specific increase in IgE+ mast cells in the tissue biopsies. These results support the concept that the amino acid switch from glutamic acid to alanine at position 375 contributes to a gain of function of the receptor. This could lead to an increase in serum IgE (as has previously been reported) but could also increase the binding of IgE to mast cells in the lung tissue where they may play a causal role in the development of severe asthma exacerbations (11, 23, 24). Interestingly, the degree of binding of IgE to mast cells in cell culture has been suggested to prolong the survival of mast cells, outlining a mechanism by which these SNPs could increase total mast cell numbers (25).
This is also the second study to address the frequency of these SNP alleles in African Americans (7). The current study confirms the previous differences in Q551R frequencies but suggests substantially larger differences by racial origin in E375A than those previously reported (7). Additional studies of SNPs in IL4Rα revealed greater overall diversity of allelic frequency in IL4Rα in African Americans as compared with that in whites, particularly in exon 12, where the SNPs of interest in this study are located (8). The authors of that study proposed that the dense distribution of coding SNPs in this single exon, associated with broad racial diversity, supported a strong environmental influence on these mutations. Allelic substitutions in these coding SNPs, which alter signaling through IL-4Rα, could almost certainly impact parasite host defense, a response of greater importance to equatorial populations as compared with populations in colder climate regions. These coding and functional SNPs likely evolved to be adaptive and advantageous traits in host responses to parasites. What is not yet clear is whether these evolutionarily advantageous parasite responses may, in fact, have negative implications for the development or progression of allergic diseases in industrialized societies.
In the replication cohort, in particular, most of the severe exacerbation events were seen in the African-American population. Although nearly 30% of the SARP cohort subjects are African Americans, their overall numbers are still relatively small. This smaller sample size, the low incidence of homozygotes for the allele common to whites, as well as the generally much higher severe asthma exacerbation rate in the African Americans studied likely diluted the ability to discern a specific difference in this subgroup by IL4Rα genotype. In contrast, the larger white sample sizes in both cohorts allowed analysis of the relevance of these SNPs to that population subgroup. In the substantially more severe NJ cohort, the removal of the African-American subjects only marginally diminished the significance of the associations for whites alone with exacerbations and tissue mast cells, especially for the E375A SNP. In the overall milder SARP cohort, the significance for the IL4Rα SNPs in relationship to the white population remained marginally present for ED exacerbations (p = 0.08), whereas the associations with lung function became stronger, consistent with previous reports (11). Although these findings continue to support the relevance of these polymorphisms to the white population, it cannot be determined from these studies whether these polymorphisms contribute to the higher morbidity and mortality rates observed in African Americans, above and beyond the contribution from the recognized disparities in health care (18). Larger cohort studies of African Americans will be required to investigate these relationships.
These studies are limited by the relatively small sample sizes, heterogeneity of allele frequencies in the racial subgroups, and heterogeneity of the disease itself. Although we analyzed small numbers of SNPs in IL4Rα (four of which were in exon 12), these SNPs were selected based on previously described associations with asthma and allergy. It is conceivable that a more important risk-associated SNP exists in exon 12 or elsewhere in this gene that is in LD with the currently genotyped SNPs. This may explain the disconnect between E375A and Q551R SNPs, with the allele substitutions at E375A associated more closely to exacerbations and IgE/mast cell processes and Q551R changes more associated with lung function abnormalities. In addition, tissue studies are limited by the small pieces of tissue obtained and the single point in time of these investigational bronchoscopic studies, such that these results in relatively small numbers are important. Questionnaire data may also be limited by recall bias and different clinical models for urgent health care utilization, especially given the mixed geographic/health systems (one site in the United Kingdom and multiple sites in the United States). However, the marked consistency of the direction of the associations supports the validity of the results and indicates that a more comprehensive analysis of the IL4Rα gene should be pursued.
In conclusion, these results confirm prior population studies, which report associations of allele substitutions in IL4Rα with asthma and decreased lung function. In addition, these observations expand our understanding by demonstrating an association with asthma exacerbations and suggest a mechanism by which variations in IL4Rα may modify asthma severity through increases in mast cells and associated local IgE expression. Finally, these studies demonstrate large racial differences in IL4Rα SNPs between racial groups, which may be related to positive evolutionary adaptations to local environments, but which may now have negative effects in industrialized countries. Further studies to determine the functional changes in IL-4Rα signaling as they relate to asthma and parasite host defense are needed.
Supplementary Material
Supported by NIH grants AI-40600, HL-69174, RR-00051, HL-69349, HL-69170, HL-69130, HL-69167, HL-69116, HL-69149, and HL-69155.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200607-909OC on December 14, 2006
Conflict of Interest Statement: S.E.W. has a financial relationship with ISIS Pharmaceuticals ($12,000 per year). S.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.A. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.A.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. W.W.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. W.J.C. has received honoraria from Genentech, and an award to support an investigator-initiated clinical trial from GlaxoSmithKline. M.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.F.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.E. is principal investigator of an industry-sponsored grant of bronchial thermoplasty for asthma from Alair/Asthmatx but receives no personal compensation for any portion of the study. B.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.I. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. W.G.T. is a member of the Merck speakers' bureau. He received $22,920 in 2005 for this activity. D.C.-E. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.A.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.R.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Moore WC, Bleecker ER, Curren-Everett D. Erzurum, SC, Ameredes BT, Bacharier L, Calhoun WJ, Castro M, Chung KF, Clark MP, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute's Severe Asthma Research Program. J Allergy Clin Immunol 2007;119:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burchard EG, Silverman EK, Rosenwasser LJ, Borish L, Yandava C, Pillari A, et al. Association between a sequence variant in the IL-4 gene promoter and FEV1 in asthma. Am J Respir Crit Care Med 1999;160:919–922. [DOI] [PubMed] [Google Scholar]
- 3.Rosa-Rosa L, Zimmermann N, Bernstein JA, Rothenberg ME, Khurana Hershey GK. The R576 IL-4 receptor alpha allele correlates with asthma severity. J Allergy Clin Immunol 1999;104:1008–1014. [DOI] [PubMed] [Google Scholar]
- 4.Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland SK, von Mutius E. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol 2006;117:269–274. [DOI] [PubMed] [Google Scholar]
- 5.Sandford AJ, Chagani T, Zhu S, Weir TD, Bai TR, Spinelli JJ, FitzGerald JM, Behbehani NA, Tan WC, Pare PD. Polymorphisms in the IL4, IL4RA, and FCERIB genes and asthma severity. J Allergy Clin Immunol 2000;106:135–140. [DOI] [PubMed] [Google Scholar]
- 6.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol 1999;17:701–738. [DOI] [PubMed] [Google Scholar]
- 7.Ober C, Leavitt SA, Tsalenko A, Howard TD, Hoki DM, Daniel R, Newman DL, Wu X, Parry R, Lester LA, et al. Variation in the interleukin 4-receptor alpha gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet 2000;66:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Di Rienzo A, Ober C. A population genetics study of single nucleotide polymorphisms in the interleukin 4 receptor alpha (IL4RA) gene. Genes Immun 2001;2:128–134. [DOI] [PubMed] [Google Scholar]
- 9.Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, Deichmann KA. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology 1999;96:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, Bleecker ER. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet 2002;70:230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med 1997;337:1720–1725. [DOI] [PubMed] [Google Scholar]
- 12.Wenzel SE, Bleecker ER. Polymorphisms in interleukin-4 receptor α (IL-4Rα) are associated with a greater risk of near-fatal events in severe asthma [abstract]. Proc Am Thorac Soc 2005;2:A31. [Google Scholar]
- 13.Wenzel SE, Fahy JV, Irvin CG, Peters SP, Spector S, Szefler SJ. Proceedings of the ATS Workshop on Refractory Asthma: current understanding, recommendations and unanswered questions. Am J Respir Crit Care Med 2000;162:2341–2351. [DOI] [PubMed] [Google Scholar]
- 14.Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, Chu HW. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med 1999;160:1001–1008. [DOI] [PubMed] [Google Scholar]
- 15.Basehore MJ, Howard TD, Lange LA, Moore WC, Hawkins GA, Marshik PL, Harkins M, Meyers D, Bleecker E. A comprehensive evaluation of IL4 variants in ethnically diverse populations: association of total serum IgE levels and asthma in white subjects. J Allergy Clin Immunol 2004;114:80–87. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 17.Schaid DJ. Relative efficiency of ambiguous vs. directly measured haplotype frequencies. Genet Epidemiol 2002;23:426–443. [DOI] [PubMed] [Google Scholar]
- 18.Gupta RS, Carrion-Carire V, Weiss KB. The widening black/white gap in asthma hospitalizations and mortality. J Allergy Clin Immunol 2006;117:351–358. [DOI] [PubMed] [Google Scholar]
- 19.Miranda C, Busacker A, Balzar S, Trudeau J, Wenzel SE. Distinguishing severe asthma phenotypes: role of age at onset and eosinophilic inflammation. J Allergy Clin Immunol 2004;113:101–108. [DOI] [PubMed] [Google Scholar]
- 20.Faffe DS, Whitehead T, Moore PE, Baraldo S, Flynt L, Bourgeois K, Panettieri RA, Shore SA. IL-13 and IL-4 promote TARC release in human airway smooth muscle cells: role of IL-4 receptor genotype. Am J Physiol Lung Cell Mol Physiol 2003;285:L907–L914. [DOI] [PubMed] [Google Scholar]
- 21.Risma KA, Wang N, Andrews RP, Cunningham CM, Ericksen MB, Bernstein JA, Chakraborty R, Khurana Hershey GK. V75R576 IL-4 receptor alpha is associated with allergic asthma and enhanced IL-4 receptor function. J Immunol 2002;169:1604–1610. [DOI] [PubMed] [Google Scholar]
- 22.Hytonen AM, Lowhagen O, Arvidsson M, Balder B, Bjork AL, Lindgren S, Hahn-Zoric M, Hanson LA, Padyukov L. Haplotypes of the interleukin-4 receptor alpha chain gene associate with susceptibility to and severity of atopic asthma. Clin Exp Allergy 2004;34:1570–1575. [DOI] [PubMed] [Google Scholar]
- 23.Balzar S, Strand M, Rhodes D, Wenzel S. Immunoglobulin (Ig)E expression pattern in lung: relation to systemic IgE and asthma phenotypes. J Allergy Clin Immunol (In press) [DOI] [PMC free article] [PubMed]
- 24.Busse W, Corren J, Lanier BQ, McAlary M, Fowler-Taylor A, Cioppa GD, van As A, Gupta N. Omalizumab, anti-IgE recombinant humanized monoclonal antibody, for the treatment of severe allergic asthma. J Allergy Clin Immunol 2001;108:184–190. [DOI] [PubMed] [Google Scholar]
- 25.Asai K, Kitaura J, Kawakami Y, Yamagata N, Tsai M, Carbone DP, Liu F, Galli S, Kawakami T. Regulation of mast cell survival by IgE. Immunity 2001;14:791–800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.