Abstract
Ciliated airway epithelial cells are critical for mucosal barrier function, including host defense against pathogens. This cell population is often the primary target and thereby the first line of defense against many common respiratory viruses. It is also the precursor for mucous cells and thereby promotes mucociliary clearance of infectious and other noxious agents. Cells with motile cilia in other organs (e.g., brain and reproductive organs) may also have roles in development and reproduction. However, definitive proof of ciliated cell function is hampered by the lack of strategies to specifically target this cell population for loss of function in vivo. To this end, cell type–specific gene promoters have been combined with the Cre/LoxP system to disrupt genes in airway and alveolar epithelial cell populations expressing surfactant protein C (SP-C) or Clara cell secretory protein (CCSP). By contrast, an analogous system to disrupt gene function in ciliated airway epithelial cells was still needed. Here we report the generation and analysis of mouse lines with a FOXJ1 promoter driving the Cre recombinase and show that this system mediates genomic recombination specifically in ciliated cells. The pattern of recombination recapitulates endogenous FOXJ1 promoter function, being restricted to ciliated cells present in pulmonary airways as well as choroid plexus, ependyma, oviduct, and testis. This transgenic mouse system thereby offers a new strategy for specific knockouts of genes in ciliated cells. It should prove extremely useful for defining ciliated cell function in airway mucosal immunity as well as development and reproduction.
Keywords: pulmonary airways, mucosal immunity, FOXJ1 transcription factor, cell-specific gene knockouts, Cre/LoxP system
CLINICAL RELEVANCE
We developed a new tool for defining the role of ciliated epithelial cells in disease. For example, the system could be used to block respiratory viral infection by down-regulating a gene that allowed virus to bind to ciliated airway epithelial cells.
Epithelial cells with motile cilia are fundamental to normal development and ongoing function of most organisms, and abnormalities in ciliated epithelial cells are the basis for several types of human disease. In the respiratory system, ciliated cells are found in the epithelium of large and small airways as well as the paranasal sinuses and Eustachian tubes of the upper airway (1). Similar ciliated cells are also present within the ependyma and choroid plexus of the brain, the testis and oviduct, and within the embryonic node, a structure within the notochord that is transiently present during early development and is critical for left-right determination (2). This distribution of ciliated cells correlates with the phenotypes for loss of ciliated cell function. Thus, inherited defects of ciliated cell function in the syndrome of primary ciliary dyskinesia can cause chronic respiratory infection based on compromised mucociliary clearance, developmental defects (e.g., situs inversus) mediated by loss of embryonic node function, and infertility due to loss of ovum and/or sperm motility in reproductive organs (3).
While a global loss of ciliary function has been at least partially characterized, other aspects of ciliated cell function remain less certain. Progress is particularly constrained by the absence of ciliated cell–specific genetic strategies. For example, ciliated airway epithelial cells are selectively infected and thereby marked for destruction during the acute phase of viral infection (4–8). However, we cannot specifically target the viral receptors on ciliated cells that might mediate this process. Similarly, ciliated airway epithelial cells are also destroyed by cigarette smoke exposure, so that smokers with chronic bronchitis have decreased levels of ciliated epithelial cells compared to healthy nonsmokers (9, 10). In this case as well, we cannot assign cell type–specific mechanisms of biochemical toxicity to ciliated cells. Immunopathologic analysis suggests that ciliated airway epithelial cells of the respiratory tract help to repair epithelial damage after injury (11). In that regard, ciliated airway epithelial cells may increase in numbers and act as mucous cell precursors during the long-term mucous cell metaplasia that develops after viral infection (12). Based on studies of isolated ciliated epithelial cells, this process appears to be mediated by EGFR-dependent ciliated cell survival and IL-13–driven ciliated-to-mucous cell transdifferentiation. However, further progress would be aided by a system to selectively modify EGFR or IL-13 signaling in ciliated epithelial cells in vivo.
Several strategies could be used to target ciliated cell function in vivo, but a particularly useful one would be the capability of altering single gene function in specific cell types in the epithelium. This approach has been developed for gain of gene function by conditional expression of transgenes under the control of the surfactant protein C (SP-C) or Clara cell secretory protein (CCSP) gene promoters (13, 14). The SP-C and CCSP promoters have also been combined with the Cre/LoxP system to achieve loss of gene function in lung epithelial cell populations expressing SP-C or CCSP, respectively (15–17). In both cases, it has been difficult to achieve cell-type specificity for the gain or loss of gene function. The human SP-C promoter is active in bronchiolar Clara cells as well as alveolar Type II cells (18). Similarly, the rat CCSP promoter appears to be active in ciliated and alveolar Type II epithelial cells as well as Clara cells (17, 18). Cell-type specificity may be decreased further in disease models, where cytokines may also influence cell lineage and corresponding markers of lineage (12). Thus, cell type–specific transgenic systems were not yet available for lung epithelial cell populations, including ciliated epithelial cells. It was therefore difficult to study the function of specific genes expressed in a cell type–specific manner.
Here we describe the use of distinct human FOXJ1 promoter elements to permit ciliated epithelial cell-specific gene targeting. We chose this approach because the corresponding mouse Foxj1 gene is expressed mainly in ciliated cells (2). Moreover, the endogenous mouse Foxj1 gene is necessary for ciliogenesis (19). In addition, an initial study of the human FOXJ1 promoter revealed that it contains sufficient sequence to drive GFP transgene expression in trachea, lung, ependyma, oviduct, and testis (20). We now develop mouse lines with a human FOXJ1 promoter driving the Cre recombinase and show that this system mediates genomic recombination specifically in ciliated cells. The pattern of recombination recapitulates endogenous FOXJ1 promoter function in ciliated cells of pulmonary airways as well as choroid plexus, ependyma, oviduct, and testis. The system should thereby prove useful for defining the function of ciliated cell genes under normal conditions and in inherited or acquired diseases with altered mucosal immunity, epithelial repair, infertility, or organ development.
MATERIALS AND METHODS
Generation of Transgenic Mice
A 1.2-kb human FOXJ1 5′ flanking sequence (−1221 to −8 nt from the ATG site) was amplified from human genomic DNA using PCR primers 5′-AGGATACGGTTCCCGAGCCTGAG-3′ and 5′-GGTGAATTCGGGACTCTCCGAGGGGGCGGTC-3′ derived from Genbank sequence accession number AC018665 (21). The resulting PCR product was digested with SalI (acting on a site in the promoter sequence) and EcoRI (acting on a site that was engineered into the 3′ primer), and then was exchanged for the CMV enhancer and β-actin promoter in pCAGG (22) using the same restriction enzymes to yield pFOXJ1-β-globin-polyA. Cre cDNA with a nuclear localization signal inserted at the N-terminus was obtained from M. Reth (Max-Planck-Institute of Immunobiology, Freiburg, Germany). This cDNA was amplified using PCR primers 5′-GCCTGAATTCGCAGACCATGCCCAAGAAGAAGAGG-3′ and 5′-GCCAGTGAATTCCTAATCGCCATCTTCCAG-3′, and then was subcloned into the EcoRI site of the pFOXJ1-β-globin-polyA vector to generate a FOXJ1-Cre cDNA construct (Figure 1a). This construct was verified by sequencing and then was linearized with SalI and PvuI digestion, purified, and microinjected into the pronuclei of C57BL/6 zygotes. The zygotes were transferred into pseudopregnant C57BL/6 mice from Taconic Farms (Germantown, NY) to generate FOXJ1-Cre mice. Transgenic founders were screened for Cre integration by PCR of genomic DNA from tail biopsies using Cre sequence-specific PCR primers 5′-ATTTGGGCCAGCTAAACATGC-3′ and 5′-GCAAAACAGGTAGTTATTCGG-3′. Cre transgene expression was assessed by real-time PCR of total RNA from trachea, lung, brain, and testis using primers 5′-TTGGGCCAGCTAAACATGCT-3′ and 5′-GCATTGCTGTCACTTGGTCG-3′. For these experiments, RNA was extracted with the RNeasy kit from Qiagen (Calenic, CA) and treated twice with DNase to eliminate DNA contamination. To assess transgene function, FOXJ1-Cre transgenic founder mice were crossed with ROSA26R (ROSA) reporter mice that carry a LacZ reporter gene with an internal stop codon flanked by LoxP sites (23). Offspring of this cross (FOXJ1-Cre x ROSA) were assessed for somatic recombination at 4–6 wk of age. All procedures and experiments were performed according to protocols approved by the University's Animal Studies Committee.
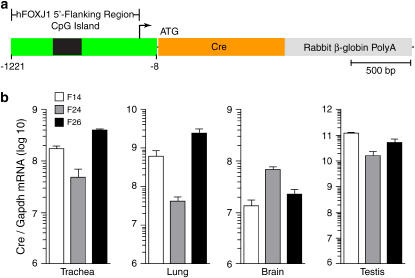
Figure 1.
Tissue localization of Cre expression in FOXJ1-Cre transgenic mice. (a) Schematic representation of DNA construct used to generate the FOXJ1-Cre transgenic mouse line. (b) Analysis of Cre transgene expression in indicated tissues isolated from FOXJ1-Cre transgenic mice and then subjected to real-time PCR for Cre mRNA levels. Values represent mean levels for two to three mice from each of three founder lines (F14, F24, and F26) and are normalized to Gapdh mRNA levels in each sample.
Histochemistry
LacZ gene expression (as a marker of somatic recombination) was assessed using X-Gal staining of target organs from double-transgenic FOXJ1-Cre/ROSA mice. For these experiments, organs were cryopreserved in OCT compound (Tissue-Tek) from Sakura Fine Tek (Torrance, CA) and then cut into 6-μm-thick sections. The sections were mounted on slides, fixed with 4% paraformaldehyde for 20 min at 25°C, washed twice with PBS containing 2 mM MgCl2, and then incubated in detergent solution (PBS, pH 7.4, containing 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% NP40) for 5 min at 4°C. The detergent solution was replaced with X-Gal staining solution (PBS, pH 7.4, containing 1 mg/ml X-Gal, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% NP40) and then incubated for 18 h at 37 °C (24). Sections were subsequently washed twice with PBS and then counterstained with eosin. Sections were imaged using a BX51 microscope with a charge-coupled device camera interfaced to MagniFire software from Olympus (Melville, NY).
Immunostaining
Organs from double-transgenic FOXJ1-Cre/ROSA mice were also subjected to immunostaining for β-Galatosidase (β-Gal) and β-Tubulin-IV (β-Tubulin). For these experiments, 6-μm frozen sections were fixed in pure acetone for 30 min at 4°C, blocked with 5% normal donkey serum, then incubated with rabbit anti–β-Gal polyclonal antibody from Chemicon International (Temecula, CA) and mouse anti–β-Tubulin mAb from Sigma (St. Louis, MO) for 18 h at 4°C. Primary antibody binding was detected using donkey anti-rabbit IgG conjugated with indocarbocyanine (Cy3) and donkey anti-mouse IgG conjugated with fluorescein isothiocyanate (FITC) from Roche (Indianapolis, IN). Sections were mounted on slides with Vectashield medium containing 4,6-diamidino-2-phenylindole (DAPI) from Vector Laboratories (Burlingame, CA). Sections were imaged by reflected fluorescent microscopy using the Olympus system described above.
Cell Culture and Immunocytochemistry
In addition to studies of isolated tissues, we extended our analysis to mouse airway epithelial cells isolated from FOXJ1-Cre/ROSA mice. Mouse tracheal epithelial cells (mTECs) were isolated, cultured under air–liquid interface conditions, and prepared for immunostaining as described previously (12, 25). For the present experiments, immunostaining for β-Gal and β-Tubulin-IV with DAPI counterstaining was performed as described above for tissue frozen sections.
RESULTS
Tissue-Specific Cre Expression
In the initial set of experiments, we developed FOXJ1-Cre transgenic mouse lines to determine whether the selected human FOXJ1 promoter sequence (−1221 to −8 nt from the ATG start site) was sufficient to direct tissue-specific gene expression of Cre recombinase. To generate transgenic mice, we used a FOXJ1-Cre plasmid vector that contained human FOXJ1 5′-flanking sequence and Cre cDNA coding sequence fused to rabbit β-globin PolyA (Figure 1a). After zygote microinjection and implantation, we identified six founders in the offspring based on PCR screening for Cre transgene integration. RNA extracted from organs of the double transgenic FOXJ1-Cre/ROSA mice were quantified for Cre expression by real-time PCR (Figure 1b). We found no amplification of RNA that was not subjected to reverse transcription, confirming that the RNA was not contaminated with genomic DNA. Cre mRNA was detected in trachea, lung, brain, and testis, but not in intestine, kidney, liver, muscle, or heart from each of three founder mouse lines (Figure 1b and data not shown). This pattern demonstrated that the human FOXJ1 5′ flanking sequence was sufficient to direct tissue-specific expression of the Cre gene. In each of the three founders exhibiting Cre expression (F14, F24, and F26), the level of Cre mRNA transcripts was highest in testis compared with that in trachea, lung, and brain. This finding may be based on the abundance of spermatogonia in testis tissue. Spermatogonia are known to express FOXJ1 and may therefore account for high-level activity of the FOXJ1 promoter in testis tissue. Cre gene expression also varied in the lung among founders. Specifically, Cre mRNA was lower in the lungs of F24 compared to F14 and F26 mice. Cre mRNA was not detectable in any organ in the remaining three founders (designated F7, F22, and F56). Variability of Cre expression is likely attributed to differences in the sites and numbers of integrated Cre transgenes among founders. Nonetheless, the overall pattern of Cre expression suggested localization to ciliated cells, since transgene expression was found only in ciliated cell-containing organs (lung, brain, and testis).
Cell-Specific Cre-Mediated Recombination
To more precisely determine whether FOXJ1-Cre mice expressed functional recombinase activity in ciliated cells, we crossed FOXJ1-Cre mice with ROSA reporter mice (as diagrammed in Figure 2a). These reporter mice carry the LacZ transgene with a LoxP-flanked stop sequence, so that Cre-mediated excision of this sequence is necessary for LacZ gene expression. Thus, in FOXJ1-Cre/ROSA double transgenic mice, Cre-mediated somatic recombination can be detected by expression and consequent activity of β-Gal using the X-Gal substrate. On the basis of X-Gal staining, we found that epithelial cells from trachea and bronchus/bronchiole expressed β-Gal (Figure 2b). This pattern of staining was found in each of the three founders that expressed Cre but in none of the Cre-negative ROSA mice.
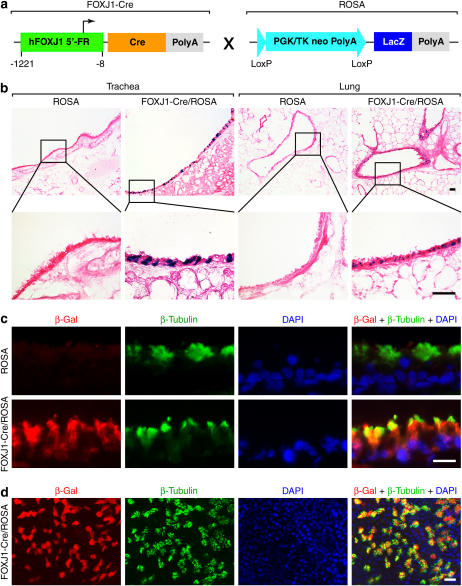
Figure 2.
Cellular localization of Cre-mediated recombination in FOXJ1-Cre/ROSA26R double-transgenic mice. (a) Schematic representation of breeding cross between FOXJ1-Cre and ROSA transgenic mice. (b) Tracheal and lung cryostat sections from FOXJ1-Cre/ROSA double-transgenic and Cre-negative ROSA control mice were stained with X-gal and counterstained with eosin. Results are representative of three FOXJ1-Cre/ROSA founder lines. Insets represent higher magnification views. Bars = 40 μm. (c) Tracheal cryostat sections from FOXJ1-Cre/ROSA mice and Cre-negative ROSA control mice were immunostained with anti–β-Gal antibody detected with Cy3-labeled secondary antibody (red fluorescence) and anti–β-Tubulin mAb detected with FITC-labeled secondary antibody (green fluorescence). Nuclear DNA was detected with DAPI (blue fluorescence). Bar = 20 μm. (d) mTEC cultures from FOXJ1-Cre/ROSA mice were immunostained using the same antibodies as in c. Cellular β-Gal staining was not detected in Cre-negative ROSA control mice (data not shown). Bar = 20 μm.
Since our goal was to use FOXJ1 transgene expression to confine Cre-mediated recombination to ciliated epithelial cells, we next assessed higher magnification of X-Gal staining as well as co-localization of Cre recombinase activity with markers of ciliated epithelial cells. Higher magnification of X-gal–stained lung tissue indicated that Cre-mediated recombination was found only in ciliated cells of the airway epithelium (Figure 2b). Similarly, expression of β-Gal was co-localized exclusively with the ciliated epithelial cell marker β-Tubulin (Figure 2c). Expression of β-Gal was undetectable in tracheal and lung sections prepared from Cre-negative ROSA transgenic mice (Figure 2c). To further establish that genetic recombination was localized to ciliated airway epithelial cells, we also isolated mouse airway epithelial cells (mTECs) from FOXJ1-Cre/RoSA mice and cultured them under air–liquid interface conditions that allow for differentiation of ciliated epithelial cells. These experiments confirmed that β-Gal expression was co-localized with β-Tubulin in ciliated epithelial cells from FOXJ1-Cre/ROSA mice but not Cre-negative ROSA control mice (Figure 2d and data not shown). Together, these findings provided evidence that the FOXJ1 promoter drives Cre expression and thereby allows for Cre-mediated recombination selectively in ciliated airway epithelial cells.
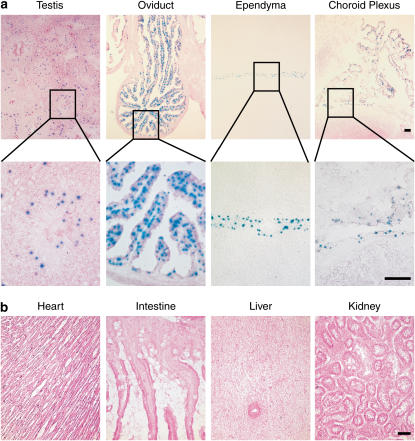
In addition to ciliated airway epithelial cells, endogenous Foxj1 is also expressed in ependyma and choroid plexus in brain, and oviduct and testis in reproductive organs. We therefore further assessed Cre functional activity at these sites using FOXJ1-Cre/ROSA double-transgenic mice. Consistent with endogenous Foxj1 promoter activity, we found that X-Gal staining was present in testis, oviduct, ependyma, and choroid plexus (Figure 3a). X-Gal staining in the testis was localized primarily to the spermatocyte precursor cells (spermatogonia) that lie next to the outer membrane of the seminiferous tubule, and was not present in mature spermatocytes. This staining pattern was identical to the distribution of expression for endogenous Foxj1 (2). X-Gal staining was not detected in heart, intestine, liver, or kidney tissues. This finding was consistent with the lack of endogenous Foxj1 expression in these organs. Again, this finding supported the fidelity of the human FOXJ1 promoter elements to direct Cre expression at the same sites as the endogenous Foxj1 promoter.
Figure 3.
Tissue localization of Cre-mediated recombination in FOXJ1-Cre/ROSA26R mice. (a) Tissue cryostat sections from testis, oviduct, ependyma, and choroid plexus of FOXJ1-Cre/ROSA double-transgenic mice were stained with X-Gal and counterstained with eosin. Insets represent higher magnification views. (b) Tissue cryostat sections from heart, intestine, liver, and kidney of mice in a were stained with X-Gal and counterstained with eosin. Bars = 40 μm.
In summary, we have developed a FOXJ1-Cre transgenic system that directs gene expression to the ciliated epithelial cells. We have further demonstrated that this system can direct somatic recombination localized to ciliated epithelial cells in combination with LoxP-flanked alleles. The system was specifically developed without the use of conditional control elements that bring with them a series of confounding variables and complications (26). Based on the utility of the Cre/LoxP system in other settings (27), the FOXJ1-Cre system will now allow for the productive analysis of single gene function in the lineage of cells with motile cilia. In the context of lung function and disease, this new tool will thereby provide the first example of gene targeting to a specific lung cell type. In the broader context of function for the whole organism, the development of FOXJ1-Cre transgenic mice also provides the capacity to determine the function of specific genes in the ciliated epithelia of ependyma, choroid plexus, oviduct, and testis. Thus, mice carrying transgenes flanked by LoxP sites are now candidates for single gene knockouts in ciliated epithelial cells simply by crossing with FOXJ-Cre transgenic mice. This system should thereby facilitate new approaches to defining the role of ciliated epithelial cells in immunity, development, and reproduction both under normal conditions and in diseases characterized by abnormal ciliated cell behavior.
Acknowledgments
The authors thank R. McCarthy in the Pulmonary Mouse Core Facility at Washington University School of Medicine for technical assistance.
This work was supported by grants from the National Institutes of Health (Heart, Lung, and Blood Institute and Institute for Allergy and Infectious Diseases), the Martin Schaeffer Fund, and the Alan A. and Edith L. Wolff Charitable Trust.
Originally Published in Press as DOI: 10.1165/rcmb.2006-0475RC on January 25, 2007
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Toskala E, Smiley-Jewell SM, Wong VJ, King D, Plopper CG. Temporal and spatial distribution of ciliogenesis in the tracheobronchial airways of mice. Am J Physiol Lung Cell Mol Physiol 2005;289:L454–L459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blatt EN, Yan XH, Wuerrfel MK, Hamilos DL, Brody SL. Forkhead transcription factor HFH-4 expression is temporally related to ciliogenesis. Am J Respir Cell Mol Biol 1999;21:168–176. [DOI] [PubMed] [Google Scholar]
- 3.Ibanez-Tallon I, Heintz N, Omran H. To beat or not to beat: roles of cilia in development and disease. Hum Mol Genet 2003;12:R27–R35. [DOI] [PubMed] [Google Scholar]
- 4.Ibricevic A, Pekosz AS, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol 2006;80:7469–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Look DC, Walter MJ, Williamson MR, Pang L, You Y, Sreshta JN, Johnson JE, Zander DS, Brody SL. Effects of paramyxoviral infection on airway epithelial cell Foxj1 expression, ciliogenesis, and mucociliary function. Am J Pathol 2001;159:2055–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 2005;79:15511–15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol 2002;76:5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Bukreyev A, Thompson CI, Watson B, Peeples ME, Collins PL, Pickles RJ. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J Virol 2005;79:1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riise GC, Larsson S, Andersson BA. A bronchoscopic brush biopsy study of large airway mucosal pathology in smokers with chronic bronchitis and in healthy nonsmokers. Eur Respir J 1992;5:382–386. [PubMed] [Google Scholar]
- 10.Sisson JH, Papi A, Beckmann JD, Leise KL, Wisecarver J, Brodersen BW, Kelling CL, Spurzem JR, Rennard SI. Smoke and viral infection cause cilia loss detectable by bronchoalveolar lavage cytology and dynein ELISA. Am J Respir Crit Care Med 1994;149:205–213. [DOI] [PubMed] [Google Scholar]
- 11.Lawson GW, Van Winkle LS, Toskala E, Senior RM, Parks WC, Plopper CG. Mouse strain modulates the role of the ciliated cell in acute tracheobronchial airway injury-distal airways. Am J Pathol 2002;160:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, Morton JD, Battaile JT, Patel AC, Patterson GA, Castro M, et al. Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J Clin Invest 2006;116:309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitsett JA, Glasser SW. Targeting gene expression to the lung. In L. A. Brigham, editor. Gene Therapy for Diseases of the Lung, Lung Biology in Health and Disease. New York: Marcel Dekker; 1997. pp. 193–208.
- 14.Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem 2000;275:11858–11864. [DOI] [PubMed] [Google Scholar]
- 15.Hokuto I, Ikegami M, Yoshida M, Takeda K, Akira S, Perl AK, Hull WM, Wert SE, Whitsett JA. Stat-3 is required for pulmonary homeostasis during hyperoxia. J Clin Invest 2004;113:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertin G, Poujeol C, Rubera I, Poujeol P, Tauc M. In vivo Cre/loxP mediated recombination in mouse Clara cells. Transgenic Res 2005;14:645–654. [DOI] [PubMed] [Google Scholar]
- 17.Perl A-KT, Wert SE, Loudy DE, Shan Z, Blair PA, Whitsett JA. Conditional recombination reveals distinct subsets of epithelial cells in trachea, bronchi, alveoli. Am J Respir Cell Mol Biol 2005;33:455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perl A-KT, Tichelaar JW, Whitsett JA. Conditional gene expression in the respiratory epithelium of the mouse. Transgenic Res 2002;11:21–29. [DOI] [PubMed] [Google Scholar]
- 19.Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. Ciliogenesis and left-right axis defects in HFH-4 null mice. Am J Respir Cell Mol Biol 2000;23:45–51. [DOI] [PubMed] [Google Scholar]
- 20.Ostrowski LE, Hutchins JR, Zakel K, O'Neal WK. Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol Ther 2003;8:637–645. [DOI] [PubMed] [Google Scholar]
- 21.Murphy DB, Seemann S, Wiese S, Kirschner R, Grzeschik KH, Thies U. The human hepatocyte nuclear factor 3/fork head gene FKHL13: genomic structure and pattern of expression. Genomics 1997;40:462–469. [DOI] [PubMed] [Google Scholar]
- 22.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991;108:193–199. [DOI] [PubMed] [Google Scholar]
- 23.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999;21:70–71. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Riesterer C, Ayrall AM, Sablitzky F, Littlewood TD, Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res 1996;24:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyner JW, Uchida O, Kajiwara N, Kim EY, Patel AC, O'Sullivan MP, Walter MJ, Schwendener RA, Cook DN, Danoff TM, et al. CCL5-CCR5 interaction provides antiapoptotic signals for macrophage survival during viral infection. Nat Med 2005;11:1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitsett JA, Perl A-KT. Conditional control of gene expression in the respiratory epithelium: a cautionary note. Am J Respir Cell Mol Biol 2006;34:519–520. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn R, Torres RM. Cre/loxP recombination system and gene targeting. Methods Mol Biol 2002;180:175–204. [DOI] [PubMed] [Google Scholar]